Synergistic Biocontrol and Growth Promotion in Strawberries by Co-Cultured Trichoderma harzianum TW21990 and Burkholderia vietnamiensis B418
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms and Plant Species
2.2. Medium and Cultivation Conditions
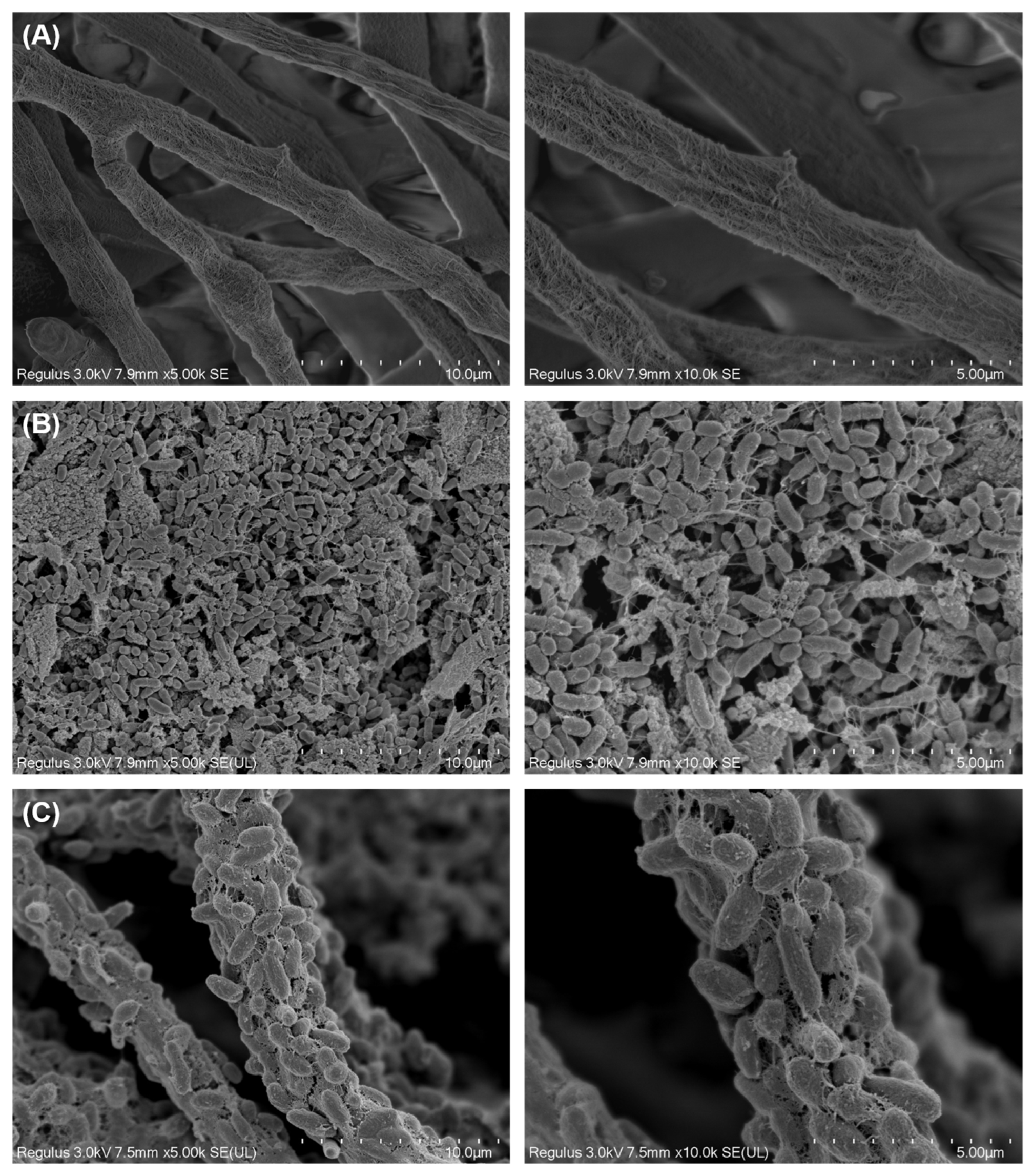
2.3. Microscopic Analysis
2.4. Antifungal Activity Assays
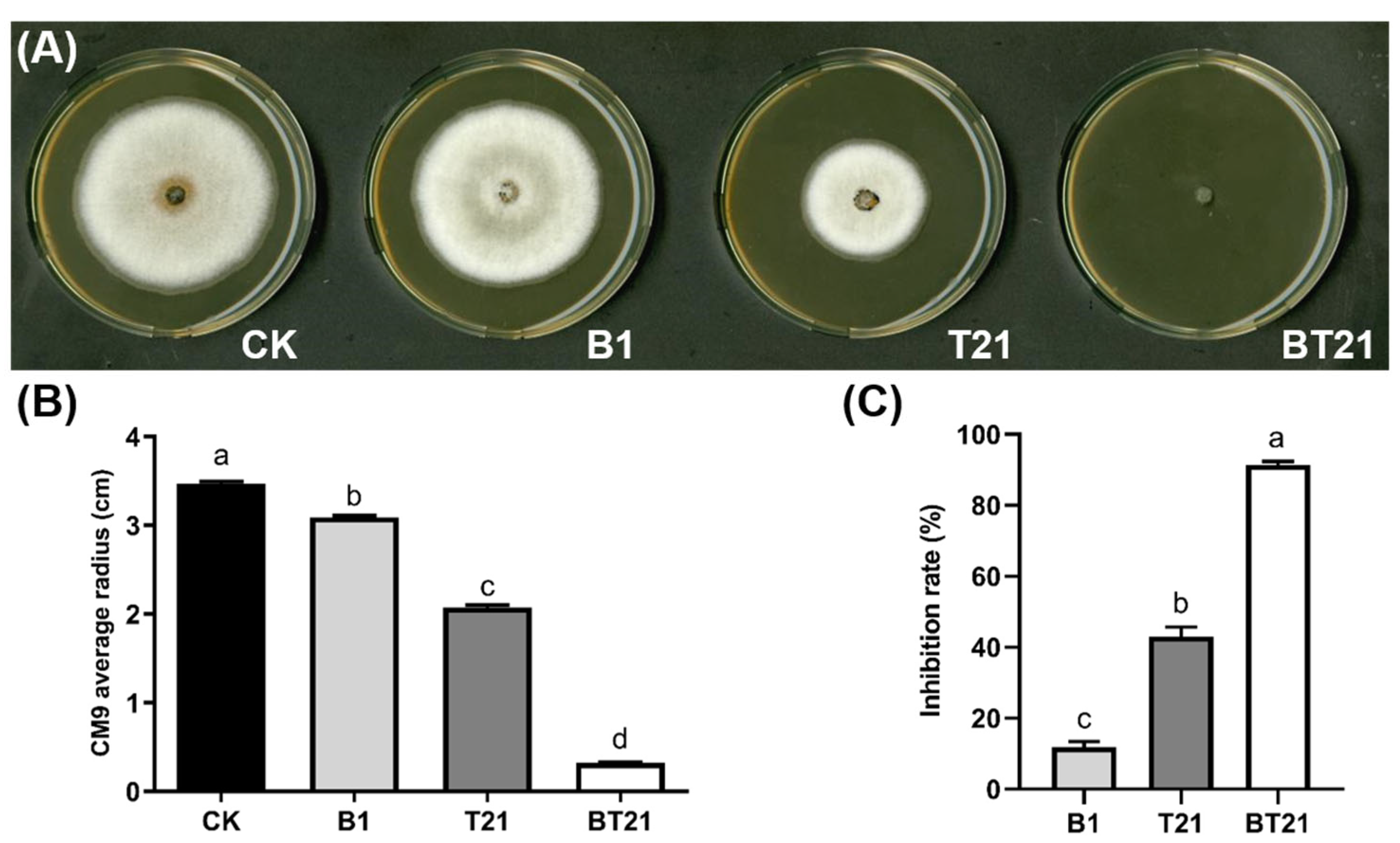
2.4.1. Inhibition Activity of Fermentation Filtrates on Mycelial Growth
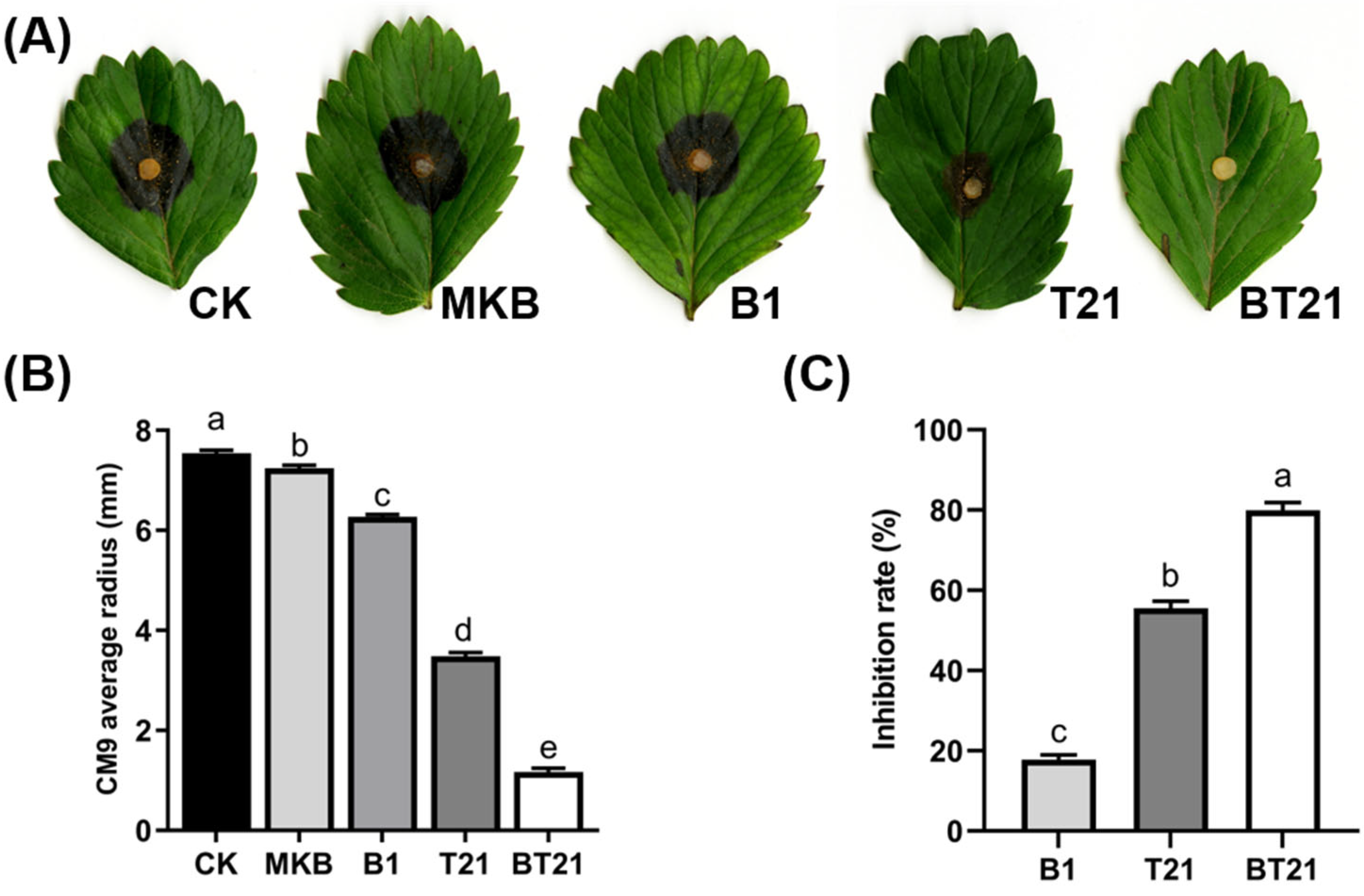
2.4.2. Antagonistic Activity of Fermentation Filtrates Using Detached Leaves
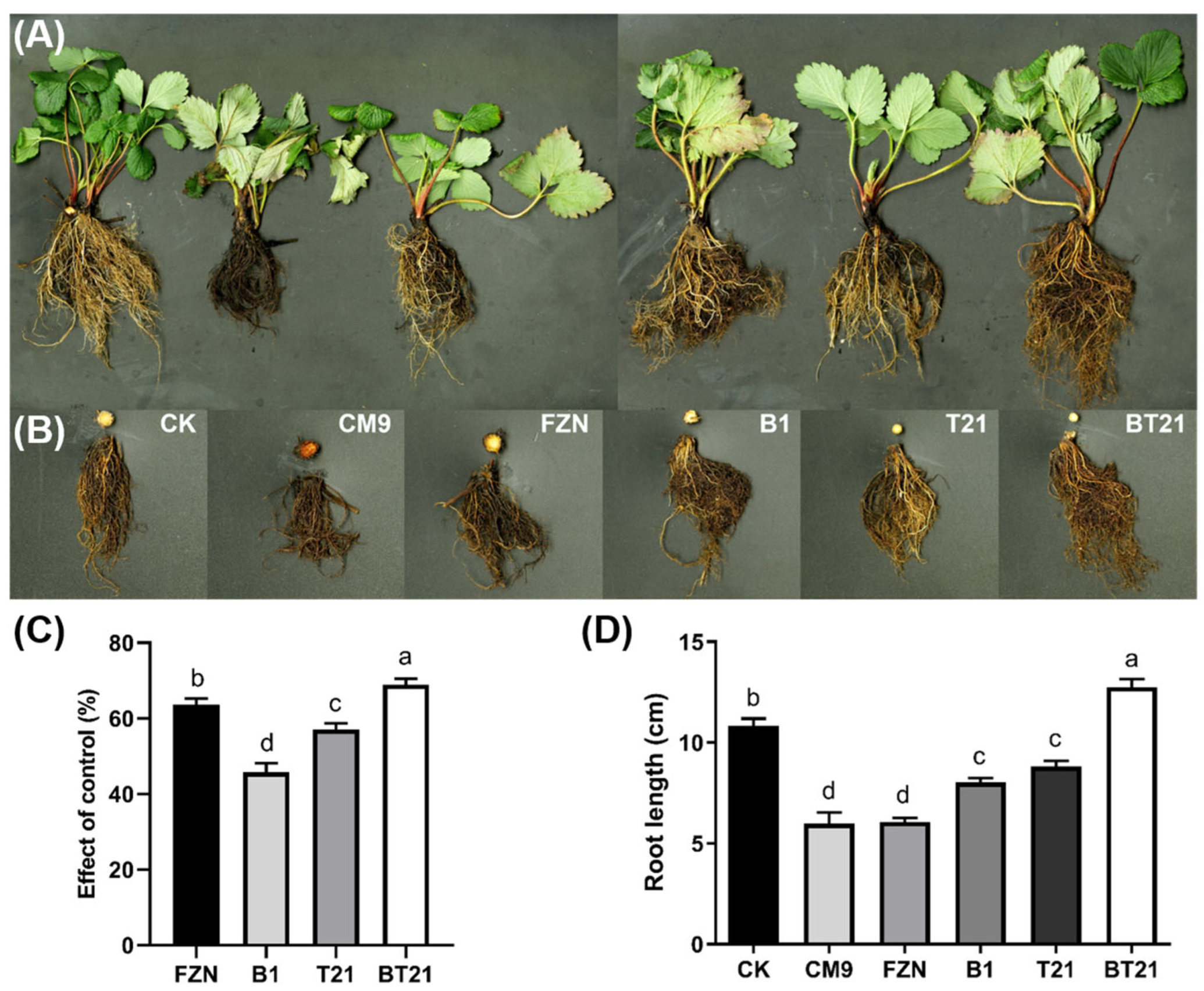
2.5. Pot Experiment in Greenhouse
2.5.1. Determination of Antioxidative Enzyme Activity
2.5.2. Determination of Total Chlorophyll Content
2.6. Metabolomic Profiles
2.6.1. Metabolites Extraction
2.6.2. UHPLC-MS/MS Analysis
2.6.3. Data Processing and Metabolite Identification
2.6.4. Data Analysis
2.7. Statistical Analysis
3. Results
3.1. Morphology of T. harzianum TW21990 with B. vietnamiensis B418 in Co-Culture
3.2. Antifungal Activity of Fermentation Filtrates of Mono- and Co-Cultures
3.2.1. Inhibition Activity of Fermentation Filtrates on Mycelial Growth
3.2.2. Antagonistic Activity of Fermentation Filtrates Using Detached Leaves
3.3. Results of Strawberry Pot Experiment in Greenhouse
3.3.1. Analysis of Antioxidative Enzyme Activity in Strawberry
3.3.2. Influence on Total Chlorophyll Content of Strawberry Leaves
3.4. Analysis of Metabolomic Profiles
3.4.1. Principal Component Analysis (PCA)
3.4.2. Partial Least Squares Discriminant Analysis (PLS-DA)
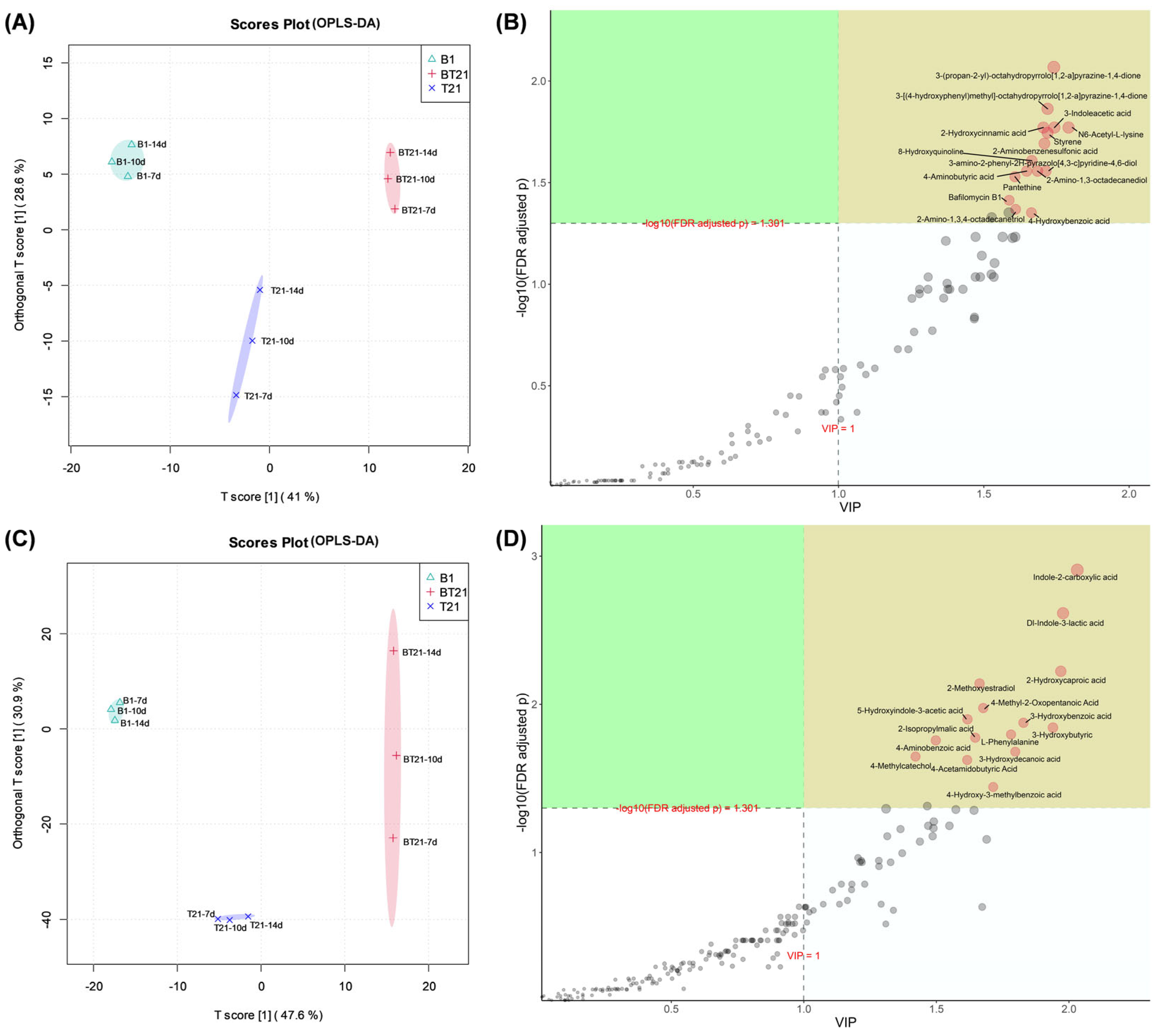
3.4.3. Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA)
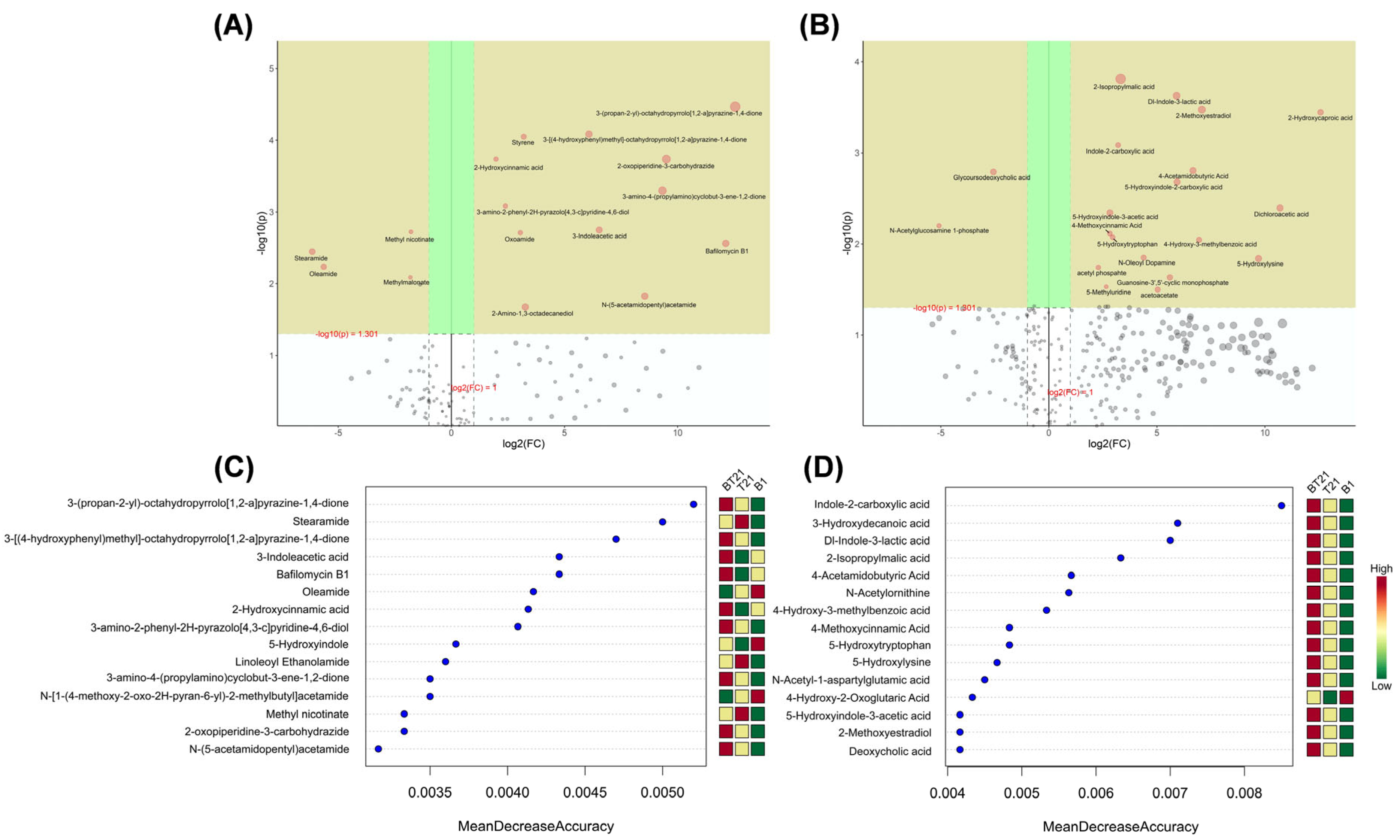
3.4.4. Univariate Analysis
3.4.5. Random Forest Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ghazanfar, M.U.; Raza, M.; Raza, W.; Qamar, M.I. Trichoderma as potential biocontrol agent, its exploitation in agriculture: A review. Plant Protect. 2018, 2, 109–135. [Google Scholar]
- Sood, M.; Kapoor, D.; Kumar, V.; Sheteiwy, M.S.; Ramakrishnan, M.; Landi, M.; Araniti, F.; Sharma, A. Trichoderma: The “secrets” of a multitalented biocontrol agent. Plants 2020, 9, 762. [Google Scholar] [CrossRef]
- Verma, M.; Brar, S.K.; Tyagi, R.D.; Surampalli, R.N.; Valero, J.R. Antagonistic fungi, Trichoderma spp.: Panoply of biological control. Biochem. Eng. J. 2007, 37, 1–20. [Google Scholar] [CrossRef]
- Gajera, H.; Domadiya, R.; Patel, S.; Kapopara, M.; Golakiya, B. Molecular mechanism of Trichoderma as bio-control agents against phytopathogen system–a review. Curr. Res. Microbiol. Biotechnol. 2013, 1, 133–142. [Google Scholar]
- Khan, R.A.A.; Najeeb, S.; Hussain, S.; Xie, B.; Li, Y. Bioactive secondary metabolites from Trichoderma spp. against phytopathogenic fungi. Microorganisms 2020, 8, 817. [Google Scholar] [CrossRef]
- Depoorter, E.; Bull, M.J.; Peeters, C.; Coenye, T.; Vandamme, P.; Mahenthiralingam, E. Burkholderia: An update on taxonomy and biotechnological potential as antibiotic producers. Appl. Microbiol. Biotechnol. 2016, 100, 5215–5229. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Nowak, J.; Coenye, T.; Clément, C.; Ait Barka, E. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 2008, 32, 607–626. [Google Scholar] [CrossRef]
- Elshafie, H.S.; Camele, I. An overview of metabolic activity, beneficial and pathogenic aspects of Burkholderia spp. Metabolites 2021, 11, 321. [Google Scholar] [CrossRef]
- Bach, E.; Passaglia, L.M.P.; Jiao, J.; Gross, H. Burkholderia in the genomic era: From taxonomy to the discovery of new antimicrobial secondary metabolites. Crit. Rev. Microbiol. 2022, 48, 121–160. [Google Scholar] [CrossRef]
- Esmaeel, Q.; Pupin, M.; Jacques, P.; Leclère, V. Nonribosomal peptides and polyketides of Burkholderia: New compounds potentially implicated in biocontrol and pharmaceuticals. Environ. Sci. Pollut. Res. 2018, 25, 29794–29807. [Google Scholar] [CrossRef]
- Shinjo, R.; Tanaka, A.; Sugiura, D.; Suzuki, T.; Uesaka, K.; Takebayashi, Y.; Kojima, M.; Sakakibara, H.; Takemoto, D.; Kondo, M. Comprehensive analysis of the mechanisms underlying enhanced growth and root N acquisition in rice by the endophytic diazotroph, Burkholderia vietnamiensis RS1. Plant Soil 2020, 450, 537–555. [Google Scholar] [CrossRef]
- Sandanakirouchenane, A.; Haque, E.; Geetha, T. Recent studies on N2 fixing Burkholderia isolates as a biofertilizer for the sustainable agriculture. Int. J. Curr. Microbiol. App. Sci. 2017, 6, 2780–2796. [Google Scholar] [CrossRef]
- Ruiz, B.; Chávez, A.; Forero, A.; García-Huante, Y.; Romero, A.; Sánchez, M.; Rocha, D.; Sánchez, B.; Rodríguez-Sanoja, R.; Sánchez, S.; et al. Production of microbial secondary metabolites: Regulation by the carbon source. Crit. Rev. Microbiol. 2010, 36, 146–167. [Google Scholar] [CrossRef] [PubMed]
- Wink, M. Introduction: Biochemistry, physiology and ecological functions of secondary metabolites. In Biochemistry of Plant Secondary Metabolism, 2nd ed.; Wink, M., Ed.; Wiley-Blackwell: Oxford, UK, 2010; Volume 1, pp. 1–19. [Google Scholar]
- Abegaz, B.M.; Kinfe, H.H. Secondary metabolites, their structural diversity, bioactivity, and ecological functions: An overview. Phys. Sci. Rev. 2019, 4, 20180100. [Google Scholar] [CrossRef]
- Dror, B.; Jurkevitch, E.; Cytryn, E. State-of-the-art methodologies to identify antimicrobial secondary metabolites in soil bacterial communities-A review. Soil Biol. Biochem. 2020, 147, 107838. [Google Scholar] [CrossRef]
- Cacho, R.A.; Tang, Y.; Chooi, Y.H. Next-generation sequencing approach for connecting secondary metabolites to biosynthetic gene clusters in fungi. Front. Microbiol. 2015, 5, 123211. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kjærbølling, I.; Mortensen, U.H.; Vesth, T.; Andersen, M.R. Strategies to establish the link between biosynthetic gene clusters and secondary metabolites. Fungal Genet. Biol. 2019, 130, 107–121. [Google Scholar] [CrossRef]
- Xu, S.; Li, M.; Hu, Z.; Shao, Y.; Ying, J.; Zhang, H. The potential use of fungal co-culture strategy for discovery of new secondary metabolites. Microorganisms 2023, 11, 464. [Google Scholar] [CrossRef] [PubMed]
- Goers, L.; Freemont, P.; Polizzi, K.M. Co-culture systems and technologies: Taking synthetic biology to the next level. J. R. Soc. Interface 2014, 11, 20140065. [Google Scholar] [CrossRef]
- Peng, X.Y.; Wu, J.T.; Shao, C.L.; Li, Z.Y.; Chen, M.; Wang, C.Y. Co-culture: Stimulate the metabolic potential and explore the molecular diversity of natural products from microorganisms. Mar. Life Sci. Technol. 2021, 3, 363–374. [Google Scholar] [CrossRef]
- Poveda, J.; Eugui, D. Combined use of Trichoderma and beneficial bacteria (mainly Bacillus and Pseudomonas): Development of microbial synergistic bio-inoculants in sustainable agriculture. Biol. Control 2022, 176, 105100. [Google Scholar] [CrossRef]
- Velmourougane, K.; Prasanna, R.; Singh, S.; Chawla, G.; Kumar, A.; Saxena, A.K. Modulating rhizosphere colonisation, plant growth, soil nutrient availability and plant defense enzyme activity through Trichoderma viride-Azotobacter chroococcum biofilm inoculation in chickpea. Plant Soil 2017, 421, 157–174. [Google Scholar] [CrossRef]
- Li, T.; Tang, J.; Karuppiah, V.; Li, Y.; Xu, N.; Chen, J. Co-culture of Trichoderma atroviride SG3403 and Bacillus subtilis 22 improves the production of antifungal secondary metabolites. Biol. Control 2020, 140, 104122. [Google Scholar] [CrossRef]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.L. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.Q.; Leow, H.Y.; Lee, D.C.W.; Karisnan, K.; Song, A.A.L.; Mai, C.W.; Yap, W.S.; Lim, S.H.E.; Lai, K.S. Co-culture systems for the production of secondary metabolites: Current and future prospects. Open Biotechnol. J. 2019, 13, 18–26. [Google Scholar] [CrossRef]
- Chen, K.; Sui, L.N.; Zhao, Z.J.; Li, L.; Hu, J.D.; Li, J.S. Control effect of co-cultured fermentation of two Trichoderma strains on cucumber Fusarium wilt (In Chinese). Chin. J. Biol. Control 2022, 1, 108–114. [Google Scholar]
- Liu, M.; Philp, J.; Wang, Y.; Hu, J.; Wei, Y.; Li, J.; Ryder, M.; Toh, R.; Zhou, Y.; Denton, M.D.; et al. Plant growth-promoting rhizobacteria Burkholderia vietnamiensis B418 inhibits root-knot nematode on watermelon by modifying the rhizosphere microbial community. Sci. Rep. 2022, 12, 8381. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L. Cross-validation of non-linear growth functions for modelling tree height–diameter relationships. Ann. Bot. 1997, 79, 251–257. [Google Scholar] [CrossRef]
- Bock, C.H.; Poole, G.H.; Parker, P.E.; Gottwald, T.R. Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Crit. Rev. Plant Sci. 2010, 29, 59–107. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, S.; Jiang, W.; Liu, D. Cadmium accumulation, activities of antioxidant enzymes, and malondialdehyde (MDA) content in Pistia stratiotes L. Environ. Sci. Pollut. Res. 2013, 20, 1117–1123. [Google Scholar] [CrossRef]
- Gewers, F.L.; Ferreira, G.R.; Arruda, H.F.D.; Silva, F.N.; Comin, C.H.; Amancio, D.R.; Costa, L.D.F. Principal component analysis: A natural approach to data exploration. ACM Comput. Surv. 2021, 54, 1–34. [Google Scholar] [CrossRef]
- Lee, L.C.; Liong, C.Y.; Jemain, A.A. Partial least squares-discriminant analysis (PLS-DA) for classification of high-dimensional (HD) data: A review of contemporary practice strategies and knowledge gaps. Analyst 2018, 143, 3526–3539. [Google Scholar] [CrossRef]
- Boccard, J.; Rutledge, D.N. A consensus orthogonal partial least squares discriminant analysis (OPLS-DA) strategy for multiblock Omics data fusion. Anal. Chim. Acta. 2013, 769, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Hoque, M.A.; Sugimoto, M. Robust volcano plot: Identification of differential metabolites in the presence of outliers. BMC Bioinf. 2018, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bénard, C.; Da Veiga, S.; Scornet, E. Mean decrease accuracy for random forests: Inconsistency, and a practical solution via the Sobol-MDA. Biometrika 2022, 109, 881–900. [Google Scholar] [CrossRef]
- Curry, K.J.; Abril, M.; Avant, J.B.; Smith, B.J. Strawberry anthracnose: Histopathology of Colletotrichum acutatum and C. fragariae. Phytopathology 2002, 92, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Cong, Y.; Feng, L.; Liu, C.; Yang, W.; Xin, Y.; Chen, K. Effects of mixed culture fermentation of Bacillus amyloliquefaciens and Trichoderma longibrachiatum on its constituent strains and the biocontrol of tomato Fusarium wilt. J. Appl. Microbiol. 2022, 132, 532–546. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, H.; Li, L.; Zhao, Z.; Hu, J.; Wei, Y.; Yang, H.; Li, J. Metabolomics reveal metabolic variation caused by co-culture of Arthrobacter ureafaciens and Trichoderma harzianum and their impacts on wheat germination. Int. Microbiol. 2023, 26, 723–739. [Google Scholar] [CrossRef]
- Karuppiah, V.; Vallikkannu, M.; Li, T.; Chen, J. Simultaneous and sequential based co-fermentations of Trichoderma asperellum GDFS1009 and Bacillus amyloliquefaciens 1841: A strategy to enhance the gene expression and metabolites to improve the bio-control and plant growth promoting activity. Microb. Cell Fact. 2019, 18, 1–16. [Google Scholar] [CrossRef]
- Covington, B.C.; McLean, J.A.; Bachmann, B.O. Comparative mass spectrometry-based metabolomics strategies for the investigation of microbial secondary metabolites. Nat. Prod. Rep. 2017, 34, 6–24. [Google Scholar] [CrossRef]
- Martins, M.B.; Carvalho, I. Diketopiperazines: Biological activity and synthesis. Tetrahedron 2007, 63, 9923–9932. [Google Scholar]
- Wang, J.H.; Quan, C.S.; Qi, X.H.; Li, X.; Fan, S.D. Determination of diketopiperazines of Burkholderia cepacia CF-66 by gas chromatography–mass spectrometry. Anal. Bioanal. Chem. 2010, 396, 1773–1779. [Google Scholar]
- Zin, N.M.; Al-Shaibani, M.M.; Jalil, J.; Sukri, A.; Al-Maleki, A.R.; Sidik, N.M. Profiling of gene expression in methicillin-resistant Staphylococcus aureus in response to cyclo-(l-Val-l-Pro) and chloramphenicol isolated from Streptomyces sp.; SUK 25 reveals gene downregulation in multiple biological targets. Arch. Microbiol. 2020, 202, 2083–2092. [Google Scholar]
- Wattana-Amorn, P.; Charoenwongsa, W.; Williams, C.; Crump, M.P.; Apichaisataienchote, B. Antibacterial activity of cyclo (L-Pro-L-Tyr) and cyclo (D-Pro-L-Tyr) from Streptomyces sp. strain 22-4 against phytopathogenic bacteria. Nat. Prod. Res. 2016, 30, 1980–1983. [Google Scholar] [PubMed]
- Sun, P.; Huang, Y.; Yang, X.; Liao, A.; Wu, J. The role of indole derivative in the growth of plants: A review. Front. Plant Sci. 2023, 13, 1120613. [Google Scholar]
- Zúñiga, A.; Poupin, M.J.; Donoso, R.; Ledger, T.; Guiliani, N.; Gutiérrez, R.A.; González, B. Quorum sensing and indole-3-acetic acid degradation play a role in colonization and plant growth promotion of Arabidopsis thaliana by Burkholderia phytofirmans PsJN. Mol. Plant Microbe Interact. 2013, 26, 546–553. [Google Scholar] [PubMed]
- Liu, W.H.; Chen, F.F.; Wang, C.E.; Fu, H.H.; Fang, X.Q.; Ye, J.R.; Shi, J.Y. Indole-3-acetic acid in Burkholderia pyrrocinia JK-SH007: Enzymatic identification of the indole-3-acetamide synthesis pathway. Front. Microbiol. 2019, 10, 2559. [Google Scholar]
- Wei, C.; Zhang, J.; Shi, J.; Gan, X.; Hu, D.; Song, B. Synthesis, antiviral activity, and induction of plant resistance of indole analogues bearing dithioacetal moiety. J. Agr. Food Chem. 2019, 67, 13882–13891. [Google Scholar]
- Wu, Q.; Ni, M.; Dou, K.; Tang, J.; Ren, J.; Yu, C.; Chen, J. Co-culture of Bacillus amyloliquefaciens ACCC11060 and Trichoderma asperellum GDFS1009 enhanced pathogen-inhibition and amino acid yield. Microb. Cell Fact. 2018, 17, 1–12. [Google Scholar]
- Zhou, Q.; Xie, Z.; Wu, D.; Liu, L.; Shi, Y.; Li, P.; Gu, Q. The effect of indole-3-lactic acid from Lactiplantibacillus plantarum ZJ316 on human intestinal microbiota in vitro. Foods 2022, 11, 3302. [Google Scholar] [CrossRef]
- Xiao, J.; Zhao, L.; Bai, Y.; Lin, R.; Ngea, G.L.N.; Dhanasekaran, S.; Li, B.; Gu, X.; Zhang, X.; Zhang, H. The biocontrol efficacy of Sporidiobolus pararoseus Y16 cultured with Gamma-aminobutyric acid and its effects on the resistant substances of postharvest grapes. Biol. Control 2022, 169, 104900. [Google Scholar] [CrossRef]
- Ricciutelli, M.; Bartolucci, G.; Campana, R.; Salucci, S.; Benedetti, S.; Caprioli, G.; Maggi, F.; Sagratini, G.; Vittori, S.; Lucarini, S. Quantification of 2-and 3-isopropylmalic acids in forty Italian wines by UHPLC-MS/MS triple quadrupole and evaluation of their antimicrobial, antioxidant activities and biocompatibility. Food Chem. 2020, 321, 126726. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, J.; Gao, X.; Shen, Y.; Qin, L.; Zhu, B. Metabolites from a co-culture of Trichoderma yunnanense and Paenibacillus peoriae improve resistance to corm rot disease in Crocus sativus. Ind. Crops Prod. 2024, 213, 118465. [Google Scholar] [CrossRef]
- Joaquim, A.R.; Gionbelli, M.P.; Gosmann, G.; Fuentefria, A.M.; Lopes, M.S.; Fernandes de Andrade, S. Novel antimicrobial 8-hydroxyquinoline-based agents: Current development, structure–activity relationships, and perspectives. J. Med. Chem. 2021, 64, 16349–16379. [Google Scholar] [CrossRef]
- Laborda, P.; Li, C.; Zhao, Y.; Tang, B.; Ling, J.; He, F.; Liu, F. Antifungal metabolite p-aminobenzoic acid (pABA): Mechanism of action and efficacy for the biocontrol of pear bitter rot disease. J. Agr. Food Chem. 2019, 67, 2157–2165. [Google Scholar] [CrossRef]
- Keman, D.; Soyer, F. Antibiotic-resistant Staphylococcus aureus does not develop resistance to vanillic acid and 2-hydroxycinnamic acid after continuous exposure in vitro. ACS Omega 2019, 4, 15393–15400. [Google Scholar] [CrossRef]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Labuschagne, N.; Dubery, I.A. Unravelling the metabolic reconfiguration of the post-challenge primed state in Sorghum bicolor responding to Colletotrichum sublineolum infection. Metabolites 2019, 9, 194. [Google Scholar] [CrossRef]
- Deng, Q.; Lei, X.; Zhang, H.; Deng, L.; Yi, L.; Zeng, K. Phenylalanine promotes biofilm formation of Meyerozyma caribbica to improve biocontrol efficacy against jujube black spot rot. J. Fungi 2022, 8, 1313. [Google Scholar] [CrossRef]


| Number | Designation | Treatment |
|---|---|---|
| 1 | CK | Blank control without pathogen inoculation |
| 2 | CM9 | 10 mL Colletotrichum siamense CM9 suspension (1 × 106 cfu/mL) |
| 3 | FZN | 10 mL CM9 suspension (1 × 106 cfu/mL) + 10 mL fungicide fluazinam 10−3 dilution (Shirlan, Syngenta) |
| 4 | B1 | 10 mL CM9 suspension (1 × 106 cfu/mL) + 10 mL 14-day B. vietnamiensis B418 monoculture (fermentation broth with bacteria cells) |
| 5 | T21 | 10 mL CM9 suspension (1 × 106 cfu/mL) + 10 mL 14-day T. harizanum TW21990 monoculture (fermentation broth with mycelia cells) |
| 6 | BT21 | 10 mL CM9 suspension (1 × 106 cfu/mL) + 10 mL 14-day B418 + TW21990 co-culture (fermentation broth with bacteria and mycelia cells) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Fu, Y.; Jiang, Y.; Hu, J.; Wei, Y.; Li, H.; Li, J.; Yang, H.; Wu, Y. Synergistic Biocontrol and Growth Promotion in Strawberries by Co-Cultured Trichoderma harzianum TW21990 and Burkholderia vietnamiensis B418. J. Fungi 2024, 10, 551. https://doi.org/10.3390/jof10080551
Li W, Fu Y, Jiang Y, Hu J, Wei Y, Li H, Li J, Yang H, Wu Y. Synergistic Biocontrol and Growth Promotion in Strawberries by Co-Cultured Trichoderma harzianum TW21990 and Burkholderia vietnamiensis B418. Journal of Fungi. 2024; 10(8):551. https://doi.org/10.3390/jof10080551
Chicago/Turabian StyleLi, Wenzhe, Yiting Fu, Yanqing Jiang, Jindong Hu, Yanli Wei, Hongmei Li, Jishun Li, Hetong Yang, and Yuanzheng Wu. 2024. "Synergistic Biocontrol and Growth Promotion in Strawberries by Co-Cultured Trichoderma harzianum TW21990 and Burkholderia vietnamiensis B418" Journal of Fungi 10, no. 8: 551. https://doi.org/10.3390/jof10080551
APA StyleLi, W., Fu, Y., Jiang, Y., Hu, J., Wei, Y., Li, H., Li, J., Yang, H., & Wu, Y. (2024). Synergistic Biocontrol and Growth Promotion in Strawberries by Co-Cultured Trichoderma harzianum TW21990 and Burkholderia vietnamiensis B418. Journal of Fungi, 10(8), 551. https://doi.org/10.3390/jof10080551






