Molecular Evaluation of the mRNA Expression of the ERG11, ERG3, CgCDR1, and CgSNQ2 Genes Linked to Fluconazole Resistance in Candida glabrata in a Colombian Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Type
2.2. Strains and Sample-Handling Conditions
2.3. Antifungal Susceptibility Testing
2.4. Fluconazole Exposure Test
2.5. RNA Extraction
2.6. Quality Criteria for RNA Extraction
2.7. Protocol Standardization
2.8. Reverse Transcription (RT) Protocol
2.9. Selection and Optimization of the Primers for the qPCR
2.10. qPCR Experiment
2.11. qPCR Reaction Analysis
2.12. Molecular Docking
2.13. Statistical Analysis
3. Results
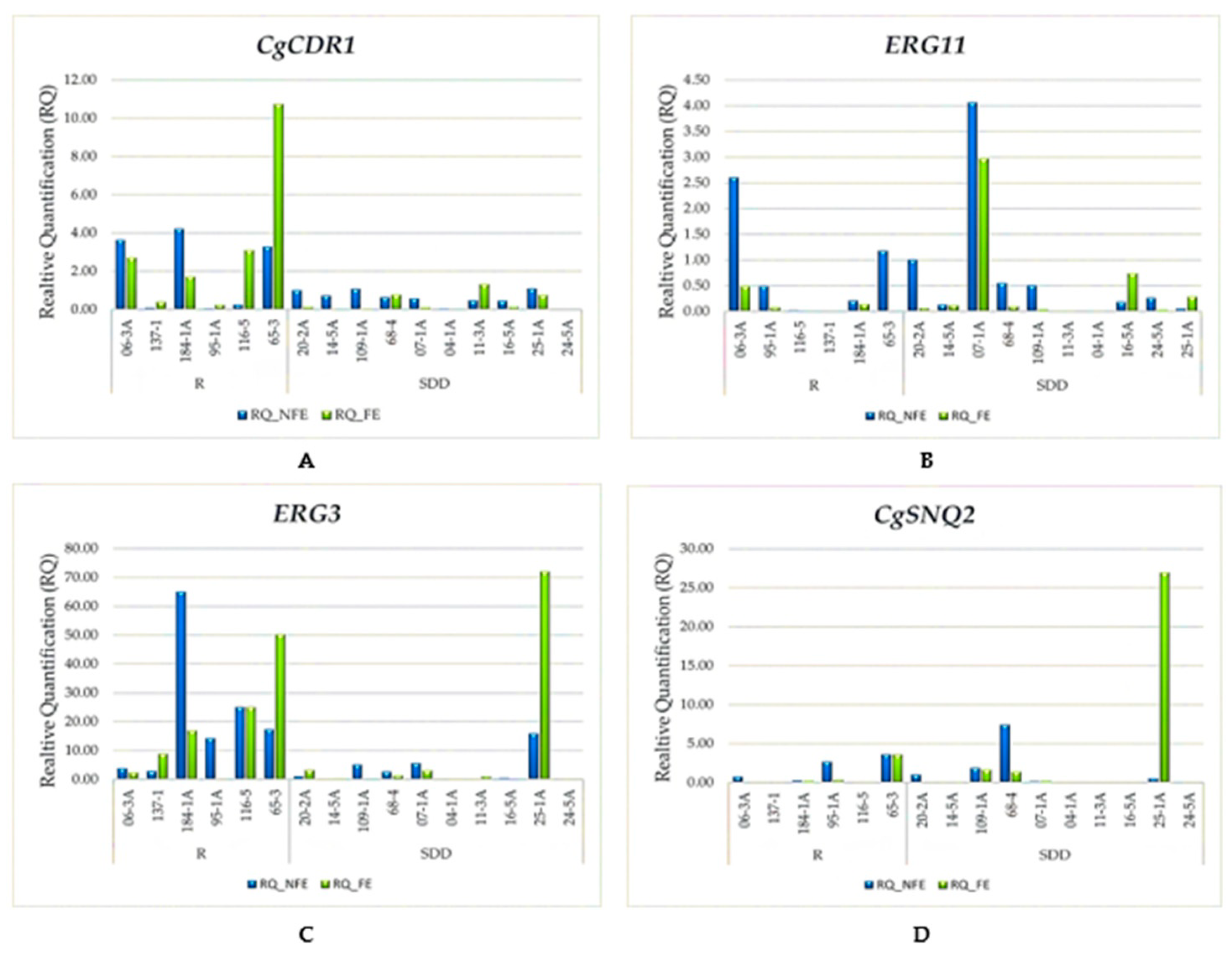
3.1. Analysis of the Relative Gene Expression
3.2. Results of the Predictive Model on the Susceptibility Profile
3.3. Molecular Docking
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Whaley, S.G.; Berkow, E.L.; Rybak, J.M.; Nishimoto, A.T.; Barker, K.S.; Rogers, P.D. Azole Antifungal Resistance in Candida albicans and Emerging Non-albicans Candida Species. Front. Microbiol. 2016, 7, 2173. [Google Scholar] [CrossRef] [PubMed]
- Arendrup, M.C.; Patterson, T.F. Multidrug-Resistant Candida: Epidemiology, Molecular Mechanisms, and Treatment. J. Infect. Dis. 2017, 216 (Suppl. S3), S445–S451. [Google Scholar] [CrossRef] [PubMed]
- Zuluaga Rodríguez, A.; de Bedout Gómez, C.; Agudelo Restrepo, C.A.; Hurtado Parra, H.; Arango Arteaga, M.; Restrepo Moreno, Á.; González Marín, Á. Sensibilidad a fluconazol y voriconazol de especies de Candida aisladas de pacientes provenientes de unidades de cuidados intensivos en Medellín, Colombia (2001–2007). Rev. Iberoam. Micol. 2010, 27, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Cortés, J.A.; Reyes, P.; Gómez, C.H.; Cuervo, S.I.; Rivas, P.; Casas, C.A.; Sánchez, R. Clinical and epidemiological characteristics and risk factors for mortality in patients with candidemia in hospitals from Bogotá, Colombia. Braz. J. Infect. Dis. 2014, 18, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, N.A.; Cano, L.E.; De Bedout, C.; Arbeláez, C.A.; Roncancio, G.; Tabares, M.; Robledo, C.G.; Robledo, J. Association of clinical and demographic factors in invasive candidiasis caused by fluconazole-resistant Candida species: A study in 15 hospitals, Medellín, Colombia 2010–2011. Diagn. Microbiol. Infect. Dis. 2014, 79, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Cortés, J.A.; Ruiz, J.F.; Melgarejo-Moreno, L.N.; Lemos, E.V. Candidemia en colombia. Biomédica 2020, 40, 195–207. [Google Scholar] [CrossRef]
- Vides-Peña, C.A.; Bolaño-Ardila, N.D.; Vides-Peña, M.V.; Córdoba, S.B. Epidemiología de la candidemia y sensibilidad in vitro frente a azoles, anfotericina B y caspofungina en una institución de salud de Valledupar, Colombia. Med. Lab. 2015, 21, 255–266. [Google Scholar] [CrossRef]
- Yao, D.; Chen, J.; Chen, W.; Li, Z.; Hu, X. Mechanisms of azole resistance in clinical isolates of Candida glabrata from two hospitals in China. Infect. Drug Resist. 2019, 12, 771–781. [Google Scholar] [CrossRef]
- Shahrokhi, S.; Noorbakhsh, F.; Rezaie, S. Quantification of CDR1 Gene Expression in Fluconazole Resistant Candida Glabrata Strains Using Real-time PCR. Iran. J. Public Health 2017, 46, 1118–1122. [Google Scholar] [PubMed]
- Tortorano, A.M.; Prigitano, A.; Morroni, G.; Brescini, L.; Barchiesi, F. Candidemia: Evolution of Drug Resistance and Novel Therapeutic Approaches. Infect. Drug Resist. 2021, 14, 5543–5553. [Google Scholar] [CrossRef]
- Whaley, S.G.; Zhang, Q.; Caudle, K.E.; Rogers, P.D. Relative Contribution of the ABC Transporters Cdr1, Pdh1, and Snq2 to Azole Resistance in Candida glabrata. Antimicrob. Agents Chemother. 2018, 62, e01070-18. [Google Scholar] [CrossRef] [PubMed]
- Maheronnaghsh, M.; Teimoori, A.; Dehghan, P.; Fatahinia, M. The evaluation of the overexpression of the ERG-11, MDR-1, CDR-1, and CDR-2 genes in fluconazole-resistant Candida albicans isolated from Ahvazian cancer patients with oral candidiasis. J. Clin. Lab. Anal. 2022, 36, e24208. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Singh, S.; Sharma, D.; Chakrabarti, A.; Rudramurthy, S.M.; Ghosh, A.K. Dynamics of in vitro development of azole resistance in Candida tropicalis. J. Glob. Antimicrob. Resist. 2020, 22, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Sakagami, T.; Kawano, T.; Yamashita, K.; Yamada, E.; Fujino, N.; Kaeriyama, M.; Fukuda, Y.; Nomura, N.; Mitsuyama, J.; Suematsu, H.; et al. Antifungal susceptibility trend and analysis of resistance mechanism for Candida species isolated from bloodstream at a Japanese university hospital. J. Infect. Chemother. 2019, 25, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Healey, K.R.; Perlin, D.S. Fungal Resistance to Echinocandins and the MDR Phenomenon in Candida glabrata. J. Fungi 2018, 4, 105. [Google Scholar] [CrossRef]
- Zare-Bidaki, M.; Maleki, A.; Ghanbarzadeh, N.; Nikoomanesh, F. Expression pattern of drug-resistance genes ERG11 and TAC1 in Candida albicans Clinical isolates. Mol. Biol. Rep. 2022, 49, 11625–11633. [Google Scholar] [CrossRef]
- Liu, Z.; Myers, L.C. Mediator Tail Module Is Required for Tac1-Activated CDR1 Expression and Azole Resistance in Candida albicans. Antimicrob. Agents Chemother. 2017, 61, e01342-17. [Google Scholar] [CrossRef]
- Feng, W.; Yang, J.; Xi, Z.; Ji, Y.; Zhu, X.; Yang, L.; Ma, Y. Regulatory Role of ERG3 and Efg1 in Azoles-Resistant Strains of Candida albicans Isolated from Patients Diagnosed with Vulvovaginal Candidiasis. Indian J. Microbiol. 2019, 59, 514–524. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Myers, L.C. Candida albicans Swi/Snf and Mediator Complexes Differentially Regulate Mrr1-Induced MDR1 Expression and Fluconazole Resistance. Antimicrob. Agents Chemother. 2017, 61, e01344-17. [Google Scholar] [CrossRef]
- Vu, B.G.; Thomas, G.H.; Moye-Rowley, W.S. Evidence that Ergosterol Biosynthesis Modulates Activity of the Pdr1 Transcription Factor in Candida glabrata. mBio 2019, 10, 1–20. [Google Scholar] [CrossRef]
- Sanguinetti, M.; Posteraro, B.; Fiori, B.; Ranno, S.; Torelli, R.; Fadda, G. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob. Agents Chemother. 2005, 49, 668–679. [Google Scholar] [CrossRef] [PubMed]
- El Said, M.; Badawi, H.; Gamal, D.; Salem, D.; Dahroug, H.; El-Far, A. Detection of ERG11 gene in fluconazole resistant urinary candida isolates. Egypt. J. Immunol. 2022, 29, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Spettel, K.; Barousch, W.; Makristathis, A.; Zeller, I.; Nehr, M.; Selitsch, B.; Lackner, M.; Rath, P.-M.; Steinmann, J.; Willinger, B. Analysis of antifungal resistance genes in Candida albicans and Candida glabrata using next generation sequencing. PLoS ONE 2019, 14, e0210397. [Google Scholar] [CrossRef] [PubMed]
- Luna-Tapia, A.; Willems, H.M.E.; Parker, J.E.; Tournu, H.; Barker, K.S.; Nishimoto, A.T.; Rogers, P.D.; Kelly, S.L.; Peters, B.M.; Palmer, G.E. Loss of Upc2p-Inducible ERG3 Transcription Is Sufficient to Confer Niche-Specific Azole Resistance without Compromising Candida albicans Pathogenicity. mBio 2018, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Robbins, N.; Cowen, L.E. Antifungal drug resistance: Deciphering the mechanisms governing multidrug resistance in the fungal pathogen Candida glabrata. Curr. Biol. 2021, 31, R1520–R1523. [Google Scholar] [CrossRef] [PubMed]
- Vu, B.G.; Moye-Rowley, W.S. Azole-Resistant Alleles of ERG11 in Candida glabrata Trigger Activation of the Pdr1 and Upc2A Transcription Factors. Antimicrob. Agents Chemother. 2022, 66, e0209821. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 4th ed.; M27-A4; CLSI: Wayne, PA, USA, 2017. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antifungal Susceptibility Testing of Yeasts M60; CLSI: Wayne, PA, USA, 2017. [Google Scholar]
- Méndez, C.C.; Sánchez, E.G.; Martín-Mazuelos, E. Actualización de los métodos de estudio de sensibilidad in vitro a los antifúngicos. Enfermedades Infecc. Microbiol. Clin. 2019, 37, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Caudle, K.E.; Barker, K.S.; Wiederhold, N.P.; Xu, L.; Homayouni, R.; Rogers, P.D. Genomewide expression profile analysis of the Candida glabrata Pdr1 regulon. Eukaryot. Cell 2011, 10, 373–383. [Google Scholar] [CrossRef]
- Silva, D.B.d.S.; Rodrigues, L.M.C.; de Almeida, A.A.; de Oliveira, K.M.P.; Grisolia, A.B. Novel point mutations in the ERG11 gene in clinical isolates of azole resistant Candida species. Mem. Inst. Oswaldo Cruz 2016, 111, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Szweda, P.; Gucwa, K.; Romanowska, E.; Dzierżanowska-Fangrat, K.; Naumiuk, Ł.; Brillowska-Dąbrowska, A.; Wojciechowska-Koszko, I.; Milewski, S. Mechanisms of azole resistance among clinical isolates of Candida glabrata in Poland. J. Med. Microbiol. 2015, 64, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Skrzypek, M.S.; Binkley, J.; Binkley, G.; Miyasato, S.R.; Simison, M.; Sherlock, G. The Candida Genome Database (CGD): Incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017, 45, D592–D596. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; A Salazar, G.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.-C.; Chen, L.-H.; Jiang, C.-R.; Deng, Y.-M.; Wang, D.-W.; Lin, C.-H.; Jou, R.; Wang, J.-K.; Wang, Y.-L. Sensible Functional Linear Discriminant Analysis Effectively Discriminates Enhanced Raman Spectra of Mycobacterium Species. Anal. Chem. 2021, 93, 2785–2792. [Google Scholar] [CrossRef] [PubMed]
- Safo, S.E.; Min, E.J.; Haine, L. Sparse linear discriminant analysis for multiview structured data. Biometrics 2022, 78, 612–623. [Google Scholar] [CrossRef] [PubMed]
- IBM SPSS Software [Internet]. 2023. Available online: https://www.ibm.com/es-es/spss (accessed on 28 June 2023).
- Castanheira, M.; Deshpande, L.M.; Davis, A.P.; Carvalhaes, C.G.; Pfaller, M.A. Azole resistance in Candida glabrata clinical isolates from global surveillance is associated with efflux overexpression. J. Glob. Antimicrob. Resist. 2022, 29, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Gohar, A.A.; Badali, H.; Shokohi, T.; Nabili, M.; Amirrajab, N.; Moazeni, M. Expression Patterns of ABC Transporter Genes in Fluconazole-Resistant Candida glabrata. Mycopathologia 2017, 182, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Won, E.J.; Choi, M.J.; Kim, M.-N.; Yong, D.; Lee, W.G.; Uh, Y.; Kim, T.S.; Byeon, S.A.; Lee, S.Y.; Kim, S.H.; et al. Fluconazole-Resistant Candida glabrata Bloodstream Isolates, South Korea, 2008–2018. Emerg. Infect. Dis. 2021, 27, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, H.O.; Arai, T.; Majima, H.; Watanabe, A.; Kamei, K. Genetic Basis of Azole and Echinocandin Resistance in Clinical Candida glabrata in Japan. Antimicrob. Agents Chemother. 2020, 64, e00783-20. [Google Scholar] [CrossRef]
- Biswas, C.; Marcelino, V.R.; Van Hal, S.; Halliday, C.; Martinez, E.; Wang, Q.; Kidd, S.; Kennedy, K.; Marriott, D.; Morrissey, C.O.; et al. Whole Genome Sequencing of Australian Candida glabrata Isolates Reveals Genetic Diversity and Novel Sequence Types. Front. Microbiol. 2018, 9, 2946. [Google Scholar] [CrossRef] [PubMed]
- Godinho, C.P.; Dias, P.J.; Ponçot, E.; Sá-Correia, I. The Paralogous Genes PDR18 and SNQ2, Encoding Multidrug Resistance ABC Transporters, Derive from a Recent Duplication Event, PDR18 Being Specific to the Saccharomyces Genus. Front. Genet. 2018, 9, 476. [Google Scholar] [CrossRef]
- Henry, K.W.; Nickels, J.T.; Edlind, T.D. Upregulation of ERG Genes in Candida Species by Azoles and Other Sterol Biosynthesis Inhibitors. Antimicrob. Agents Chemother. 2000, 44, 2693–2700. [Google Scholar] [CrossRef] [PubMed]
- Iyer, K.R.; Robbins, N.; Cowen, L.E. The role of Candida albicans stress response pathways in antifungal tolerance and resistance. iScience 2022, 25, 103953. [Google Scholar] [CrossRef] [PubMed]
- Berman, J.; Krysan, D.J. Drug resistance and tolerance in fungi. Nat. Rev. Microbiol. 2020, 18, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.A.; O’meara, T.R.; Cowen, L.E. Fitness Trade-Offs Associated with the Evolution of Resistance to Antifungal Drug Combinations. Cell Rep. 2015, 10, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Song, J.L.; Harry, J.B.; Eastman, R.T.; Oliver, B.G.; White, T.C. The Candida albicans lanosterol 14-α-demethylase (ERG11) gene promoter is maximally induced after prolonged growth with antifungal drugs. Antimicrob. Agents Chemother. 2004, 48, 1136–1144. [Google Scholar] [CrossRef] [PubMed]

| Sample | Fluconazole Concentration |
|---|---|
| 06-3A | 8 µg/mL |
| 95-1A | 4 µg/mL |
| 116-5 | 8 µg/mL |
| 137-1 | 4 µg/mL |
| 184-1A | 8 µg/mL |
| 65-3 | 16 µg/mL |
| 11-3A | 16 µg/mL |
| 04-1A | 32 µg/mL |
| 16-5A | 8 µg/mL |
| 24-5A | 8 µg/mL |
| 25-1A | 16 µg/mL |
| 20-2A | 16 µg/mL |
| 14-5A | 32 µg/mL |
| 07-1A | 8 µg/mL |
| 68-4 | 8 µg/mL |
| 109-1A | 8 µg/mL |
| Gene | Primer Sequence (5′–3′) | Reference | |
|---|---|---|---|
| CgCDR1 | Forward | CATACAAGAAACACCAAAGTCGGT | [30] |
| Reverse | GAGACACGCTAACGTTCACCAC | ||
| ERG11 | Forward | TCGGTCCATCTCTGTTTCTT | [31] |
| Reverse | GAACACTGGGGTGGTCAAGT | ||
| ERG 3 | Forward | AAGCGTGTGAACAAGGAC | [30] |
| Reverse | GCGTAGGTCTTCTCTGTGA | ||
| CgSNQ2 | Forward | CGTCCTATGTCTTCCTTACACCATT | [30] |
| Reverse | TTTGAACCGCTTTTGTCTCTGA | ||
| CgURA3 | Forward | GAAAACCAATCTTTGTGCTTCTCT | [32] |
| Reverse | CATGAGTCTTAAGCAAGCAAATGT |
| Analyzed Gene | Exposure to the Antifungal | Resistant One-Tailed Analysis | Susceptible and Dose-Dependent One-Tailed Analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Median | Minimum | Maximum | DE | p | Mean | Median | Minimum | Maximum | DE | p | ||
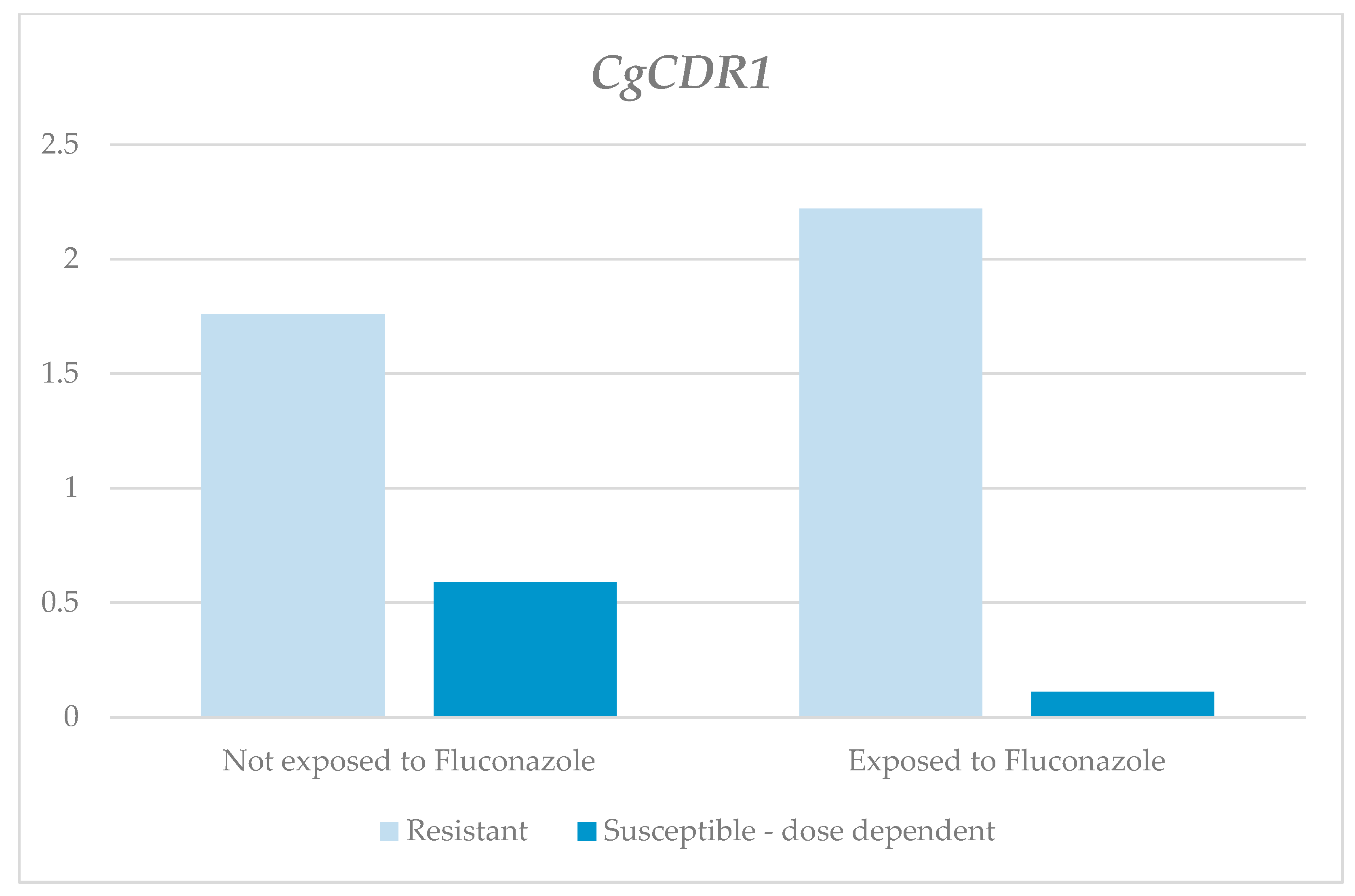
| CgCDR1 | Without Fluconazole | 1.91 | 1.76 | 0.05 | 4.22 | 1.99 | 0.231 1 | 0.6 | 0.59 | 0.04 | 1.09 | 0.38 | 0.046 1 |
| With Fluconazole | 3.15 | 2.22 | 0.24 | 10.75 | 3.89 | 0.33 | 0.11 | 0.01 | 1.33 | 0.45 | |||
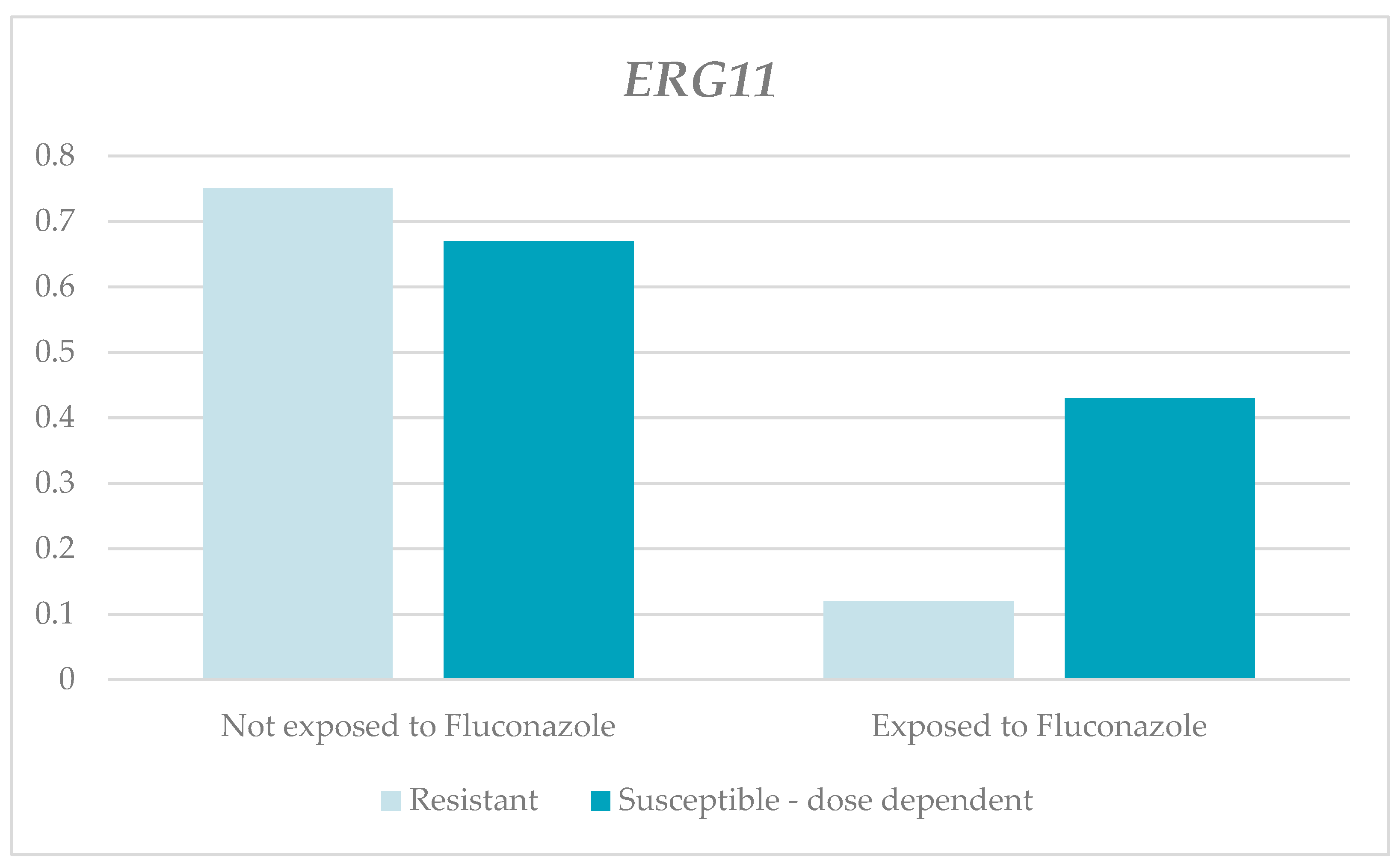
| ERG11 | Without fluconazole | 0.75 | 0.35 | 0.01 | 2.6 | 1.00 | 0.065 2 | 0.67 | 0.22 | 0 | 4.06 | 1.23 | 0.086 2 |
| With fluconazole | 0.12 | 0.04 | 0.01 | 0.49 | 0.18 | 0.43 | 0.08 | 0 | 2.97 | 0.91 | |||
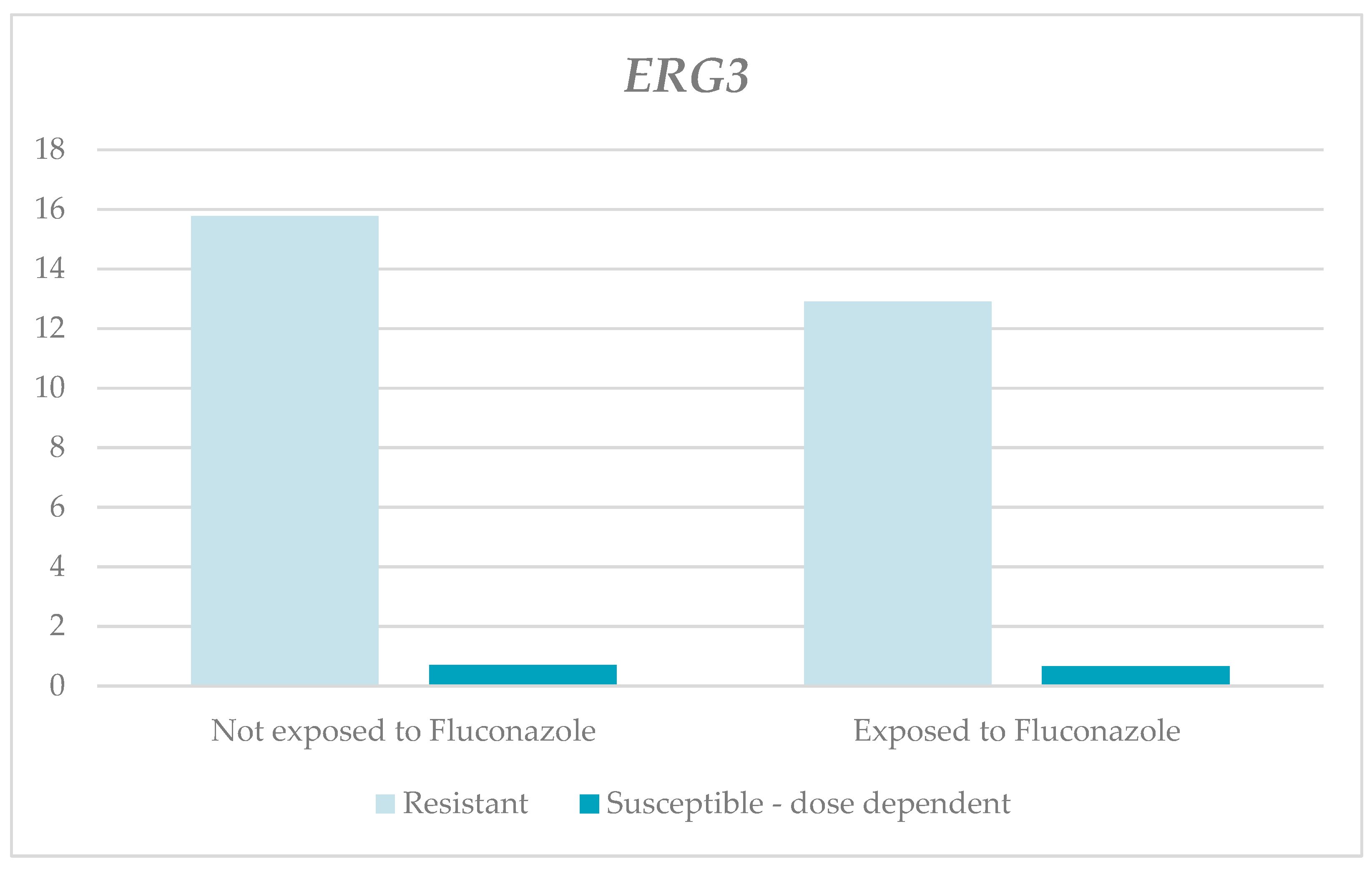
| ERG3 | Without fluconazole | 21.39 | 15.77 | 2.93 | 65 | 22.9 | 0.343 1 | 3.09 | 0.71 | 0 | 15.9 | 4.97 | 0.399 1 |
| With fluconazole | 17.28 | 12.94 | 0.01 | 50.3 | 18.64 | 8.1 | 0.66 | 0.01 | 72.12 | 22.51 | |||
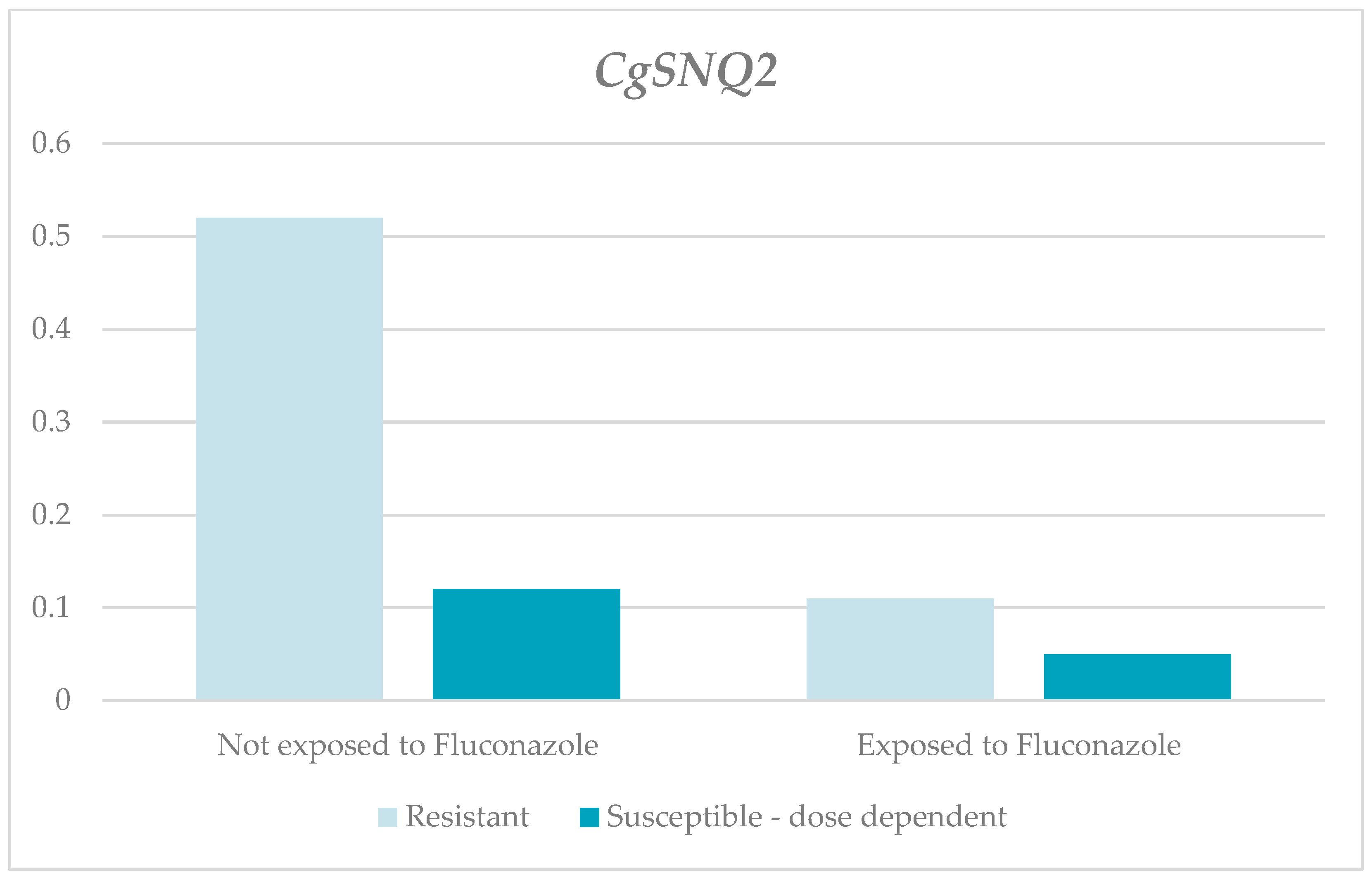
| CgSNQ2 | Without fluconazole | 1.22 | 0.52 | 0.01 | 3.59 | 1.53 | 0.014 1 | 1.1 | 0.12 | 0 | 7.4 | 2.29 | 0.166 1 |
| With fluconazole | 0.69 | 0.11 | 0 | 3.59 | 1.42 | 3.03 | 0.05 | 0 | 26.95 | 8.42 | |||
| General Susceptibility Profile | ||
|---|---|---|
| Variables in the Model | Resistant | SDD |
| Gene analyzed | 4.141 | 3.730 |
| Categorized strain | 0.369 | 0.276 |
| RQ mean without fluconazole | 0.363 | 0.302 |
| RQ mean with fluconazole | 0.111 | 0.042 |
| Constant | −8.009 | −5.820 |
| Susceptibility Profile | Forecasted Profile | |||
|---|---|---|---|---|
| Variables in the Model | Resistant | SDD | Resistant | SDD |
| Gene analyzed | 4.141 | 3.730 | 2 × 4.141 | 2 × 3.730 |
| Categorized strain | 0.369 | 0.276 | 2 × 0.369 | 2 × 0.276 |
| RQ mean without fluconazole | 0.363 | 0.302 | 0.57 × 0.363 | 0.57 × 0.302 |
| RQ mean with fluconazole | 0.111 | 0.042 | 0.22 × 0.111 | 0.22 × 0.042 |
| Constant | −8.009 | −5.820 | 1 × (−8.009) | 1 × (−5.820) |
| Total | 1.2425 | 2.3738 | ||
| Fluconazole | |||
|---|---|---|---|
| Protein | Model | ΔG (Kcal/mol) | Number of Contacts |
| ERG3 | 1. | −6.2 | 19 |
| 2. | −6.1 | 13 | |
| 3. | −6.0 | 6 | |
| 4. | −5.9 | 11 | |
| 5. | −5.8 | 6 | |
| ERG11 | 1. | −2.8 | 12 |
| 2. | −2.6 | 11 | |
| 3. | −2.5 | 11 | |
| 4. | −2.4 | 10 | |
| 5. | −2.2 | 9 | |
| CgCDR1 | 1. | −7.0 | 14 |
| 2. | −6.4 | 14 | |
| 3. | −6.4 | 15 | |
| 4. | −6.2 | 14 | |
| 5. | −6.2 | 11 | |
| CgSNQ2 | 1. | −7.6 | 17 |
| 2. | −7.5 | 12 | |
| 3. | −7.4 | 12 | |
| 4. | −7.2 | 11 | |
| 5. | −7.1 | 11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cárdenas Parra, L.Y.; Rojas Rodríguez, A.E.; Pérez Cárdenas, J.E.; Pérez-Agudelo, J.M. Molecular Evaluation of the mRNA Expression of the ERG11, ERG3, CgCDR1, and CgSNQ2 Genes Linked to Fluconazole Resistance in Candida glabrata in a Colombian Population. J. Fungi 2024, 10, 509. https://doi.org/10.3390/jof10070509
Cárdenas Parra LY, Rojas Rodríguez AE, Pérez Cárdenas JE, Pérez-Agudelo JM. Molecular Evaluation of the mRNA Expression of the ERG11, ERG3, CgCDR1, and CgSNQ2 Genes Linked to Fluconazole Resistance in Candida glabrata in a Colombian Population. Journal of Fungi. 2024; 10(7):509. https://doi.org/10.3390/jof10070509
Chicago/Turabian StyleCárdenas Parra, Leidy Yurany, Ana Elisa Rojas Rodríguez, Jorge Enrique Pérez Cárdenas, and Juan Manuel Pérez-Agudelo. 2024. "Molecular Evaluation of the mRNA Expression of the ERG11, ERG3, CgCDR1, and CgSNQ2 Genes Linked to Fluconazole Resistance in Candida glabrata in a Colombian Population" Journal of Fungi 10, no. 7: 509. https://doi.org/10.3390/jof10070509





