Epidemiology of Candidemia in Mashhad, Northeast Iran: A Prospective Multicenter Study (2019–2021)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Sample Processing
2.2. In Vitro Antifungal Susceptibility Testing (AFST)
2.3. Multiplex Short Tandem Repeat (STR) Genotyping
3. Results
3.1. Patients’ Characteristics and Species Distribution
3.2. Resistance Investigation
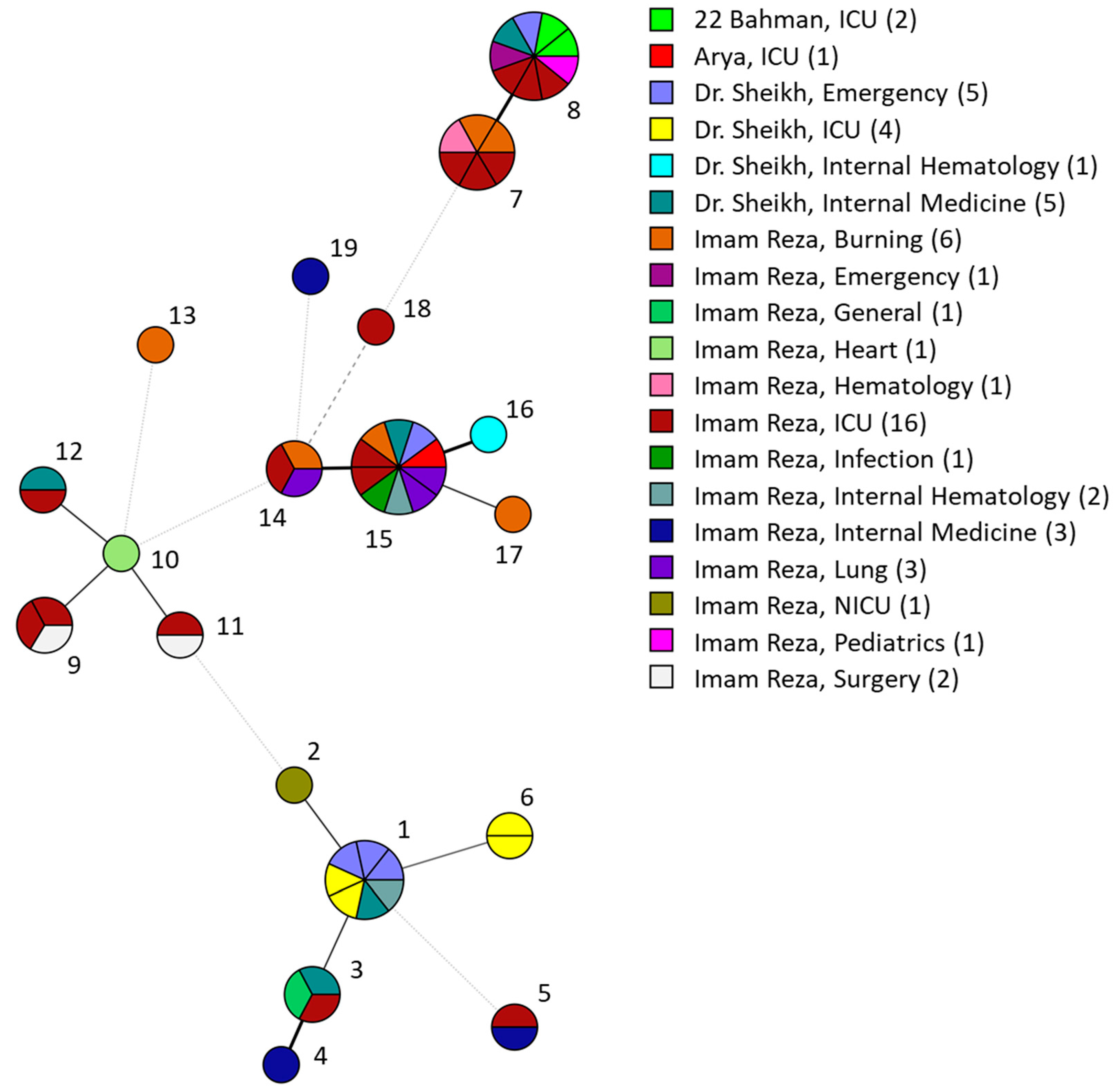
3.3. C. parapsilosis STR Genotyping Shows Clusters
4. Discussion
4.1. Epidemiology
4.2. Resistance Investigation
4.3. Outbreak Investigation with STR Genotyping
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lamoth, F.; Lockhart, S.R.; Berkow, E.L.; Calandra, T. Changes in the epidemiological landscape of invasive candidiasis. J. Antimicrob. Chemother. 2018, 73, i4–i13. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.W. Global incidence and mortality of severe fungal disease. Lancet Infect. Dis. 2024, 23, S1473–S3099. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Ahmad, S.; Al-Sweih, N.; Mokaddas, E.; Al-Banwan, K.; Alfouzan, W.; Al-Obaid, I.; Al-Obaid, K.; Asadzadeh, M.; Jeragh, A.; et al. Changing trends in epidemiology and antifungal susceptibility patterns of six bloodstream Candida species isolates over a 12-year period in Kuwait. PLoS ONE 2019, 14, e0216250. [Google Scholar] [CrossRef] [PubMed]
- Astvad, K.M.T.; Johansen, H.K.; Røder, B.L.; Rosenvinge, F.S.; Knudsen, J.D.; Lemming, L.; Schønheyder, H.C.; Hare, R.K.; Kristensen, L.; Nielsen, L.; et al. Update from a 12-Year Nationwide Fungemia Surveillance: Increasing Intrinsic and Acquired Resistance Causes Concern. J. Clin. Microbiol. 2018, 56, e01564-17. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Iqbal, N.; Cleveland, A.A.; Farley, M.M.; Harrison, L.H.; Bolden, C.B.; Baughman, W.; Stein, B.; Hollick, R.; Park, B.J.; et al. Species identification and antifungal susceptibility testing of Candida bloodstream isolates from population-based surveillance studies in two U.S. cities from 2008 to 2011. J. Clin. Microbiol. 2012, 50, 3435–3442. [Google Scholar] [CrossRef] [PubMed]
- Guinea, J. Global trends in the distribution of Candida species causing candidemia. Clin. Microbiol. Infect 2014, 20, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Doi, A.M.; Pignatari, A.C.C.; Edmond, M.B.; Marra, A.R.; Camargo, L.F.A.; Siqueira, R.A.; da Mota, V.P.; Colombo, A.L. Epidemiology and Microbiologic Characterization of Nosocomial Candidemia from a Brazilian National Surveillance Program. PLoS ONE 2016, 11, e0146909. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Miyazaki, T.; Ito, Y.; Wakayama, M.; Shibuya, K.; Yamashita, K.; Takazono, T.; Saijo, T.; Shimamura, S.; Yamamoto, K.; et al. Virulence assessment of six major pathogenic Candida species in the mouse model of invasive candidiasis caused by fungal translocation. Sci. Rep. 2020, 10, 3814. [Google Scholar] [CrossRef] [PubMed]
- Andes, D.R.; Safdar, N.; Baddley, J.W.; Playford, G.; Reboli, A.C.; Rex, J.H.; Sobel, J.D.; Pappas, P.G.; Kullberg, J.B.; Mycoses Study Group. Impact of treatment strategy on outcomes in patients with candidemia and other forms of invasive candidiasis: A patient-level quantitative review of randomized trials. Clin. Infect. Dis. 2012, 54, 1110–1122. [Google Scholar] [CrossRef]
- Hoenigl, M.; Salmanton-García, J.; Egger, M.; Gangneux, J.P.; Bicanic, T.; Arikan-Akdagli, S.; Alastruey-Izqueirdo, A.; Klimko, N.; Barac, A.; Özenci, V.; et al. Guideline adherence and survival of patients with candidaemia in Europe: Results from the ECMM Candida III multinational European observational cohort study. Lancet Infect. Dis. 2023, 23, 751–761. [Google Scholar] [CrossRef]
- Farmakiotis, D.; Kontoyiannis, D.P. Epidemiology of antifungal resistance in human pathogenic yeasts: Current viewpoint and practical recommendations for management. Int. J. Antimicrob. Agents 2017, 50, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Daneshnia, F.; de Almeida Júnior, J.N.; Ilkit, M.; Lombardi, L.; Perry, A.M.; Gao, M.; Nobile, C.J.; Egger, M.; Perlin, D.S.; Zhai, B.; et al. Worldwide emergence of fluconazole-resistant Candida parapsilosis: Current framework and future research roadmap. Lancet Microbe 2023, 4, e470–e480. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Daneshnia, F.; Najafzadeh, M.J.; Hagen, F.; Mahmoudi, S.; Salehi, M.; Zarrinfar, H.; Namvar, Z.; Zareshahrabadi, Z.; Khodavaisy, S.; et al. Evaluation of Molecular Epidemiology, Clinical Characteristics, Antifungal Susceptibility Profiles, and Molecular Mechanisms of Antifungal Resistance of Iranian Candida parapsilosis Species Complex Blood Isolates. Front. Cell Infect. Microbiol. 2020, 10, 206. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A.; Sood, P.; Rudramurthy, S.M.; Chen, S.; Kaur, H.; Capoor, M.; Chhina, D.; Rao, R.; Eshwara, V.K.; Xess, I.; et al. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med. 2015, 41, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Spruijtenburg, B.; Baqueiro, C.C.S.Z.; Colombo, A.L.; Meijer, E.F.J.; de Almeida, J.N., Jr.; Berrio, I.; Fernández, N.B.; Chaves, G.M.; Meis, J.F.; de Groot, T.; et al. Short Tandem Repeat Genotyping and Antifungal Susceptibility Testing of Latin American Candida tropicalis Isolates. J. Fungi 2023, 9, 207. [Google Scholar] [CrossRef] [PubMed]
- Ahangarkani, F.; Shokohi, T.; Rezai, M.S.; Ilkit, M.; Nesheli, H.M.; Karami, H.; Tamaddoni, A.; Alizadeh-Navaei, R.; Khodavaisy, S.; Meis, J.F.; et al. Epidemiological features of nosocomial candidaemia in neonates, infants and children: A multicentre study in Iran. Mycoses 2020, 63, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Najafzadeh, M.J.; Shaban, T.; Zarrinfar, H.; Sedaghat, A.; Hosseinikargar, N.; Berenji, F.; Jalali, M.; Lackner, M.; James, J.E.; Ilkit, M.; et al. COVID-19 associated candidemia: From a shift in fungal epidemiology to a rise in azole drug resistance. Med. Mycol. 2024, 62, myae031. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Shaban, T.; Zarrinfar, H.; Roudbary, M.; Ghazanfari, M.; Hedayati, M.T.; Sedaghat, A.; Ilkit, M.; Najafzadeh, M.J.; Perlin, D.S. Candidemia among Iranian Patients with Severe COVID-19 Admitted to ICUs. J. Fungi 2021, 7, 280. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Wickes, B.L.; Ilkit, M.; Pincus, D.H.; Daneshnia, F.; Pan, W.; Fang, W.; Boekhout, T. Identification of Mycoses in Developing Countries. J. Fungi 2019, 5, 90. [Google Scholar] [CrossRef]
- Thomaz, D.Y.; de Almeida, J.N., Jr.; Lima, G.M.E.; Nunes, M.O.; Camargo, C.H.; Grenfell, R.C.; Benard, G.; Del Negro, G.M.B. An Azole-Resistant Candida parapsilosis Outbreak: Clonal Persistence in the Intensive Care Unit of a Brazilian Teaching Hospital. Front. Microbiol. 2018, 9, 2997. [Google Scholar] [CrossRef]
- Stielow, J.B.; Lévesque, C.A.; Seifert, K.A.; Meyer, W.; Iriny, L.; Smits, D.; Renfurm, R.; Verkley, G.J.M.; Groenewald, M.; Chaduli, D.; et al. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia 2015, 35, 242–263. [Google Scholar] [CrossRef]
- CLSI standards M27; Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 4th ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017.
- CLSI supplement M57S; Epidemiological Cutoff Values for Antifungal Susceptibility Testing, 4th ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022.
- de Groot, T.; Spruijtenburg, B.; Parnell, L.A.; Chow, N.A.; Meis, J.F. Optimization and Validation of Candida auris Short Tandem Repeat Analysis. Microbiol. Spectr. 2022, 10, e0264522. [Google Scholar] [CrossRef]
- Diab-Elschahawi, M.; Forstner, C.; Hagen, F.; Meis, J.F.; Lassnig, A.M.; Presterl, E.; Klaassen, C.H.W. Microsatellite genotyping clarified conspicuous accumulation of Candida parapsilosis at a cardiothoracic surgery intensive care unit. J. Clin. Microbiol. 2012, 50, 3422–3426. [Google Scholar] [CrossRef]
- Vaezi, A.; Fakhim, H.; Khodavaisy, S.; Alizadeh, A.; Nazeri, M.; Soleimani, A.; Boekhout, T.; Badali, H. Epidemiological and mycological characteristics of candidemia in Iran: A systematic review and meta-analysis. J. Mycol. Med. 2017, 27, 146–152. [Google Scholar] [CrossRef]
- Charsizadeh, A.; Mirhendi, H.; Nikmanesh, B.; Eshaghi, H.; Makimura, K. Microbial epidemiology of candidaemia in neonatal and paediatric intensive care units at the Children’s Medical Center, Tehran. Mycoses 2018, 61, 22–29. [Google Scholar] [CrossRef]
- Arastehfar, A.; Yazdanpanah, S.; Bakhtiari, M.; Fang, W.; Pan, W.; Mahmoudi, S.; Pakshir, K.; Daneshnia, F.; Boekhout, T.; Ilkit, M.; et al. Epidemiology of candidemia in Shiraz, southern Iran: A prospective multicenter study (2016–2018). Med. Mycol. 2021, 59, 422–430. [Google Scholar] [CrossRef]
- Kord, M.; Salehi, M.; Khodavaisy, S.; Hashemi, S.J.; Ghazvini, R.D.; Rezaei, S.; Maleki, A.; Elmimoghaddam, A.; Alijani, N.; Abdollahi, A.; et al. Epidemiology of yeast species causing bloodstream infection in Tehran, Iran (2015–2017); superiority of 21-plex PCR over the Vitek 2 system for yeast identification. J. Med. Microbiol. 2020, 69, 712–720. [Google Scholar] [CrossRef]
- Kord, M.; Salehi, M.; Hashemi, S.J.; Abdollahi, A.; Alijani, N.; Maleki, A.; Mahmoudi, S.; Ahmadikia, K.; Parsameher, N.; Moradi, M.; et al. Clinical, epidemiological, and mycological features of patients with candidemia: Experience in two tertiary referral centers in Iran. Curr. Med. Mycol. 2022, 8, 9–17. [Google Scholar] [CrossRef]
- Karakoyun, A.S.; Spruijtenburg, B.; Unal, N.; Meijer, E.F.J.; Sucu, M.; Hilmioğlu-Polat, S.; Meis, J.F.; de Groot, T.; Ilkit, M. Molecular typing and antifungal susceptibility profile of Candida krusei bloodstream isolates from Turkiye. Med. Mycol. 2024, 62, myae005. [Google Scholar] [CrossRef]
- Sharma, M.; Chakrabarti, A. Candidiasis and Other Emerging Yeasts. Curr. Fungal Infect. Rep. 2023, 17, 15–24. [Google Scholar] [CrossRef]
- Lass-Flörl, C.; Kanj, S.S.; Govender, N.P.; Thompson III, G.R.; Ostrosky-Zeichner, L.; Govrins, M.A. Invasive candidiasis. Nat. Rev. Dis. Primers 2024, 10, 20. [Google Scholar] [CrossRef]
- Asner, S.A.; Giulieri, S.; Diezi, M.; Marchetti, O.; Sanglard, D. Acquired Multidrug Antifungal Resistance in Candida lusitaniae during Therapy. Antimicrob. Agents Chemother. 2015, 59, 7715–7722. [Google Scholar] [CrossRef]
- Coste, A.T.; Kritikos, A.; Li, J.; Khanna, N.; Goldenberger, D.; Garzoni, C.; Zehnder, C.; Boggian, K.; Neofytos, D.; Riat, A.; et al. Emerging echinocandin-resistant Candida albicans and glabrata in Switzerland. Infection 2020, 48, 761–766. [Google Scholar] [CrossRef]
- Spruijtenburg, B.; Ahmad, S.; Asadzadeh, M.; Alfouzan, W.; Al-Obaid, I.; Mokaddas, E.; Meijer, E.F.J.; Meis, J.F.; de Groot, T. Whole genome sequencing analysis demonstrates therapy-induced echinocandin resistance in Candida auris isolates. Mycoses 2023, 66, 1079–1086. [Google Scholar] [CrossRef]
- Spruijtenburg, B.; Rudramurthy, S.M.; Meijer, E.F.J.; van Haren, M.H.I.; Kaur, H.; Chakrabarti, A.; Meis, J.F.; de Groot, T. Application of Novel Short Tandem Repeat Typing for Wickerhamomyces anomalus Reveals Simultaneous Outbreaks within a Single Hospital. Microorganisms 2023, 11, 1525. [Google Scholar] [CrossRef]
- Spruijtenburg, B.; Meijer, E.F.J.; Xiao, M.; Shawky, S.M.; Meis, J.F.; de Groot, T.; El-Kholy, M.A. Genotyping and susceptibility testing uncovers large azole-resistant Candida tropicalis clade in Alexandria, Egypt. J. Glob. Antimicrob. Resist 2023, 34, 99–105. [Google Scholar] [CrossRef]
- Alcoceba, E.; Gómez, A.; Lara-Esbrí, P.; Oliver, A.; Ferre Beltrán, A.; Ayestarán, I.; Muñoz, P.; Escribano, P.; Guinea, J. Fluconazole-resistant Candida parapsilosis clonally related genotypes: First report proving the presence of endemic isolates harbouring the Y132F ERG11 gene substitution in Spain. Clin. Microbiol. Infect 2022, 28, 1113–1119. [Google Scholar] [CrossRef]
- Misas, E.; Witt, L.S.; Farley, M.M.; Thomas, S.; Jenkins, E.N.; Gade, L.; Peterson, J.G.; Mesa Restrepo, A.; Fridkin, S.; Lockhart, S.R.; et al. Molecular and Epidemiological Investigation of Fluconazole-resistant Candida parapsilosis-Georgia, United States, 2021. Open Forum Infect Dis. 2024, 11, ofae264. [Google Scholar] [CrossRef]
- Guinea, J.; Mezquita, S.; Gómez, A.; Padilla, B.; Zamora, E.; Sánchez-Luna, M.; Sánchez-Carrillo, C.; Muñoz, P.; Escribano, P. Whole genome sequencing confirms Candida albicans and Candida parapsilosis microsatellite sporadic and persistent clones causing outbreaks of candidemia in neonates. Med. Mycol. 2022, 60, myab068. [Google Scholar] [CrossRef]
- De Carolis, E.; Posteraro, B.; Falasca, B.; Spruijtenburg, B.; Meis, J.F.; Sanguinetti, M. The Fourier-transform infrared spectroscopy-based method as a new typing tool for Candida parapsilosis clinical isolates. Microbiol. Spectr. 2023, 11, e0238823. [Google Scholar] [CrossRef]
- Díaz-García, J.; Gómez, A.; Alcalá, L.; Reigadas, E.; Sánchez-Carrillo, C.; Pérez-Ayala, A.; Gómez-García de la Pedrosa, E.; González-Romo, F.; Merino-Amador, P.; Soledad Cuétara, M.; et al. Evidence of Fluconazole-Resistant Candida parapsilosis Genotypes Spreading across Hospitals Located in Madrid, Spain and Harboring the Y132F ERG11p Substitution. Antimicrob. Agents Chemother. 2022, 66, e00710-22. [Google Scholar] [CrossRef]
- McTaggart, L.R.; Eshaghi, A.; Hota, S.; Poutanen, S.M.; Johnstone, J.; De Luca, D.G.; Bharat, A.; Patel, S.N.; Kus, J.V. First Canadian report of transmission of fluconazole-resistant Candida parapsilosis within two hospital networks confirmed by genomic analysis. J. Clin. Microbiol. 2024, 62, e0116123. [Google Scholar] [CrossRef]

| Characteristic | Overall (n = 160, 100%) | C. parapsilosis (n = 58, 36%) | C. albicans (n = 52, 33%) | C. tropicalis (n = 20, 13%) | C. glabrata (n = 15, 10%) | C. krusei (n = 7, 4%) | Other Yeasts 1 (n = 8, 5%) |
|---|---|---|---|---|---|---|---|
| Age | |||||||
| <16 years | 63 (39) | 28 (48) | 23 (44) | 5 (25) | 3 (20) | - | 4 (50) |
| ≥16 years | 96 (60) | 30 (52) | 29 (56) | 15 (75) | 12 (80) | 7 (100) | 3 (38) |
| Unknown | 1 (1) | - | - | - | - | - | 1 (13) |
| Sex | |||||||
| Male | 83 (52) | 28 (48) | 33 (63) | 7 (35) | 8 (53) | 4 (57) | 3 (38) |
| Female | 77 (48) | 30 (52) | 19 (37) | 13 (65) | 7 (47) | 3 (43) | 5 (63) |
| Hospital | |||||||
| Emam Reza | 96 (60) | 41 (70) | 27 (52) | 8 (40) | 13 (87) | 4 (57) | 3 (38) |
| Doctor Sheikh | 42 (26) | 14 (24) | 20 (38) | 4 (20) | 1 (7) | - | 3 (38) |
| Ghaem | 15 (9) | - | 5 (10) | 5 (25) | - | 3 (42) | 2 (25) |
| 22 Bahman | 5 (3) | 2 (3) | - | 2 (10) | 1 (7) | - | - |
| Arya | 2 (1) | 1 (2) | - | 1 (5) | - | - | - |
| Wards | |||||||
| ICU | 79 (49) | 25 (43) | 30 (58) | 6 (30) | 9 (60) | 6 (86) | 3 (38) |
| Internal | 51 (32) | 24 (41) | 16 (30) | 3 (15) | 4 (27) | - | 4 (50) |
| Emergency | 13 (8) | 5 (9) | 4 (8) | 1 (5) | 2 (13) | 1 (14) | - |
| Surgery | 5 (3) | 3 (5) | 1 (2) | 1 (5) | - | - | - |
| Other | 12 (8) | 1 (2) | 1 (2) | 9 (45) | - | - | 1 (13) |
| Underlying conditions | |||||||
| Cardiovascular complications | 13 (8) | 5 (9) | 3 (6) | 4 (20) | - | - | 1 (14) |
| Malignancy | 25 (16) | 8 (14) | 10 (19) | 4 (20) | 2 (13) | 1 (14) | - |
| Diabetes | 35 (22) | 13 (22) | 9 (17) | 4 (20) | 5 (33) | 3 (43) | 1 (14) |
| Internal complications | 67 (42) | 26 (45) | 20 (38) | 8 (20) | 7 (47) | 2 (29) | 4 (57) |
| Cerebrospinal complications | 5 (3) | 1 (2) | 3 (6) | - | - | 1 (14) | - |
| None | 14 (9) | 5 (9) | 7 (13) | - | 1 (7) | - | 1 (14) |
| Antifungal treatment | |||||||
| Fluconazole | 48 (30) | 10 (17) | 16 (31) | 3 (15) | 12 (80) | 3 (43) | 4 (50) |
| Liposomal Amphotericin B | 40 (25) | 7 (12) | 20 (38) | 9 (45) | 1 (7) | 3 (43) | - |
| Caspofungin | 3 (2) | 1 (2) | 1 (2) | 1 (5) | - | - | - |
| Clotrimazole | 3 (2) | - | 3 (6) | - | - | - | - |
| Nystatin | 2 (1) | 2 (3) | - | - | - | - | |
| Fluconazole and nystatin | 2 (1) | 2 (3) | - | - | - | - | - |
| Fluconazole and amphotericin B | 7 (4) | 2 (3) | 3 (6) | - | - | 1 (14) | 1 (13) |
| Fluconazole and caspofungin | 1 (1) | - | 1 (2) | - | - | - | - |
| Not treated | 54 (34) | 34 (59) | 8 (15) | 7 (35) | 2 (13) | - | 3 (38) |
| Outcome | |||||||
| Died | 82 (51) | 33 (57) | 20 (39) | 12 (60) | 10 (67) | 5 (61) | 2 (25) |
| Survived | 74 (46) | 25 (43) | 31 (60) | 8 (40) | 5 (33) | 1 (14) | 4 (50) |
| Unknown | 4 (3) | - | 1 (2) | - | - | 1 (14) | 2 (25) |
| Antifungal Drug | Species | Range | GM | MIC50 | MIC90 | n Resistant/Non-WT (%) |
|---|---|---|---|---|---|---|
| Amphotericin B | C. albicans | 0.125–4 | 0.545 | 0.5 | 1 | 1 (2) |
| C. parapsilosis | 0.031–1 | 0.500 | 0.5 | 1 | 0 | |
| C. tropicalis | 0.25–1 | 0.569 | 0.5 | 1 | 0 | |
| C. glabrata | 0.5–1 | 0.730 | 1 | 1 | 0 | |
| C. krusei | 1 | 1 | 1 | N/A | 0 | |
| C. lusitaniae | 0.5–1 | 0.630 | 0.5 | N/A | 0 | |
| Fluconazole | C. albicans | 0.125–4 | 0.380 | 0.25 | 1 | 0 |
| C. parapsilosis | 0.125–4 | 0.412 | 0.5 | 1 | 0 | |
| C. tropicalis | 0.25–1 | 0.595 | 1 | 1 | 0 | |
| C. glabrata | 0.25–4 | 2.416 | 4 | 4 | 0 | |
| C. krusei | 8–32 | 17.665 | 16 | N/A | N/A | |
| C. lusitaniae | 0.5–2 | 0.707 | 0.5 | N/A | 1 (17) | |
| Voriconazole | C. albicans | 0.032–0.125 | 0.036 | 0.032 | 0.064 | 0 |
| C. parapsilosis | 0.032–0.125 | 0.035 | 0.032 | 0.064 | 0 | |
| C. tropicalis | 0.032–0.125 | 0.051 | 0.064 | 0.125 | 0 | |
| C. glabrata | 0.032–0.125 | 0.050 | 0.064 | 0.125 | 0 | |
| C. krusei | 0.064–0.25 | 0.125 | 0.125 | N/A | 0 | |
| C. lusitaniae | 0.032 | 0.032 | 0.032 | N/A | N/A | |
| Micafungin | C. albicans | 0.016–0.5 | 0.022 | 0.016 | 0.064 | 0 |
| C. parapsilosis | 0.016–1 | 0.255 | 0.5 | 1 | 0 | |
| C. tropicalis | 0.016–0.032 | 0.017 | 0.016 | 0.032 | 0 | |
| C. glabrata | 0.016 | 0.016 | 0.016 | 0.016 | 0 | |
| C. krusei | 0.064–0.125 | 0.103 | 0.125 | N/A | 0 | |
| C. lusitaniae | 0.016–0.064 | 0.029 | 0.032 | N/A | 0 | |
| Anidulafungin | C. albicans | 0.016–1 | 0.027 | 0.016 | 0.25 | 1 (2) |
| C. parapsilosis | 0.016–2 | 0.376 | 1 | 1 | 0 | |
| C. tropicalis | 0.016–0.064 | 0.029 | 0.032 | 0.064 | 0 | |
| C. glabrata | 0.016–0.064 | 0.032 | 0.032 | 0.064 | 0 | |
| C. krusei | 0.032–0.125 | 0.070 | 0.064 | N/A | 0 | |
| C. lusitaniae | 0.032–0.064 | 0.057 | 0.064 | N/A | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dolatabadi, S.; Najafzadeh, M.J.; Raeisabadi, A.; Zarrinfar, H.; Jalali, M.; Spruijtenburg, B.; Meijer, E.F.J.; Meis, J.F.; Lass-Flörl, C.; de Groot, T. Epidemiology of Candidemia in Mashhad, Northeast Iran: A Prospective Multicenter Study (2019–2021). J. Fungi 2024, 10, 481. https://doi.org/10.3390/jof10070481
Dolatabadi S, Najafzadeh MJ, Raeisabadi A, Zarrinfar H, Jalali M, Spruijtenburg B, Meijer EFJ, Meis JF, Lass-Flörl C, de Groot T. Epidemiology of Candidemia in Mashhad, Northeast Iran: A Prospective Multicenter Study (2019–2021). Journal of Fungi. 2024; 10(7):481. https://doi.org/10.3390/jof10070481
Chicago/Turabian StyleDolatabadi, Somayeh, Mohammad Javad Najafzadeh, Abbas Raeisabadi, Hossein Zarrinfar, Mahsa Jalali, Bram Spruijtenburg, Eelco F. J. Meijer, Jacques F. Meis, Cornelia Lass-Flörl, and Theun de Groot. 2024. "Epidemiology of Candidemia in Mashhad, Northeast Iran: A Prospective Multicenter Study (2019–2021)" Journal of Fungi 10, no. 7: 481. https://doi.org/10.3390/jof10070481






