New Diagnostic Strategy for Onychomycosis: First-Line Utilization of DermaGenius® PCR and Calcofluor Microscopy Combined with Selective Culturing
Abstract
:1. Introduction
2. Materials and Methods
- Direct examination with calcofluor (DEC)
- Culture
- PCR DermaGenius® 2.0 (PathoNostics B.V., Maastricht, The Netherlands)
- Mixed algorithm (MA)
- Positive DEC with hyphae and detection of a dermatophyte on PCR;
- Positive DEC with pseudohyphae or yeast and detection of C. albicans on PCR;
- Both DEC and PCR are negative;
- Negative DEC and detection of a dermatophyte and/or C. albicans on PCR (uncommon situation; if applicable, as with C. albicans, response is provided with a comment on the fungus’s colonization potential).
- Positive DEC (whatever the type of fungal elements observed) and negative PCR;
- Positive DEC with yeasts/pseudohyphae and detection of only a dermatophyte on PCR (uncommon situation);
- Positive DEC with hyphae and detection of only C. albicans on PCR (uncommon situation).
- 1.
- Interpretation of results
- 2.
- Statistical analysis
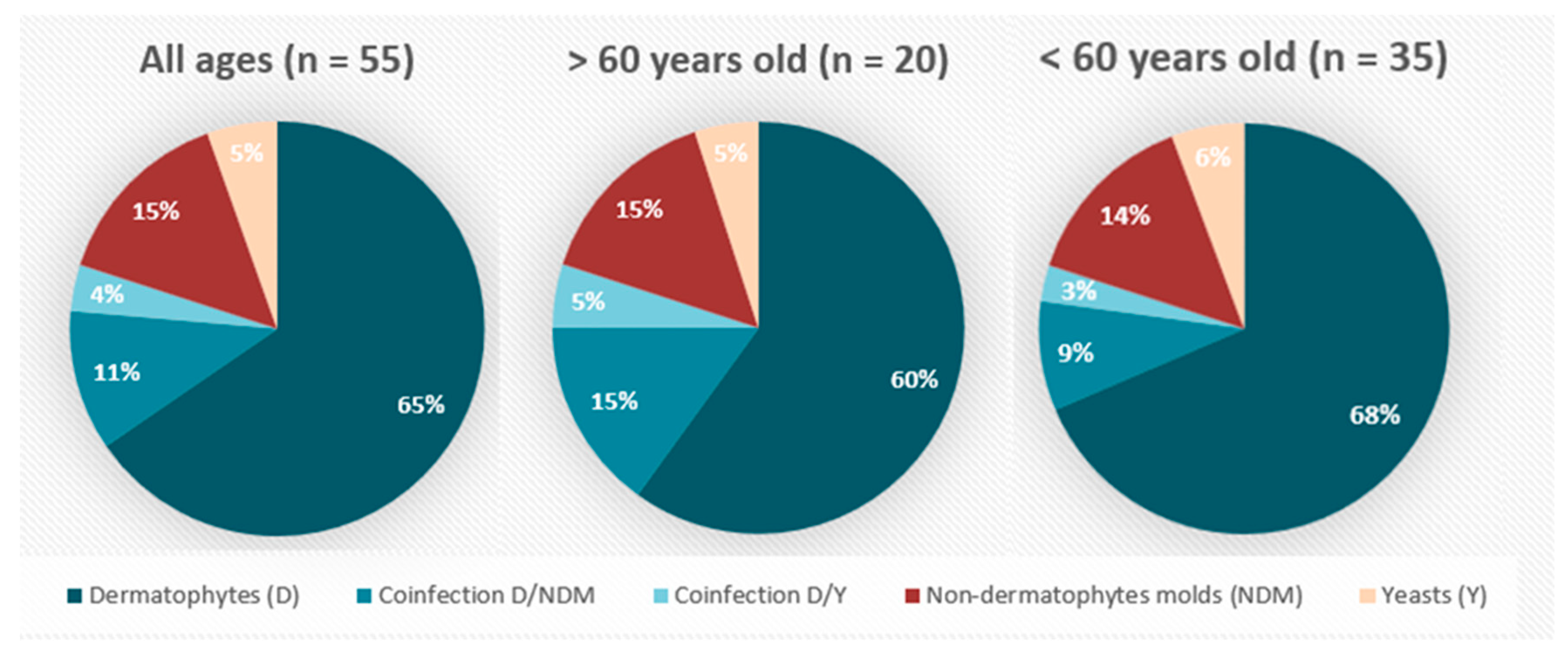
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Species Detected (Occurrence in Brackets) | Literature: Clinical Significance in OM | Identification Method | Direct Examination + (Compatible Image) | Direct Examination (or Uncompatible Image) | ||
|---|---|---|---|---|---|---|
| Detection by Culture | Detection by DG | Without Coinfection | With Co-Detection of a Fungus of Greater Clinical Significance | |||
| Dermatophytes | ||||||
| Trichophyton rubrum (38) | undeniable | Macro/microscopy (22) | DermaGenius (36) | Proven pathogen (35) | Proven pathogen (3) | |
| Trichophyton interdigitale (6) | undeniable | Macro/microscopy (4) | DermaGenius (5) | Proven pathogen (4) | Proven pathogen (2) | |
| Molds | ||||||
| Acremonium (3) | moderate | Macro/microscopy (2) MALDI (1): Sarocladium kiliense | - | Possible pathogen (3) | - | |
| Alternaria (1) | weak | Reference center (1) | - | Probable contaminant (1) | - | |
| Arthrinium sp. (1) | none | Reference center (1) | - | - | Contaminant (1) | |
| Arthrographis kalrae (1) | weak | Reference center (1) | - | - | Contaminant (1) | |
| Aspergillus flavus (1) | weak (considering contaminant potential) | Macro/microscopy (1) | Possible pathogen (1) | - | - | |
| Aspergillus fumigatus (3) | faible (considering contaminant potential) | Macro/microscopy (3) | Possible pathogen (1) | Probable contaminant (1) | Contaminant (1) | |
| Aspergillus versicolor (1) | moderate | Macro/microscopy (1) | Probable pathogen (1) | - | - | |
| Bipolaris sp. (1) | none | Macro/microscopy (1) | - | - | Contaminant (1) | |
| Chaetonium sp. (2) | weak | Macro/microscopy (1) Reference center: sequencing (1) | - | Probable contaminant (1) | Probable contaminant (1) | |
| Fusarium sp. (2) | strong | Macro/microscopy (2) | Proven pathogen (1) | - | Possible pathogen (1) | |
| Neopestalotiopsis (1) | none | Reference center: sequencing (1) | - | - | Contaminant (1) | |
| Pénicillium sp. (12) | none | Macro/microscopy (12) | - | Contaminant (7) | Contaminant (5) | |
| Phialophora sp. (2) | none | Macro/microscopy (2) (+1 RNC confirmation) | - | Contaminant (1) | Contaminant (1) | |
| Scedosporium sp. (2) | none | Macro/microscopy (2) | - | Contaminant (1) | Contaminant (1) | |
| Scytalidium dimidiatum (1) | Strong | Macro/microscopy (1) | Proven pathogen (1) | - | - | |
| Scopulariopsis (5) | Strong | Macro/microscopy (5) | Proven pathogen (2) | Probable pathogen (3) | - | |
| Trichoderma sp. (1) | none | RNC (1) | - | Contaminant (1) | - | |
| Ulocladium (1) | weak | Macro/microscopy (1) | - | Probable contaminant (1) | - | |
| Yeasts | ||||||
| Candida albicans (2) | Strong | MALDI-TOF (1) | DermaGenius (2) | Proven pathogen (2) | - | - |
| Candida parapsilosis cplx (9): C. parapsilosis (6) C. metapsilosis (2) C. orthopsilosis (1) | moderate | MALDI-TOF: C. parapsilosis (6), C. metapsilosis (2), C. orthopsilosis (1) | Probable pathogen (3: C. parapsilosis) | - | Probable contaminant (6: 3 C. parapsilosis + 2 C. metapsilosis + 1 C. orthopsilosis) | |
| Candida pararugosa (1) | moderate (by extrapolation to other Candida) | MALDI-TOF (1) | - | - | Probable contaminant (1) | |
| Debaryomyces kansenii (1) | none | MALDI-TOF (1) | - | - | Contaminant (1) | |
| Yeast not identifiable by MALDI-TOF (2) | moderate (by default) | Macro/microscopy (2) | - | - | Probable contaminant (2) | |
| Rhodotorula (4) | none | MALDI-Tof (4) | - | - | Contaminant (4) | |
| Trichosporon sp. (7) | weak | MALDI-Tof (7) | - | Probable contaminant (2) | Contaminant (5) | |
References
- Moreno, G.; Arenas, R. Other fungi causing onychomycosis. Clin. Dermatol. 2010, 28, 160–163. [Google Scholar] [CrossRef] [PubMed]
- Piraccini, B.M.; Alessandrini, A. Onychomycosis: A review. J. Fungi 2015, 1, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Nouripour-Sisakht, S.; Mirhendi, H.; Shidfar, M.; Ahmadi, B.; Rezaei-Matehkolaei, A.; Geramishoar, M.; Zarei, F.; Jalalizand, N. Aspergillus species as emerging causative agents of onychomycosis. J. Med. Mycol. 2015, 25, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Kuvandik, G.; Çetin, M.; Genctoy, G.; Horoz, M.; Duru, M.; Akcali, C.; Satar, S.; Kiykim, A.; Kaya, H. The prevalance, epidemiology and risk factors for onychomycosis in hemodialysis patients. BMC Infect. Dis. 2007, 7, 102. [Google Scholar] [CrossRef] [PubMed]
- Eenglish, M.P. Nails and fungi. Br. J. Dermatol. 1976, 94, 697–701. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Nakrieko, K.-A. Molecular determination of mixed infections of dermatophytes and nondermatophyte moulds in individuals with onychomycosis. J. Am. Podiatr. Med. Assoc. 2014, 104, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Jayatilake, J.A.M.S.; Tilakaratne, W.M.; Panagoda, G.J. Candidal onychomycosis: A mini-review. Mycopathologia 2009, 168, 165–173. [Google Scholar] [CrossRef] [PubMed]
- De Berker, D. Fungal nail disease. N. Engl. J. Med. 2009, 360, 2108–2116. [Google Scholar] [CrossRef] [PubMed]
- Trovato, L.; Domina, M.; Calvo, M.; De Pasquale, R.; Scalia, G.; Oliveri, S. Use of real time multiplex PCR for the diagnosis of dermatophytes onychomycosis in patients with empirical antifungal treatments. J. Infect. Public Health 2022, 15, 539–544. [Google Scholar] [CrossRef]
- Hayette, M.-P.; Seidel, L.; Adjetey, C.; Darfouf, R.; Wéry, M.; Boreux, R.; Sacheli, R.; Melin, P.; Arrese, J. Clinical evaluation of the DermaGenius® Nail real-time PCR assay for the detection of dermatophytes and Candida albicans in nails. Med. Mycol. 2019, 57, 277–283. [Google Scholar] [CrossRef]
- Gupta, A.K.; Versteeg, S.G.; Shear, N.H. Confirmatory testing prior to initiating onychomycosis therapy is cost-effective. J. Cutan. Med. Surg. 2018, 22, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Koshnick, R.L.; Lilly, K.K.; Clair, K.S.; Finnegan, M.T.; Warshaw, E.M. Use of diagnostic tests by dermatologists, podiatrists and family practitioners in the United States: Pilot data from a cross-sectional survey. Mycoses 2007, 50, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Effendy, I.; Lecha, M.; de Chauvin, M.F.; Di Chiacchio, N.; Baran, R. Epidemiology and clinical classification of onychomycosis. J. Eur. Acad. Dermatol. Venereol. 2005, 19, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Barzilai, A.; Greenbaum, H.; Huszar, M.; Ikher, S.; Shemer, A.; Baum, S. The role of histological examination of nail clipping in the diagnosis of onychomycosis. Isr. Med. Assoc. J. IMAJ 2023, 25, 117–121. [Google Scholar] [PubMed]
- Brillowska-Dąbrowska, A.; Saunte, D.M.; Arendrup, M.C. Five-hour diagnosis of dermatophyte nail infections with specific detection of Trichophyton rubrum. J. Clin. Microbiol. 2007, 45, 1200–1204. [Google Scholar] [CrossRef] [PubMed]
- Koo, S.H.; Teoh, Y.L.; Koh, W.L.; Ochi, H.; Tan, S.K.; Sim, D.M.F.; Jiang, B.; Tan, A.L.; Tan, T.Y.; Lim, S.P.R. Development and validation of a real-time multiplex PCR assay for the detection of dermatophytes and Fusarium spp. J. Med. Microbiol. 2019, 68, 1641–1648. [Google Scholar] [CrossRef] [PubMed]
- Petinataud, D.; Berger, S.; Ferdynus, C.; Debourgogne, A.; Contet-Audonneau, N.; Machouart, M. Optimising the diagnostic strategy for onychomycosis from sample collection to fungal identification evaluation of a diagnostic kit for real-time PCR. Mycoses 2016, 59, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Mehlig, L.; Garve, C.; Ritschel, A.; Zeiler, A.; Brabetz, W.; Weber, C.; Bauer, A. Clinical evaluation of a novel commercial multiplex-based PCR diagnostic test for differential diagnosis of dermatomycoses. Mycoses 2014, 57, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Lecerf, P.; Abdy, S.; Vollono, L.; Pastushenko, I.; Richert, B.; André, J. Direct examination, histopathology and fungal culture for the diagnosis of onychomycosis: A retrospective, comparative study on 2245 specimens. Mycoses 2021, 64, 187–193. [Google Scholar] [CrossRef]
- Monod, M.; Méhul, B. Recent findings in onychomycosis and their application for appropriate treatment. J. Fungi 2019, 5, 20. [Google Scholar] [CrossRef]
- Gupta, A.K.; Stec, N.; Summerbell, R.C.; Shear, N.H.; Piguet, V.; Tosti, A.; Piraccini, B.M. Onychomycosis: A review. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 1972–1990. [Google Scholar] [CrossRef]
- Ghannoum, M.A.; Hajjeh, R.A.; Scher, R.; Konnikov, N.; Gupta, A.K.; Summerbell, R.; Sullivan, S.; Daniel, R.; Krusinski, P.; Fleckman, P.; et al. A large-scale North American study of fungal isolates from nails: The frequency of onychomycosis, fungal distribution, and antifungal susceptibility patterns. J. Am. Acad. Dermatol. 2000, 43, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Sacheli, R.; Cuypers, L.; Seidel, L.; Darfouf, R.; Adjetey, C.; Lagrou, K.; Hayette, M.-P. Epidemiology of dermatophytes in Belgium: A 5 Years’ Survey. Mycopathologia 2021, 186, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Dolenc-Voljč, M. Dermatophyte infections in the Ljubljana region, Slovenia, 1995–2002. Mycoses 2005, 48, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Asticcioli, S.; Di Silverio, A.; Sacco, L.; Fusi, I.; Vincenti, L.; Romero, E. Dermatophyte infections in patients attending a tertiary care hospital in northern Italy. New Microbiol. 2008, 31, 543–548. [Google Scholar]
- Drakensjö, I.T.; Chryssanthou, E. Epidemiology of dermatophyte infections in Stockholm, Sweden: A retrospective study from 2005–2009. Med. Mycol. 2010, 49, 484–488. [Google Scholar] [CrossRef]
- Navarro-Pérez, D.; García-Oreja, S.; Tardáguila-García, A.; León-Herce, D.; Álvaro-Afonso, F.J.; Lázaro-Martínez, J.L. Microbiological culture combined with PCR for the diagnosis of onychomycosis: Descriptive analysis of 121 patients. Mycoses 2023, 66, 1045–1049. [Google Scholar] [CrossRef]
- Pospischil, I.; Reinhardt, C.; Bontems, O.; Salamin, K.; Fratti, M.; Blanchard, G.; Chang, Y.-T.; Wagner, H.; Hermann, P.; Monod, M.; et al. Identification of dermatophyte and non-dermatophyte agents in onychomycosis by PCR and DNA Sequencing—A retrospective comparison of diagnostic tools. J. Fungi 2022, 8, 1019. [Google Scholar] [CrossRef]
- Bin Lin, B.; Pattle, N.; Kelley, P.; Jaksic, A.S. Multiplex RT-PCR provides improved diagnosis of skin and nail dermatophyte infections compared to microscopy and culture: A laboratory study and review of the literature. Diagn. Microbiol. Infect. Dis. 2021, 101, 115413. [Google Scholar] [CrossRef]
- Paugam, A.; L’Ollivier, C.; Viguié, C.; Anaya, L.; Mary, C.; de Ponfilly, G.; Ranque, S. Comparison of real-time PCR with conventional methods to detect dermatophytes in samples from patients with suspected dermatophytosis. J. Microbiol. Methods 2013, 95, 218–222. [Google Scholar] [CrossRef]
- Cuchí-Burgos, E.; Rubio-Casino, R.; Ballestero-Téllez, M.; Pariente-Jiménez, F.; Pérez-Jové, J.; Blanco-Suárez, A. Commercial real time PCR implementation for rapid diagnosis of onychomycosis: A new workflow in a clinical laboratory. Enfermedades Infecc. Microbiol. Clín. 2021, 39, 326–329. [Google Scholar] [CrossRef]
- Bao, F.; Fan, Y.; Sun, L.; Yu, Y.; Wang, Z.; Pan, Q.; Yu, C.; Liu, H.; Zhang, F. Comparison of fungal fluorescent staining and ITS rDNA PCR-based sequencing with conventional methods for the diagnosis of onychomycosis. J. Eur. Acad. Dermatol. Venereol. 2018, 32, 1017–1021. [Google Scholar] [CrossRef] [PubMed]
- Motamedi, M.; Lari, M.S.; Pakshir, K.; Zomorodian, K. Comparing real-time PCR and calcofluor-white with conventional methods for rapid detection of dermatophytes: Across-sectional study. J. Microbiol. Methods 2019, 161, 84–86. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Cooper, E.A.; Wang, T.; Lincoln, S.A.; Bakotic, W.L. Single-point nail sampling to diagnose onychomycosis caused by non-dermatophyte molds: Utility of polymerase chain reaction (PCR) and histopathology. J. Fungi 2023, 9, 671. [Google Scholar] [CrossRef] [PubMed]
- Finch, J.; Arenas, R.; Baran, R. Fungal melanonychia. J. Am. Acad. Dermatol. 2012, 66, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Hamer, E.; Moore, C.; Denning, D. Comparison of two fluorescent whiteners, calcofluor and blankophor, for the detection of fungal elements in clinical specimens in the diagnostic laboratory. Clin. Microbiol. Infect. 2006, 12, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, J.M.; Koestenblatt, E.K.; Tutrone, W.D.; Tishler, H.R.; Najarian, L. Comparison of diagnostic methods in the evaluation of onychomycosis. J. Am. Acad. Dermatol. 2003, 49, 193–197. [Google Scholar] [CrossRef]
- Yadav, S.; Saxena, A.; Capoor, M.R.; Ramesh, V. Comparison of direct microscopic methods using potassium hydroxide, periodic acid schiff, and calcofluor white with culture in the diagnosis of onychomycosis. Indian J. Dermatol. Venereol. Leprol. 2013, 79, 242–243. [Google Scholar] [CrossRef]
| Definition of Clinical Categories | Literature: Involvement in OM (Balanced against Commensal Nature and Contaminant Potential) | DEC Positivity (Compatible Image) | Codetection with a Fungus of Greater Clinical Significance |
|---|---|---|---|
| Proven pathogen | Undeniable (dermatophytes) | +/- | yes/no |
| Strong | + | no | |
| Probable pathogen | Strong | + | yes |
| Moderate | + | no | |
| Possible pathogen | Strong | - | yes/no |
| Moderate | + | yes | |
| Weak | + | no | |
| Probable contaminant | Moderate | - | yes/no |
| Weak | + | yes | |
| None | + | no | |
| Contaminant | Weak | - | yes/no |
| None | + | yes | |
| None | - | yes/no |
| Sample Categorization (n = 106) | Strategies Assessed | Gold Standard | ||
|---|---|---|---|---|
| PCR | Culture | MA | Combinaison | |
| At least one definite pathogen * | 43 | 31 | 50 | 50 |
| At least one probable pathogen * | 0 | 4 | 2 | 2 |
| At least one possible pathogen * | 0 | 6 | 2 | 3 |
| At least one definite, probable or possible pathogen * | 43 | 41 | 54 | 55 |
| At least one probable contaminant * | 0 | 10 | 0 | 6 |
| At least one definite contaminant | 0 | 19 | 0 | 14 |
| Contaminants (probables ou certains) | 0 | 29 | 0 | 20 |
| Sterile/not detected | 63 | 36 | 52 | 31 |
| Total | 106 | 106 | 106 | 106 |
| Sensitivity (definite, probable or possible pathogens) | 78.18% | 74.55% | 98.18% | |
| Specificity (definite, probable or possible pathogens) | 100% | 38.60% | 100% | |
| Cohen’s Kappa | 0.78 | 0.74 | 0.98 | |
| Sensitivity (exclusion of possible pathogens) | 82.69% | 67.31% | 100% | |
| Specificity (exclusion of possible pathogens) | 100% | 35.19% *2 | 96.3% *2 | |
| Detection of species included in multiplex PCR DG | 43 | 27 | 46 | 46 |
| Sensitivity on species included in multiplex PCR DG | 93.48% *3 | 58.70% | 100% | |
| Detection of coinfected samples | 0 | 4 | 3 | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Evrard, S.; Minon, C.; Lamtiri Laarif, M.; De Backer, B.; Paridaens, H.; Hayette, M.-P.; Frère, J.; Senterre, J.-M.; Minon, J.-M. New Diagnostic Strategy for Onychomycosis: First-Line Utilization of DermaGenius® PCR and Calcofluor Microscopy Combined with Selective Culturing. J. Fungi 2024, 10, 515. https://doi.org/10.3390/jof10080515
Evrard S, Minon C, Lamtiri Laarif M, De Backer B, Paridaens H, Hayette M-P, Frère J, Senterre J-M, Minon J-M. New Diagnostic Strategy for Onychomycosis: First-Line Utilization of DermaGenius® PCR and Calcofluor Microscopy Combined with Selective Culturing. Journal of Fungi. 2024; 10(8):515. https://doi.org/10.3390/jof10080515
Chicago/Turabian StyleEvrard, Séverine, Caroline Minon, Mouhsine Lamtiri Laarif, Benjamin De Backer, Henry Paridaens, Marie-Pierre Hayette, Julie Frère, Jean-Marc Senterre, and Jean-Marc Minon. 2024. "New Diagnostic Strategy for Onychomycosis: First-Line Utilization of DermaGenius® PCR and Calcofluor Microscopy Combined with Selective Culturing" Journal of Fungi 10, no. 8: 515. https://doi.org/10.3390/jof10080515
APA StyleEvrard, S., Minon, C., Lamtiri Laarif, M., De Backer, B., Paridaens, H., Hayette, M.-P., Frère, J., Senterre, J.-M., & Minon, J.-M. (2024). New Diagnostic Strategy for Onychomycosis: First-Line Utilization of DermaGenius® PCR and Calcofluor Microscopy Combined with Selective Culturing. Journal of Fungi, 10(8), 515. https://doi.org/10.3390/jof10080515






