The Expanding Mycovirome of Aspergilli
Abstract
1. Introduction
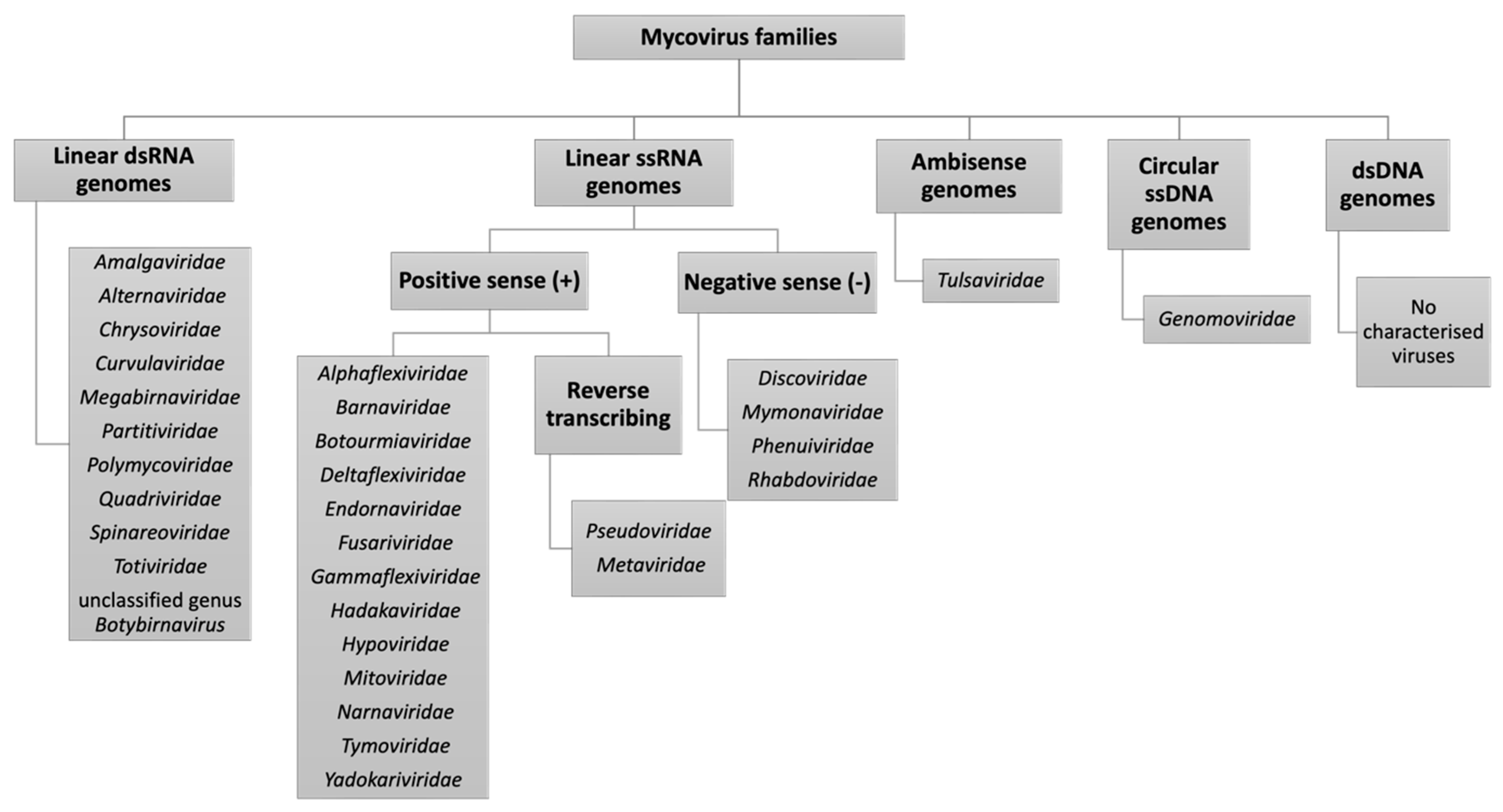
2. Characterisation of Novel Mycoviruses in Aspergilli
2.1. The First Observations of ssRNA Viruses in Aspergilli
2.1.1. Narnaviridae
2.1.2. Mitoviridae
2.1.3. Botourmiaviridae
2.2. Expansion of the dsRNA Virome
2.2.1. Partitiviridae
2.2.2. Chrysoviridae
2.2.3. Totiviridae
2.2.4. Polymycoviridae
| Family | Genome Type | Genome Length (kbp) | Segmentation * | Encapsidation | Example of Mycovirus in Aspergillus |
|---|---|---|---|---|---|
| Narnaviridae | Linear (+) ssRNA | 2.3–2.9 | Unsegmented | Unencapsidated | Aspergillus fumigatus narnavirus 1 (AfuNV1) [62] |
| Mitoviridae | Linear (+) ssRNA | 2.5–2.9 | Mono-segmented | Unencapsidated | Aspergillus fumigatus mitovirus 1 (AfuMV1) [62] |
| Botourmiaviridae | Linear (+) ssRNA | 2.9–5 | Monosegmented ** | Unencapsidated ** | Aspergillus fumigatus botourmiavirus 1 (AfuBOV1) [28] |
| Partitiviridae | Linear dsRNA | 3–4.8 | Bi- or tri-segmented | Encapsidated | Aspergillus fumigatus partitivirus 1 (AfuPV1) [80,81] |
| Totiviridae | Linear dsRNA | 4.6–6.7 | Mono-segmented | Encapsidated | Aspergillus niger victorivirus 1 (AnV1) [102] |
| Chrysoviridae | Linear dsRNA | 8.9–16 | Four segments | Encapsidated | Aspergillus fumigatus chrysovirus 41,362 (AfuCV41362) [99] |
| Polymycoviridae | Linear dsRNA | 7.5–12.5 | Four segments | Unconventionally encapsidated | Aspergillus fumigatus polymycovirus 1 (AfuPMV1) [60,105] |
| Alternaviridae | Linear dsRNA | 8.4–10.7 | Three to four segments | Encapsidated | Aspergillus foetidus alternavirus (AfAV) [108] |
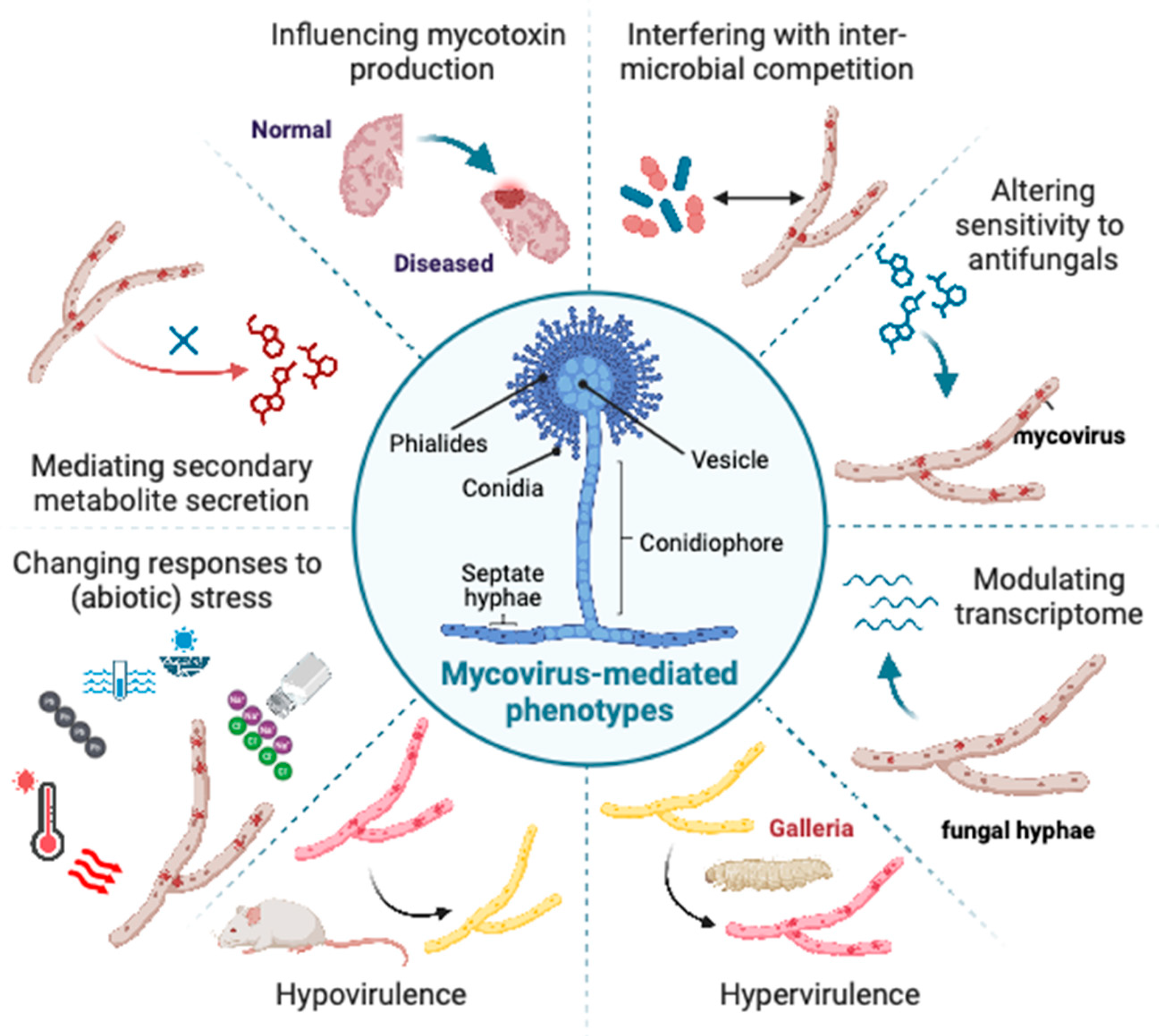
3. Mycovirus-Mediated Phenotypes
3.1. Aspergillus fumigatus
3.2. Aspergillus flavus
3.3. Factors Affecting Manifestation of Mycovirus-Mediated Phenotypes
4. Molecular Mechanisms Underpinning Mycovirus-Mediated Phenotypes
4.1. RNA Silencing: Antiviral Defence in Fungi
4.1.1. Mycoviruses Are Triggers, Targets and Suppressors of RNA Silencing
4.1.2. RNA Silencing Suppression Mechanisms
4.2. Function of Individual Viral Proteins
5. Mycovirus-Based Applications: A Role in Therapeutics?
6. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hollings, M. Viruses associated with a die-back disease of cultivated mushroom. Nature 1962, 196, 962–965. [Google Scholar] [CrossRef]
- Ghabrial, S.A.; Castón, J.R.; Jiang, D.; Nibert, M.L.; Suzuki, N. 50-plus years of fungal viruses. Virology 2015, 479, 356–368. [Google Scholar] [CrossRef]
- Myers, J.M.; James, T.Y. Mycoviruses. Curr. Biol. 2022, 32, R150–R155. [Google Scholar] [CrossRef] [PubMed]
- Villan Larios, D.C.; Diaz Reyes, B.M.; Pirovani, C.P.; Loguercio, L.L.; Santos, V.C.; Góes-Neto, A.; Fonseca, P.L.C.; Aguiar, E.R.G.R. Exploring the mycovirus universe: Identification, diversity, and biotechnological applications. J. Fungi 2023, 9, 361. [Google Scholar] [CrossRef] [PubMed]
- Kondo, H.; Botella, L.; Suzuki, N. Mycovirus diversity and evolution revealed/inferred from recent studies. Annu. Rev. Phytopathol. 2022, 60, 307–336. [Google Scholar] [CrossRef]
- Llorens, C.; Soriano, B.; Krupovic, M. ICTV Virus Taxonomy Profile: Metaviridae. J. Gen. Virol. 2020, 101, 1131–1132. [Google Scholar] [CrossRef] [PubMed]
- Llorens, C.; Soriano, B.; Krupovic, M. ICTV Virus Taxonomy Profile: Pseudoviridae. J. Gen. Virol. 2021, 102, 1563. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Adkins, S.; Brown, K.; de la Torre, J.C.; Digiaro, M.; Hughes, H.R.; Junglen, S.; Lambert, A.J.; Maes, P.; Marklewitz, M.; et al. ICTV Virus Taxonomy Profile: Tulasviridae 2023. J. Gen. Virol. 2023, 104, 001933. [Google Scholar] [CrossRef]
- Varsani, A.; Krupovic, M. Family Genomoviridae: 2021 taxonomy update. Arch. Virol. 2021, 166, 2911–2926. [Google Scholar] [CrossRef]
- Wang, X.; Kotta-Loizou, I.; Coutts, R.H.; Deng, H.; Han, Z.; Hong, N.; Shafik, K.; Wang, L.; Guo, Y.; Yang, M.; et al. A circular single-stranded DNA mycovirus infects plants and confers broad-spectrum fungal resistance. Mol. Plant. 2024, 17, 955–971. [Google Scholar] [CrossRef] [PubMed]
- Dolja, V.V.; Koonin, E.V. Metagenomics reshapes the concepts of RNA virus evolution by revealing extensive horizontal virus transfer. Virus Res. 2018, 244, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Marzano, S.Y.L.; Nelson, B.D.; Ajayi-Oyetunde, O.; Bradley, C.A.; Hughes, T.J.; Hartman, G.L.; Eastburn, D.M.; Domier, L.L. Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J. Virol. 2016, 90, 6846–6863. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lin, X.D.; Tian, J.H.; Chen, L.J.; Chen, X.; Li, C.X.; Qin, X.C.; Li, J.; Cao, J.P.; Eden, J.S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Shi, M.; Holmes, E.C. Using metagenomics to characterize an expanding virosphere. Cell 2018, 172, 1168–1172. [Google Scholar] [CrossRef] [PubMed]
- Mata, C.P.; Rodríguez, J.M.; Suzuki, N.; Castón, J.R. Structure and assembly of double-stranded RNA mycoviruses. Adv. Virus Res. 2020, 108, 213–247. [Google Scholar] [PubMed]
- Hough, B.; Steenkamp, E.; Wingfield, B.; Read, D. Fungal viruses unveiled: A comprehensive review of mycoviruses. Viruses 2023, 15, 1202. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Turina, M.; Chiba, S.; Okada, R.; Bhatti, M.F.; Kotta-Loizou, I.; Coutts, R.H.; Kondo, H.; Sabanadzovic, S.; Suzuki, N. ICTV Virus Taxonomy Profile: Hadakaviridae 2023. J. Gen. Virol. 2023, 104, 001820. [Google Scholar] [CrossRef]
- Hillman, B.I.; Cai, G. The family Narnaviridae: Simplest of RNA viruses. Adv. Virus Res. 2013, 86, 149–176. [Google Scholar]
- Venkataraman, S.; Prasad, B.V.L.S.; Selvarajan, R. RNA dependent RNA polymerases: Insights from structure, function and evolution. Viruses 2018, 10, 76. [Google Scholar] [CrossRef]
- Khalifa, M.E.; MacDiarmid, R.M. A mechanically transmitted DNA mycovirus is targeted by the defence machinery of its host, Botrytis cinerea. Viruses 2021, 13, 1315. [Google Scholar] [CrossRef]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Adriaenssens, E.M.; Zerbini, F.M.; Abrescia, N.G.A.; Aiewsakun, P.; Alfenas-Zerbini, P.; Bao, Y.; Barylski, J.; Drosten, C.; Duffy, S.; et al. Four principles to establish a universal virus taxonomy. PLoS Biol. 2023, 21, e3001922. [Google Scholar] [CrossRef] [PubMed]
- Buck, K.W. (Ed.) Fungal Virology; CRC Press: Boca Raton, FL, USA, 1986; pp. 1–84. [Google Scholar]
- Nuss, D.L. Hypovirulence: Mycoviruses at the fungal–plant interface. Nat. Rev. Microbiol. 2005, 3, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Hillman, B.I.; Suzuki, N. Viruses of the chestnut blight fungus, Cryphonectria parasitica. Adv. Virus Res. 2004, 63, 423–472. [Google Scholar]
- Rigling, D.; Prospero, S. Cryphonectria parasitica, the causal agent of chestnut blight: Invasion history, population biology and disease control. Mol. Plant Pathol. 2018, 19, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Jiang, D. New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu. Rev. Phytopathol. 2014, 52, 45–68. [Google Scholar] [CrossRef]
- Kinsella, C.M.; Deijs, M.; Gittelbauer, H.; van der Hoek, L.; van Dijk, K. Human clinical isolates of pathogenic fungi are host to diverse mycoviruses. Microbiol. Spectr. 2022, 10, e01610-22. [Google Scholar] [CrossRef]
- Ahn, I.P.; Lee, Y.H. A viral double-stranded RNA up regulates the fungal virulence of Nectria radicicola. Mol. Plant Microbe Interact. 2001, 14, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Shah, U.A.; Kotta-Loizou, I.; Fitt, B.D.L.; Coutts, R.H.A. Mycovirus induced hypervirulence of Leptosphaeria biglobosa enhances systemic acquired resistance to Leptosphaeria maculans in Brassica napus. Mol. Plant Microbe Interact. 2020, 33, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Olivé, M.; Campo, S. The dsRNA mycovirus ChNRV1 causes mild hypervirulence in the fungal phytopathogen Colletotrichum higginsianum. Arch. Microbiol. 2021, 203, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Ejmal, M.A.; Holland, D.J.; MacDiarmid, R.M.; Pearson, M.N. A novel chrysovirus from a clinical isolate of Aspergillus thermomutatus affects sporulation. PLoS ONE 2018, 13, e0209443. [Google Scholar] [CrossRef]
- Filippou, C.; Diss, R.M.; Daudu, J.O.; Coutts, R.H.A.; Kotta-Loizou, I. The polymycovirus-mediated growth enhancement of the entomopathogenic fungus Beauveria bassiana is dependent on carbon and nitrogen metabolism. Front. Microbiol. 2021, 12, 606366. [Google Scholar] [CrossRef] [PubMed]
- Shah, U.A.; Kotta-Loizou, I.; Fitt, B.D.L.; Coutts, R.H.A. Identification, molecular characterization, and biology of a novel quadrivirus infecting the phytopathogenic fungus Leptosphaeria biglobosa. Viruses 2019, 11, 9. [Google Scholar] [CrossRef]
- Craven, M.G.; Pawlyk, D.M.; Choi, G.H.; Nuss, D.L. Papain-like protease p29 as a symptom determinant encoded by a hypovirulence associated virus of the chestnut blight fungus. J. Virol. 1993, 67, 6513–6521. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, A.; Urayama, S.I.; Suo, R.; Itoi, S.; Fuji, S.I.; Moriyama, H.; Hagiwara, D. Mycovirus-induced tenuazonic acid production in a rice blast fungus Magnaporthe oryzae. Front Microbiol. 2020, 11, 1641. [Google Scholar] [CrossRef]
- Lee, K.M.; Cho, W.K.; Yu, J.; Son, M.; Choi, H.; Min, K.; Lee, Y.-W.; Kim, K.-H. A comparison of transcriptional patterns and mycological phenotypes following infection of Fusarium graminearum by four mycoviruses. PLoS ONE 2014, 9, e100989. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Liu, D.; Pan, X.; Yan, S.; Song, J.; Liu, D.; Wang, Z.; Xie, Y.; Dai, J.; Liu, J.; et al. Deoxynivalenol Biosynthesis in Fusarium pseudograminearum Significantly Repressed by a Megabirnavirus. Toxins 2022, 14, 503. [Google Scholar] [CrossRef] [PubMed]
- Bevan, E.A.; Herring, A.J.; Mitchell, D.J. Preliminary characterisation of two species of dsRNA in yeast and their relationship to the “killer” character. Nature 1973, 245, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Patil, R.H.; Kotta-Loizou, I.; Palyzová, A.; Pluháček, T.; Coutts, R.H.A.; Stevens, D.A.; Havlíček, V. Freeing Aspergillus fumigatus of polymycovirus infection renders it more resistant to competition with Pseudomonas aeruginosa due to altered iron-acquiring tactics. J. Fungi 2021, 7, 497. [Google Scholar] [CrossRef]
- Sass, G.; Kotta-Loizou, I.; Martinez, M.; Larwood, D.J.; Stevens, D.A. Polymycovirus Infection Sensitizes Aspergillus fumigatus for Antifungal Effects of Nikkomycin Z. Viruses 2023, 15, 197. [Google Scholar] [CrossRef]
- Niu, Y.; Yuan, Y.; Mao, J.; Yang, Z.; Cao, Q.; Zhang, T.; Wang, S.; Liu, D. Characterization of two novel mycoviruses from Penicillium digitatum and the related fungicide resistance analysis. Sci Rep. 2018, 8, 5513. [Google Scholar] [CrossRef]
- Zhang, H.; Xie, J.; Fu, Y.; Cheng, J.; Qu, Z.; Zhao, Z.; Cheng, S.; Chen, T.; Li, B.; Wang, Q.; et al. A 2-kb mycovirus converts a pathogenic fungus into a beneficial endophyte for brassica protection and yield enhancement. Mol. Plant. 2020, 13, 1420–1433. [Google Scholar] [CrossRef]
- Zhou, L.; Li, X.; Kotta-Loizou, I.; Dong, K.; Li, S.; Ni, D.; Hong, N.; Wang, G.; Xu, W. A mycovirus modulates the endophytic and pathogenic traits of a plant associated fungus. ISME J. 2021, 15, 1893–1906. [Google Scholar] [CrossRef]
- Nazik, H.; Kotta-Loizou, I.; Sass, G.; Coutts, R.H.A.; Stevens, D.A. Virus infection of Aspergillus fumigatus compromises the fungus in intermicrobial competition. Viruses 2021, 13, 686. [Google Scholar] [CrossRef]
- Sass, G.; Martinez, M.; Kotta-Loizou, I.; Stevens, D. AfuPmV-1-infected Aspergillus fumigatus is more susceptible to stress than virus-free fungus. J. Fungi 2023, 9, 750. [Google Scholar] [CrossRef]
- Stevens, D.A.; Kotta-Loizou, I.; Martinez, M.; Coutts, R.H.A.; Sass, G. Virus infection impairs fungal response to stress: Effect of salt. Viruses 2023, 15, 718. [Google Scholar] [CrossRef] [PubMed]
- Kotta-Loizou, I. Mycoviruses and their role in fungal pathogenesis. Curr. Opin. Microbiol. 2021, 63, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Jacobson, D.J.; Shiu, P.K.T. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu. Rev. Genet. 2000, 34, 165–186. [Google Scholar] [CrossRef] [PubMed]
- Biella, S.; Smith, M.L.; Aist, J.R.; Cortesi, P.; Milgroom, M.G. Programmed cell death correlates with virus transmission in a filamentous fungus. Proc. R. Soc. B 2002, 269, 2269–2276. [Google Scholar] [CrossRef] [PubMed]
- García-Pedrajas, M.; Cañizares, M.; Sarmiento-Villamil, J.L.; Jacquat, A.G.; Dambolena, J.S. Mycoviruses in biological control: From basic research to field implementation. Phytopathology 2019, 109, 1828–1839. [Google Scholar] [CrossRef]
- Anagnostakis, S.L. Biological control of chestnut blight. Science 1982, 215, 466–471. [Google Scholar] [CrossRef]
- Hai, D.; Li, J.; Jiang, D.; Cheng, J.; Fu, Y.; Xiao, X.; Yin, H.; Lin, Y.; Chen, T.; Li, B.; et al. Plants interfere with non-self recognition of a phytopathogenic fungus via proline accumulation to facilitate mycovirus transmission. Nat. Commun. 2024, 15, 4748. [Google Scholar] [CrossRef]
- Rokas, A. Aspergillus. Curr. Biol. 2013, 23, R187–R188. [Google Scholar] [CrossRef]
- Latgé, J.P.; Chamilos, G. Aspergillus fumigatus and aspergillosis in 2019. Clin. Microbiol. Rev. 2019, 33, e00140-18. [Google Scholar] [CrossRef]
- Klich, M.A. Aspergillus flavus: The major producer of aflatoxin. Mol. Plant Pathol. 2007, 8, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Tevz, G.; Bencina, M.; Legisa, M. Enhancing itaconic acid production by Aspergillus terreus. Appl. Microbiol. Biotechnol. 2010, 87, 1657–1664. [Google Scholar] [CrossRef]
- Li, C.; Zhou, J.; Du, G.; Chen, J.; Takahashi, S.; Liu, S. Developing Aspergillus niger as a cell factory for food enzyme production. Biotechnol. Adv. 2020, 44, 107630. [Google Scholar] [CrossRef] [PubMed]
- Banks, G.T.; Buck, K.W.; Chain, E.B.; Darbyshire, J.E.; Himmelweit, F.; Ratti, G.; Sharpe, T.J.; Planterose, D.N. Antiviral activity of double stranded RNA from a virus isolated from Aspergillus foetidus. Nature 1970, 227, 505–507. [Google Scholar] [CrossRef]
- Kotta-Loizou, I.; Coutts, R.H.A. Mycoviruses in Aspergilli: A comprehensive review. Front. Microbiol. 2017, 8, 1699. [Google Scholar] [CrossRef] [PubMed]
- Okada, R.; Kiyota, E.; Moriyama, H. A simple and rapid method to purify viral dsRNA from plant tissue. J. Gen. Plant Pathol. 2015, 81, 103–107. [Google Scholar] [CrossRef]
- Zoll, J.; Verweij, P.E.; Melchers, W.J.G. Discovery and characterization of novel Aspergillus fumigatus mycoviruses. PLoS ONE 2018, 13, e0200511. [Google Scholar] [CrossRef] [PubMed]
- Chiba, Y.; Oiki, S.; Yaguchi, T.; Urayama, S.I.; Hagiwara, D. Discovery of divided RdRp sequences and a hitherto unknown genomic complexity in fungal viruses. Virus Evol. 2021, 7, veaa101. [Google Scholar] [CrossRef] [PubMed]
- Chiba, Y.; Oiki, S.; Zhao, Y.; Nagano, Y.; Urayama, S.I.; Hagiwara, D. Splitting of RNA-dependent RNA polymerase is common in Narnaviridae: Identification of a type II divided RdRp from deep-sea fungal isolates. Virus Evol. 2021, 7, veab095. [Google Scholar] [CrossRef] [PubMed]
- Sutela, S.; Forgia, M.; Vainio, E.J.; Chiapello, M.; Daghino, S.; Vallino, M.; Martino, E.; Girlanda, M.; Perotto, S.; Turina, M. The virome from a collection of endomycorrhizal fungi reveals new viral taxa with unprecedented genome organization. Virus Evol. 2020, 6, veaa076. [Google Scholar] [CrossRef]
- Degola, F.; Spadola, G.; Forgia, M.; Turina, M.; Dramis, L.; Chitarra, W.; Nerva, L. Aspergillus goes viral: Ecological insights from the geographical distribution of the mycovirome within an Aspergillus flavus population and its possible correlation with aflatoxin biosynthesis. J. Fungi 2021, 7, 833. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.K.; Liu, H.; Jia, X.; Xu, M.; Lu, Y.; Zou, X.; Li, Q. A novel narnavirus from the entomogenous fungus Beauveria bassiana Vuillemin. 23 November 2023. PREPRINT (Version 1). Available online: https://www.researchsquare.com/article/rs-3465422/v1 (accessed on 5 June 2024).
- Sadiq, S.; Chen, Y.M.; Zhang, Y.Z.; Holmes, E.C. Resolving deep evolutionary relationships within the RNA virus phylum Lenarviricota. Virus Evol. 2022, 8, veac055. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Silas, S.; Wang, Y.; Wu, S.; Bocek, M.; Kazlauskas, D.; Krupovic, M.; Fire, A.; Dolja, V.V.; Koonin, E.V. Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome. Nat. Microbiol. 2020, 5, 1262–1270. [Google Scholar] [CrossRef] [PubMed]
- Ayllón, M.A.; Turina, M.; Xie, J.; Nerva, L.; Marzano, S.L.; Donaire, L.; Jiang, D.; Consortium, I.R. ICTV Virus Taxonomy Profile: Botourmiaviridae. J. Gen. Virol. 2020, 101, 454–455. [Google Scholar] [CrossRef]
- Lutz, T.; Langer, G.; Heinze, C. Complete genome sequence of a novel alternavirus infecting the fungus Ilyonectria crassa. Arch. Virol. 2023, 168, 34. [Google Scholar] [CrossRef]
- Aoki, N.; Moriyama, H.; Kodama, M.; Arie, T.; Teraoka, T.; Fukuhara, T. A novel mycovirus associated with four double-stranded RNAs affects host fungal growth in Alternaria alternata. Virus Res. 2009, 140, 179–187. [Google Scholar] [CrossRef]
- Hammond, T.M.; Andrewski, M.D.; Roossinck, M.J.; Keller, N.P. Aspergillus mycoviruses are targets and suppressors of RNA silencing. Eukaryot. Cell 2008, 7, 350–357. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Chen, X.; Li, P.; Qiu, D.; Guo, L. Complete genome sequence of a Fusarium graminearum double-stranded RNA virus in a newly proposed family, Alternaviridae. Genome Announc. 2018, 6, e00064-18. [Google Scholar] [CrossRef]
- Lutz, T.; Japić, E.; Bien, S.; Langer, G.J.; Heinze, C. Characterization of a novel alternavirus infecting the fungal pathogen Fusarium solani. Virus Res. 2022, 317, 198817. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.F.; Aoki, N.; Takeshita, N.; Fukuhara, T.; Chiura, H.X.; Arie, T.; Kotta-Loizou, I.; Okada, R.; Komatsu, K.; Moriyama, H. Unique terminal regions and specific deletions of the segmented double-stranded RNA genome of Alternaria alternata virus 1, in the proposed family Alternaviridae. Front. Microbiol. 2021, 12, 773062. [Google Scholar] [CrossRef]
- Zhang, X.; Xie, Y.; Zhang, F.; Sun, H.; Zhai, Y.; Zhang, S.; Yuan, H.; Zhou, L.; Gao, F.; Li, H. Complete genome sequence of an alternavirus from the phytopathogenic fungus Fusarium incarnatum. Arch. Virol. 2019, 164, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Vainio, E.J.; Chiba, S.; Ghabrial, S.A.; Maiss, E.; Roossinck, M.; Sabanadzovic, S.; Suzuki, N.; Xie, J.; Nibert, M.; Ictv Report Consortium. ICTV Virus Taxonomy Profile: Partitiviridae. J. Gen. Virol. 2018, 99, 17–18. [Google Scholar] [CrossRef]
- Liu, W.; Duns, G.; Chen, J. Genomic characterization of a novel partitivirus infecting Aspergillus ochraceus. Virus Genes 2008, 37, 322–327. [Google Scholar] [CrossRef]
- Bhatti, M.F.; Bignell, E.M.; Coutts, R.H.A. Complete nucleotide sequences of two dsRNAs associated with a new partitivirus infecting Aspergillus fumigatus. Arch. Virol. 2011, 156, 1677–1680. [Google Scholar] [CrossRef]
- Filippou, C.; Coutts, R.H.A.; Stevens, D.A.; Sabino, R.; Kotta-Loizou, I. Completion of the sequence of the Aspergillus fumigatus partitivirus 1 genome. Arch. Virol. 2020, 165, 1891–1894. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, J.; Yang, B.; Wang, Q.; Zhou, J.; Yu, W. Molecular characterization of a debilitation-associated partitivirus infecting the pathogenic fungus Aspergillus flavus. Front. Microbiol. 2019, 10, 626. [Google Scholar] [CrossRef]
- Kuroki, M.; Yaguchi, T.; Urayama, S.I.; Hagiwara, D. Experimental verification of strain-dependent relationship between mycovirus and its fungal host. Iscience 2023, 26, 107337. [Google Scholar] [CrossRef] [PubMed]
- Gnanasekaran, P.; Chakraborty, S. Biology of viral satellites and their role in pathogenesis. Curr. Opin. Virol. 2018, 33, 96–105. [Google Scholar] [CrossRef]
- Shah, U.A.; Kotta-Loizou, I.; Coutts, R.H.A. Sequence determination of a satellite RNA isolated from Aspergillus foetidus. Arch. Virol. 2015, 160, 883–885. [Google Scholar] [CrossRef]
- Han, Z.; Liu, J.; Kong, L.; He, Y.; Wu, H.; Xu, W. A special satellite-like RNA of a novel hypovirus from Pestalotiopsis fici broadens the definition of fungal satellite. PLoS Pathog. 2023, 19, e1010889. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Q.; Cheng, J.; Fu, Y.; Jiang, D.; Xie, J. Molecular characterization of a bipartite double-stranded RNA virus and its satellite-like RNA co-infecting the phytopathogenic fungus Sclerotinia sclerotiorum. Front. Microbiol. 2015, 6, 406. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, B.; Liu, X.; Tian, X.; Wang, Q.; Wang, B.; Zhang, Q.; Yu, W.; Qi, X.; Jiang, Y.; et al. attenuates the induction of helper virus-mediated symptoms in Aspergillus flavus. Front. Microbiol. 2022, 13, 895844. [Google Scholar]
- Nibert, M.L.; Ghabrial, S.A.; Maiss, E.; Lesker, T.; Vainio, E.J.; Jiang, D.; Suzuki, N. Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res. 2014, 188, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Ghabrial, S.A.; Suzuki, N. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 2009, 47, 353–384. [Google Scholar] [CrossRef]
- Pinheiro, A.; Piontkivska, D.; Sequeira, P.; Martins, T.M.; Silva Pereira, C. Aspergillus nidulans . Trends Microbiol. 2023, 31, 212–213. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, B.; Liu, J.; Wang, Q.; Zhang, Q.; Yu, W.; Hsiang, T. A novel mycovirus infecting Aspergillus nidulans that is closely related to viruses in a new genus of the family Partitiviridae. Arch. Virol. 2021, 166, 659–664. [Google Scholar] [CrossRef]
- Kotta-Loizou, I.; Castón, J.R.; Coutts, R.H.A.; Hillman, B.I.; Jiang, D.; Kim, D.H.; Moriyama, H.; Suzuki, N.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Chrysoviridae. J. Gen. Virol. 2020, 101, 143–144. [Google Scholar] [CrossRef] [PubMed]
- Lutz, T.; Langer, G.J.; Heinze, C. A virus from Aspergillus cibarius with features of alpha- and betachrysoviruses. Virus Genes. 2024, 60, 71–79. [Google Scholar] [CrossRef]
- Wu, Z.; Tian, X.; Liu, X.; Zhou, J.; Yu, W.; Qi, X.; Peng, J.; Hsiang, T.; Wang, Q.; Wu, N.; et al. Complete genome sequence of a novel chrysovirus infecting Aspergillus terreus. Arch. Virol. 2023, 168, 209. [Google Scholar] [CrossRef]
- Huang, X.; Men, P.; Tang, S.; Lu, X. Aspergillus terreus as an industrial filamentous fungus for pharmaceutical biotechnology. Curr. Opin. Biotechnol. 2021, 69, 273–280. [Google Scholar] [CrossRef]
- Tortorano, A.M.; Prigitano, A.; Dho, G.; Biraghi, E.; Stevens, D.A.; Ghannoum, M.; Nolard, N.; Viviani, M.A. In vitro activity of amphotericin B against Aspergillus terreus isolates from different countries. J. Chemother. 2008, 20, 756–757. [Google Scholar] [CrossRef] [PubMed]
- Gallie, D.R.; Walbot, V. Identification of the motifs within the tobacco mosaic virus 5′-leader responsible for enhancing translation. Nucleic Acids Res. 1992, 20, 4631–4638. [Google Scholar] [CrossRef] [PubMed]
- Takahashi-Nakaguchi, A.; Shishido, E.; Yahara, M.; Urayama, S.I.; Sakai, K.; Chibana, H.; Kamei, K.; Moriyama, H.; Gonoi, T. Analysis of an intrinsic mycovirus associated with reduced virulence of the human pathogenic fungus Aspergillus fumigatus. Front. Microbiol. 2020, 10, 3045. [Google Scholar] [CrossRef]
- Mertens, P.P.; Sangar, D.V. Analysis of the terminal sequences of the genome segments of four orbiviruses. Prog. Clin. Biol. Res. 1985, 178, 371–387. [Google Scholar] [CrossRef]
- Ayllon, M.A.; Vainio, E.J. Mycoviruses as a part of the global virome: Diversity, evolutionary links and lifestyle. Adv. Virus Res. 2023, 115, 1–86. [Google Scholar]
- Jiang, Y.; Liu, X.; Yang, B.; Tian, X.; Liu, J.; Wang, Q.; Zhang, Q.; Yu, W.; Qi, X.; Hsiang, T. Complete genome sequence of a novel victorivirus infecting Aspergillus niger. Arch. Virol. 2022, 167, 1475–1479. [Google Scholar] [CrossRef]
- Li, H.; Havens, W.; Nibert, M.; Ghabrial, S. RNA sequence determinants of a coupled termination-reinitiation strategy for downstream open reading frame translation in Helminthosporium victoriae virus 190S and other victoriviruses (Family Totiviridae). J. Virol. 2011, 85, 7343–7352. [Google Scholar] [CrossRef] [PubMed]
- Kotta-Loizou, I.; Coutts, R.H.A.; ICGV Report Consortium. ICTV Virus Taxonomy Profile: Polymycoviridae 2022. J. Gen. Virol 2022, 103, 1747 . [Google Scholar] [CrossRef]
- Kanhayuwa, L.; Kotta-Loizou, I.; Özkan, S.; Gunning, A.P.; Coutts, R.H. A novel mycovirus from Aspergillus fumigatus contains four unique dsRNAs as its genome and is infectious as dsRNA. Proc. Natl. Acad. Sci. USA 2015, 112, 9100–9105. [Google Scholar] [CrossRef] [PubMed]
- Nerva, L.; Forgia, M.; Ciuffo, M.; Chitarra, W.; Chiapello, M.; Vallino, M.; Varese, G.C.; Turina, M. The mycovirome of a fungal collection from the sea cucumber Holothuria polii. Virus Res. 2019, 273, 197737. [Google Scholar] [CrossRef]
- Takahashi-Nakaguchi, A.; Shishido, E.; Yahara, M.; Urayama, S.I.; Ninomiya, A.; Chiba, Y.; Sakai, K.; Hagiwara, D.; Chibana, H.; Moriyama, H.; et al. Phenotypic and molecular biological analysis of polymycovirus AfuPmV-1M from Aspergillus fumigatus: Reduced fungal virulence in a mouse infection model. Front. Microbiol. 2020, 11, 607795. [Google Scholar] [CrossRef]
- Kozlakidis, Z.; Herrero, N.; Özkan, S.; Kanhayuwa, L.; Jamal, A.; Bhatti, M.F.; Coutts, R.H.A. Sequence determination of a quadripartite dsRNA virus isolated from Aspergillus foetidus. Arch. Virol. 2013, 158, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Sinden, J.; Hauser, E. Report on two new mushroom diseases. Mushroom Sci. 1950, 1, 96–100. [Google Scholar]
- van der Lende, T.R.; Harmsen, M.C.; Wessels, J.G. Double-stranded RNAs and proteins associated with the 34 nm virus particles of the cultivated mushroom Agaricus bisporus. J. Gen. Virol. 1994, 75, 2533–2536. [Google Scholar] [CrossRef] [PubMed]
- Berry, E.A.; Bevan, E.A. A new species of double-stranded RNA from yeast. Nature 1972, 239, 279–280. [Google Scholar] [CrossRef]
- Maske, B.L.; Neto, D.P.D.C.; da Silva, G.B.; Lindner, J.D.D.; Soccol, C.R.; de Melo Pereira, G.V. Yeast viruses and their implications in fermented foods and beverages. Curr. Opin. Food Sci. 2022, 47, 100879. [Google Scholar] [CrossRef]
- Mannazzu, I.; Domizio, P.; Carboni, G.; Zara, S.; Zara, G.; Comitini, F.; Budroni, M.; Ciani, M. Yeast killer toxins: From ecological significance to application. Crit. Rev. Biotechnol. 2019, 39, 603–617. [Google Scholar] [CrossRef]
- Hillman, B.I.; Turina, M. Viruses that affect phenotype and fitness of fungi. In Fungal Associations; Springer International Publishing: Cham, Switzerland, 2024; pp. 113–144. [Google Scholar]
- Ikeda, A.; Chiba, Y.; Kuroki, M.; Urayama, S.I.; Hagiwara, D. Efficient elimination of RNA mycoviruses in Aspergillus species using RdRp-inhibitors ribavirin and 2’-C-methylribonucleoside derivatives. Front. Microbiol. 2022, 13, 1024933. [Google Scholar] [CrossRef]
- Khan, H.A.; Baig, D.I.; Bhatti, M.F. An overview of mycoviral curing strategies used in evaluating fungal host fitness. Mol. Biotechnol. 2023, 65, 1547–1564. [Google Scholar] [CrossRef]
- Bhatti, M.F.; Jamal, A.; Petrou, M.A.; Cairns, T.C.; Bignell, E.M.; Coutts, R.H.A. The effects of dsRNA mycoviruses on growth and murine virulence of Aspergillus fumigatus. Fungal Genet. Biol. 2011, 48, 1071–1075. [Google Scholar] [CrossRef] [PubMed]
- Herrero, N.; Zabalgogeazcoa, I. Mycoviruses infecting the endophytic and entomopathogenic fungus Tolypocladium cylindrosporum. Virus Res. 2011, 160, 409–413. [Google Scholar] [CrossRef] [PubMed]
- van Diepeningen, A.D.; Debets, A.J.M.; Hoekstra, R.F. Dynamics of dsRNA mycoviruses in black Aspergillus populations. Fungal Genet. Biol. 2006, 43, 446–452. [Google Scholar] [CrossRef]
- Ferreira, J.A.G.; Penner, J.; Moss, R.B.; Haagensen, J.A.J.; Clemons, K.V.; Spormann, A.M.; Nazik, H.; Cohen, K.; Banaei, N.; Carolino, E.; et al. Inhibition of Aspergillus fumigatus and its biofilm by Pseudomonas aeruginosa is dependent on the source, phenotype and growth conditions of the bacterium. PLoS ONE 2015, 10, e0134692. [Google Scholar] [CrossRef]
- Özkan, S.; Coutts, R.H.A. Aspergillus fumigatus mycovirus causes mild hypervirulent effect on pathogenicity when tested on Galleria mellonella. Fungal Genet. Biol. 2015, 76, 20–26. [Google Scholar] [CrossRef]
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet Infect. Dis. 2017, 17, e383–e392. [Google Scholar] [CrossRef]
- Brown, N.A.; Goldman, G.H. The contribution of Aspergillus fumigatus stress responses to virulence and antifungal resistance. J. Microbiol. 2016, 54, 243–253. [Google Scholar] [CrossRef]
- Grahl, N.; Puttikamonkul, S.; Macdonald, J.M.; Gamcsik, M.P.; Ngo, L.Y.; Hohl, T.M.; Cramer, R.A. In vivo hypoxia and a fungal alcohol dehydrogenase influence the pathogenesis of invasive pulmonary Aspergillosis. PLoS Pathog. 2011, 7, e1002145. [Google Scholar] [CrossRef] [PubMed]
- Grahl, N.; Shepardson, K.M.; Chung, D.; Cramer, R.A. Hypoxia and fungal pathogenesis: To air or not to air? Eukaryot Cell. 2012, 11, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Cánovas, D.; Marcos, J.F.; Marcos, A.T.; Strauss, J. Nitric oxide in fungi: Is there NO light at the end of the tunnel? Curr Genet. 2016, 62, 513–518. [Google Scholar] [CrossRef]
- Orciuolo, E.; Stanzani, M.; Canestraro, M.; Galimberti, S.; Carulli, G.; Lewis, R.; Petrini, M.; Komanduri, K.V. Effects of Aspergillus fumigatus gliotoxin and methylprednisolone on human neutrophils: Implications for the pathogenesis of invasive aspergillosis. J. Leukoc Biol. 2007, 82, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Sugui, J.A.; Pardo, J.; Chang, Y.C.; Zarember, K.A.; Nardone, G.; Galvez, E.M.; Müllbacher, A.; Gallin, J.I.; Simon, M.M.; Kwon-Chung, K.J. Gliotoxin is a virulence factor of Aspergillus fumigatus: GliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot. Cell. 2007, 6, 1562–1569. [Google Scholar] [CrossRef] [PubMed]
- Guruceaga, X.; Ezpeleta, G.; Mayayo, E.; Sueiro-Olivares, M.; Abad-Diaz-De-Cerio, A.; Aguirre Urízar, J.M.; Liu, H.G.; Wiemann, P.; Bok, J.W.; Filler, S.G.; et al. A possible role for fumagillin in cellular damage during host infection by Aspergillus fumigatus. Virulence 2018, 9, 1548–1561. [Google Scholar] [CrossRef] [PubMed]
- Hatinguais, R.; Pradhan, A.; Brown, G.D.; Brown, A.J.P.; Warris, A.; Shekhova, E. Mitochondrial Reactive Oxygen Species Regulate Immune Responses of Macrophages to Aspergillus fumigatus. Front. Immunol. 2021, 12, 641495. [Google Scholar] [CrossRef]
- Meyling, N.V.; Eilenberg, J. Ecology of the entomopathogenic fungi Beauveria bassiana and Metarhizium anisopliae in temperate agroecosystems: Potential for conservation biological control. Biol. Control 2007, 43, 145–155. [Google Scholar] [CrossRef]
- Bukhari, T.; Takken, W.; Koenraadt, C.J. Development of Metarhizium anisopliae and Beauveria bassiana formulations for control of malaria mosquito larvae. Parasites Vectors 2011, 4, 1–14. [Google Scholar] [CrossRef]
- Sabino, R.; Ferreira, J.A.; Moss, R.B.; Valente, J.; Veríssimo, C.; Carolino, E.; Clemons, K.V.; Everson, C.; Banaei, N.; Penner, J.; et al. Molecular epidemiology of Aspergillus collected from cystic fibrosis patients. J. Cyst Fibros. 2015, 14, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Williams, H.D.; Davies, J.C. Basic science for the chest physician: Pseudomonas aeruginosa and the cystic fibrosis airway. Thorax 2012, 67, 465–467. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yu, W. Interaction between Pseudomonas aeruginosa and Aspergillus fumigatus in cystic fibrosis. Peer J. 2018, 6, e5931. [Google Scholar] [CrossRef] [PubMed]
- Sass, G.; Nazik, H.; Penner, J.; Shah, H.; Ansari, S.R.; Clemons, K.V.; Groleau, M.C.; Dietl, A.M.; Visca, P.; Haas, H.; et al. Studies of Pseudomonas aeruginosa mutants indicate pyoverdine as the central factor in inhibition of Aspergillus fumigatus biofilm. J Bacteriol. 2017, 200, e00345-17. [Google Scholar] [CrossRef]
- Lin, J.; Cheng, J.; Wang, Y.; Shen, X. The Pseudomonas quinolone signal (PQS): Not just for quorum sensing anymore. Front. Cell Infect. Microbiol. 2018, 8, 230. [Google Scholar] [CrossRef]
- Sass, G.; Ansari, S.R.; Dietl, A.-M.; Deziel, E.; Haas, H.; Stevens, D.A. Intermicrobial interaction: Aspergillus fumigatus siderophores protect against competition by Pseudomonas aeruginosa. PLoS ONE 2019, 14, e0216085. [Google Scholar] [CrossRef]
- Secor, P.R.; Sweere, J.M.; Michaels, L.A.; Malkovskiy, A.V.; Rajadas, J.; Lazzareschi, D.; Katznelson, E.; Arrigoni, A.; Braun, K.R.; Evanko, S.P.; et al. Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe 2015, 18, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Penner, J.C.; Ferreira, J.A.G.; Secor, P.R.; Sweere, J.; Birukova, M.; Joubert, L.-M.; Haagensen, J.A.J.; Garcia, O.; Malkovskiy, A.V.; Kaber, G.; et al. Pf4 bacteriophage produced by Pseudomonas aeruginosa inhibits Aspergillus fumigatus metabolism via iron sequestration. Microbiology 2016, 162, 1583–1594. [Google Scholar] [CrossRef] [PubMed]
- Nazik, H.; Joubert, L.-M.; Secor, P.R.; Sweere, J.; Bollyky, P.L.; Sass, G.; Cegelski, L.; Stevens, D.A. Pseudomonas phage inhibition of Candida albicans. Microbiology 2017, 163, 1568–1577. [Google Scholar] [CrossRef] [PubMed]
- Abrashev, R.I.; Pashova, S.B.; Stefanova, L.N.; Vassilev, S.V.; Dolashka-Angelova, P.A.; Angelova, M.B. Heat-shock-induced oxidative stress and antioxidant response in Aspergillus niger 26. Can. J. Microbiol. 2008, 54, 977–983. [Google Scholar] [CrossRef]
- Methneni, N.; Ezdini, K.; Ben Abdeljelil, N.; Van Loco, J.; Van den Houwe, K.; Jabeur, R.; Fekih Sallem, O.; Jaziri, A.; Fernandez-Serrano, M.; Khdary, N.H.; et al. Occurrence of textile dyes and metals in tunisian textile dyeing effluent: Effects on oxidative stress status and histological changes in Balb/c Mice. Int. J. Mol. Sci. 2021, 22, 12568. [Google Scholar] [CrossRef]
- Sharwani, A.A.; Narayanan, K.B.; Khan, M.E.; Han, S.S. Photocatalytic degradation activity of goji berry extract synthesized silver-loaded mesoporous zinc oxide (Ag@ZnO) nanocomposites under simulated solar light irradiation. Sci. Rep. 2022, 12, 10017. [Google Scholar] [CrossRef] [PubMed]
- Nuskern, L.; Tkalec, M.; Ježi´c, M.; Katani´c, Z.; Krstin, L.; Curkovíc-Perica, M. Cryphonectria hypovirus 1-induced changes of stress enzyme activity in transfected phytopathogenic fungus Cryphonectria parasitica. Microb. Ecol. 2017, 74, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Manavathu, E.K.; Chandrasekar, P.H. Aspergillus flavus: An emerging non-fumigatus Aspergillus species of significance. Mycoses 2009, 52, 206–222. [Google Scholar] [CrossRef]
- Shabeer, S.; Asad, S.; Jamal, A.; Ali, A. Aflatoxin contamination, its impact and management strategies: An updated review. Toxins 2022, 14, 307. [Google Scholar] [CrossRef]
- Luo, S.; Du, H.; Kebede, H.; Liu, Y.; Xing, F. Contamination status of major mycotoxins in agricultural product and food stuff in Europe. Food Control 2021, 127, 108120. [Google Scholar] [CrossRef]
- Benkerroum, N. Chronic and Acute Toxicities of Aflatoxins: Mechanisms of Action. Int. J. Environ. Res. Public Health 2020, 17, 423. [Google Scholar] [CrossRef]
- Schmidt, F.R.; Lemke, P.A.; Esser, K. Viral influences on aflatoxin formation by Aspergillus flavus. Appl. Microbiol. Biotechnol. 1986, 24, 248–252. [Google Scholar] [CrossRef]
- Silva, V.N.; Durigon, E.L.; de Fátima Costa Pires, M.; Lourenço, A.; de Faria, M.J.; Corrêa, B. Time course of virus-like particles (VLPs) double-stranded rna accumulation in toxigenic and non-toxigenic strains of Aspergillus flavus. Braz. J. Microbiol. 2001, 32, 56–60. [Google Scholar] [CrossRef]
- Nerva, L.; Chitarra, W.; Siciliano, I.; Gaiotti, F.; Ciuffo, M.; Forgia, M.; Varese, G.C.; Turina, M. Mycoviruses mediate mycotoxin regulation in Aspergillus ochraceus. Environ. Microbiol. 2019, 21, 1957–1968. [Google Scholar] [CrossRef]
- Claeys, L.; Romano, C.; De Ruyck, K.; Wilson, H.; Fervers, B.; Korenjak, M.; Zavadil, J.; Gunter, M.J.; De Saeger, S.; De Boevre, M.; et al. Mycotoxin exposure and human cancer risk: A systematic review of epidemiological studies. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1449–1464. [Google Scholar] [CrossRef] [PubMed]
- Tebbi, C.K. Mycoviruses in Fungi: Carcinogenesis of Fungal Agents May Not Always Be Mycotoxin Related. J. Fungi 2023, 9, 368. [Google Scholar] [CrossRef] [PubMed]
- Tebbi, C.K.; Badiga, A.; Sahakian, E.; Powers, J.J.; Achille, A.N.; Patel, S.; Migone, F. Exposure to a mycovirus containing Aspergillus flavus reproduces acute lymphoblastic leukemia cell surface and genetic markers in cells from patients in remission and not controls. Cancer Treat. Res. Commun. 2021, 26, 100279. [Google Scholar] [CrossRef]
- Tebbi, C.K.; Kotta-Loizou, I.; Coutts, R.H.A. Mycovirus containing Aspergillus flavus and acute lymphoblastic leukemia: Carcinogenesis beyond mycotoxin production. In The Genus Aspergillus-Pathogenicity, Mycotoxin Production and Industrial Applications; IntechOpen: London, UK, 2021. [Google Scholar]
- Wang, M.B.; Smith, N.A. Satellite RNA pathogens of plants: Impacts and origins-an RNA silencing perspective. Wiley Interdiscip. Rev. RNA 2015, 7, 5–16. [Google Scholar] [CrossRef]
- Alizon, S.; Hurford, A.; Mideo, N.; Van Baalen, M. Virulence evolution and the trade-off hypothesis: History, current state of affairs and the future. J. Evol. Biol. 2009, 22, 245–259. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Dasaradhi, P.V.N.; Mohmmed, A.; Malhotra, P.; Bhatnagar, R.K.; Mukherjee, S.K. RNA Interference: Biology, mechanism, and applications. Microbiol. Mol. Biol. R. 2003, 67, 657–685. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, L. Current advances in antiviral RNA interference in mammals. FEBS J. 2024, 291, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Chen, M.; Xiang, M.; Guo, Z. RNAi-based antiviral innate immunity in plants. Viruses 2022, 14, 432. [Google Scholar] [CrossRef] [PubMed]
- Özkan, S.; Mohorianu, I.; Xu, P.; Dalmay, T.; Coutts, R.H. Profile and functional analysis of small RNAs derived from Aspergillus fumigatus infected with double-stranded RNA mycoviruses. BMC Genom. 2017, 18, 1–13. [Google Scholar] [CrossRef]
- Segers, G.C.; Zhang, X.; Deng, F.; Sun, Q.; Nuss, D.L. Evidence that RNA silencing functions as an antiviral defence mechanism in fungi. Proc. Natl. Acad. Sci. USA 2007, 104, 12902–12906. [Google Scholar] [CrossRef]
- Sun, Q.; Choi, G.H.; Nuss, D.L. A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc. Natl. Acad. Sci. USA 2009, 106, 17927–17932. [Google Scholar] [CrossRef]
- Varga, J.; Toth, B.; Vagvolgyi, C. Recent advances in mycovirus research. Acta Microbiol. Immunol. Hung. 2003, 50, 77–94. [Google Scholar] [CrossRef]
- Nakayashiki, H. RNA silencing in fungi: Mechanisms and applications. FEBS Lett. 2005, 579, 5950–5957. [Google Scholar] [CrossRef] [PubMed]
- Nuss, D.L. Mycoviruses, RNA silencing, and viral RNA recombination. Adv. Virus Res. 2011, 80, 25–48. [Google Scholar] [PubMed]
- Chiba, S.; Suzuki, N. Highly activated RNA silencing via strong induction of dicer by one virus can interfere with the replication of an unrelated virus. Proc. Natl. Acad. Sci. USA 2015, 112, E4911–E4918. [Google Scholar] [CrossRef]
- Andika, I.B.; Jamal, A.; Kondo, H.; Suzuki, N. SAGA complex mediates the transcriptional up-regulation of antiviral RNA silencing. Proc. Natl. Acad. Sci. USA 2017, 114, E3499–E3506. [Google Scholar] [CrossRef]
- Andika, I.B.; Kondo, H.; Suzuki, N. Dicer functions transcriptionally and post-transcriptionally in a multilayer antiviral defense. Proc. Natl. Acad. Sci. USA 2019, 116, 2274–2281. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, X.; Tian, X.; Zhou, J.; Wang, Q.; Wang, B.; Yu, W.; Jiang, Y.; Hsiang, T.; Qi, X. RNA interference of Aspergillus flavus in response to Aspergillus flavus partitivirus 1 infection. Front. Microbiol. 2023, 14, 1252294. [Google Scholar] [CrossRef]
- Segers, G.C.; van Wezel, R.; Zhang, X.; Hong, Y.; Nuss, D.L. Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot. Cell 2006, 5, 896–904. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, J.; Nzabanita, C.; Zhang, M.; Nie, J. ; Guo, L Fungal virus, FgHV1-encoded p20 suppresses RNA silencing through single-strand small RNA binding. J. Fungi 2022, 8, 1171. [Google Scholar] [CrossRef]
- Aulia, A.; Hyodo, K.; Hisano, S.; Kondo, H.; Hillman, B.I.; Suzuki, N. Identification of an RNA silencing suppressor encoded by a symptomless fungal hypovirus, Cryphonectria hypovirus 4. Biology 2021, 10, 100. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Park, J.Y.; Heo, J.I.; Kim, K.H. The ORF2 protein of Fusarium graminearum virus 1 suppresses the transcription of FgDICER2 and FgAGO1 to limit host antiviral defences. Mol. Plant Pathol. 2020, 21, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Yaegashi, H.; Yoshikawa, N.; Ito, T.; Kanematsu, S.A. Mycoreovirus suppresses RNA silencing in the white root rot fungus, Rosellinia necatrix. Virology 2013, 444, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Patil, R.H.; Kotta-Loizou, I.; Palyzová, A.; Pluháček, T.; Coutts, R.; Stevens, D.A.; Havlíček, V. Mycotoxin secretion by Aspergillus fumigatus as a response to mycovirus infection. In Proceedings of the 10th Advances Against Aspergillosis and Mucormycosis Meeting, Virtual, 2–3 February 2022. [Google Scholar]
- Patil, R.; Abd El-Hafeez, A.A.; Sass, G.; Stevens, D.A.; Palyzová, A.; Hsu, J.; Kotta-Loizou, I.; Havlicek, V. Aspergillus fumigatus Secretes Gliotoxin in Response to Polymycovirus Infection; American Society for Mass Spectrometry: Anaheim, CA, USA, 2024. [Google Scholar]
- Avendano, C.; Nguyen, S.; Kotta-Loizou, I.; Sass, G.; Roth, D.; Stevens, D.A.; Kalkum, M. Polymycovirus Alteration of Virulence Factors in Aspergillus fumigatus: A Mass Spectrometric Proteomics Study; American Society for Mass Spectrometry: Anaheim, CA, USA, 2024. [Google Scholar]
- Abd El-Hafeez, A.A.; Sass, G.; Stevens, D.A.; Havlicek, V.; Kotta-Loizou, I.; Hsu, J. Mycovirus infection disrupts Aspergillus fumigatus iron acquisition through down regulation of HapX. In Proceedings of the 11th Advances Against Aspergillosis and Mucormycosis, Milan, Italy, 25–27 January 2024. [Google Scholar]
- Lass-Flörl, C.; Steixner, S. The changing epidemiology of fungal infections. Mol. Aspects Med. 2023, 94, 101215. [Google Scholar] [CrossRef]
- Lehrnbecher, T.; Frank, C.; Engels, K.; Kriener, S.; Groll, A.H.; Schwabe, D. Trends in the post-mortem epidemiology of invasive fungal infections at a university hospital. J. Infect. 2010, 61, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, A.; Ferrara, F.; Boccellino, M.; Ponzo, A.; Cimmino, C.; Comberiati, E.; Zovi, A.; Clemente, S.; Sabbatucci, M. Antifungal drug resistance: An emergent health threat. Biomedicines 2023, 11, 1063. [Google Scholar] [CrossRef]
- Loc-Carrillo, C.; Abedon, S.T. Pros and cons of phage therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Waters, E.M.; Neill, D.R.; Kaman, B.; Sahota, J.S.; Clokie, M.R.J.; Winstanley, C.; Kadioglu, A. Phage therapy is highly effective against chronic lung infections with Pseudomonas aeruginosa. Thorax 2017, 72, 666–667. [Google Scholar] [CrossRef]
- Alemayehu, D.; Casey, P.G.; McAuliffe, O.; Guinane, C.M.; Martin, J.G.; Shanahan, F.; Coffey, A.; Ross, R.P.; Hill, C. Bacteriophages φMR299-2 and φNH-4 can eliminate Pseudomonas aeruginosa in the murine lung and on cystic fibrosis lung airway cells. mBio 2012, 3, e00029-12. [Google Scholar] [CrossRef]
- Chadha, P.; Katare, O.P.; Chhibber, S. In vivo efficacy of single phage versus phage cocktail in resolving burn wound infection in BALB/c mice. Microb. Pathog. 2016, 99, 68–77. [Google Scholar] [CrossRef]
- Green, S.I.; Kaelber, J.T.; Ma, L.; Trautner, B.W.; Ramig, R.F.; Maresso, A.W. Bacteriophages from ExPEC reservoirs kill pandemic multidrug-resistant strains of clonal group ST131 in animal models of bacteremia. Sci. Rep. 2017, 7, 46151. [Google Scholar] [CrossRef]
- Hoyle, N.; Zhvaniya, P.; Balarjishvili, N.; Bolkvadze, D.; Nadareishvili, L.; Nizharadze, D.; Wittmann, J.; Rohde, C.; Kutateladze, M. Phage therapy against Achromobacter xylosoxidans lung infection in a patient with cystic fibrosis: A case report. Res. Microbiol. 2018, 169, 540–542. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Van Belleghem, J.D.; de Vries, C.R.; Burgener, E.; Chen, Q.; Manasherob, R.; Aronson, J.R.; Amanatullah, D.F.; Tamma, P.D.; Suh, G.A. The safety and toxicity of phage therapy: A review of animal and clinical studies. Viruses 2021, 13, 1268. [Google Scholar] [CrossRef] [PubMed]
- van de Sande, W.W.J.; Vonk, A.G. Mycovirus therapy for invasive pulmonary aspergillosis? Med. Mycol. 2019, 57, S179–S188. [Google Scholar] [CrossRef]
- Casadevall, A.; Nosanchuk, J.D.; Williamson, P.; Rodrigues, M.L. Vesicular transport across the fungal cell wall. Trends Microbiol. 2009, 17, 158–162. [Google Scholar] [CrossRef]
- Yu, X.; Li, B.; Fu, Y.; Jiang, D.; Ghabrial, S.A.; Li, G.; Peng, Y.; Xie, J.; Cheng, J.; Huang, J.; et al. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. USA 2010, 107, 8387–8392. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, B.; Fu, Y.; Xie, J.; Cheng, J.; Ghabrial, S.A.; Li, G.; Yi, X.; Jiang, D. Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc. Natl. Acad. Sci. USA 2013, 110, 1452–1457. [Google Scholar] [CrossRef] [PubMed]
- van de Sande, W.W.J.; Lo-Ten-Foe, J.R.; van Belkum, A.; Netea, M.G.; Kullberg, B.J.; Vonk, A.G. Mycoviruses: Future therapeutic agents of invasive fungal infections in humans? Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 755–763. [Google Scholar] [CrossRef]
- Liu, G.L.; Chi, Z.; Wang, G.Y.; Wang, Z.P.; Li, Y.; Chi, Z.M. Yeast killer toxins, molecular mechanisms of their action and their applications. Crit. Rev. Biotechnol. 2015, 35, 222–234. [Google Scholar] [CrossRef]
- Polonelli, L.; Conti, S.; Campani, L.; Fanti, F. Biotyping of Aspergillus fumigatus and related taxa by the yeast killer system. In Modern Concepts in Penicillium and Aspergillus Classification; Springer: Boston, MA, USA, 1990; pp. 225–233. [Google Scholar]
- Polonelli, L.; Lorenzini, R.; De Bernardis, F.; Morace, G. Potential therapeutic effect of yeast killer toxin. Mycopathologia 1986, 96, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Stevens, D.A.; Brummer, E.; Clemons, K.V. Interferon-gamma as an antifungal. J. Infect. Dis. 2006, 194, S33–S37. [Google Scholar] [CrossRef]
- Eren, R.O.; Reverte, M.; Rossi, M.; Hartley, M.A.; Castiglioni, P.; Prevel, F.; Martin, R.; Desponds, C.; Lye, L.F.; Drexler, S.K.; et al. Mammalian innate immune response to a Leishmania-resident RNA virus increases macrophage survival to promote parasite persistence. Cell Host Microbe 2016, 20, 318–328. [Google Scholar] [CrossRef]
- Ives, A.; Ronet, C.; Prevel, F.; Ruzzante, G.; Fuertes-Marraco, S.; Schutz, F.; Zangger, H.; Revaz-Breton, M.; Lye, L.F.; Hickerson, S.M.; et al. Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Science 2011, 331, 775–778. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Cho, Y.J.; Kim, D.; Yang, C.S.; Lee, S.M.; Dawson, T.L.; Nakamizo, S.; Kabashima, K.; Lee, Y.W.; Jung, W.H. A novel virus alters gene expression and vacuolar morphology in Malassezia cells and induces a TLR3-mediated inflammatory immune response. mBio 2020, 11, e01521-20. [Google Scholar] [CrossRef]
- Pettoello-Mantovani, M.; Nocerino, A.; Polonelli, L.; Morace, G.; Conti, S.; di Martino, L.; de Ritis, G.; Iafusco, M.; Guandalini, S. Hansenula anomala killer toxin induces secretion and severe acute injury in the rat intestine. Gastroenterology 1995, 109, 1900–1906. [Google Scholar] [CrossRef]
- Cenci, E.; Mencacci, A.; Spreca, A.; Montagnoli, C.; Bacci, A.; Perruccio, K.; Velardi, A.; Magliani, W.; Conti, S.; Polonelli, L.; et al. Protection of killer anti-idiotypic antibodies against early invasive aspergillosis in a murine model of allogeneic T-cell-depleted bone marrow transplantation. Infect. Immun. 2002, 70, 2375–2382. [Google Scholar] [CrossRef] [PubMed]
- Córdoba, L.; Ruiz-Padilla, A.; Rodríguez-Romero, J.; Ayllón, M.A. Construction and characterization of a Botrytis virus F infectious clone. J. Fungi 2022, 8, 459. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, S.; Ruan, S.; Nzabanita, C.; Wang, Y.; Guo, L. A mycovirus VIGS vector confers hypovirulence to a plant pathogenic fungus to control wheat FHB. Adv. Sci. 2023, 10, e2302606. [Google Scholar] [CrossRef] [PubMed]
- Marzano, S.Y.; Hobbs, H.A.; Nelson, B.D.; Hartman, G.L.; Eastburn, D.M.; McCoppin, N.K.; Domier, L.L. Transfection of Sclerotinia sclerotiorum with in vitro transcripts of a naturally occurring interspecific recombinant of Sclerotinia sclerotiorum hypovirus 2 significantly reduces virulence of the fungus. J. Virol. 2015, 89, 5060–5071. [Google Scholar] [CrossRef]
- Choi, G.H.; Nuss, D.L. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 1992, 257, 800–803. [Google Scholar] [CrossRef] [PubMed]
- Moleleki, N.; van Heerden, S.W.; Wingfield, M.J.; Wingfield, B.D.; Preisig, O. Transfection of Diaporthe perjuncta with Diaporthe RNA virus. Appl. Environ. Microbiol. 2003, 69, 3952–3956. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, S.; Zhang, L.; Qiu, D.; Zhou, X.; Guo, L. A tripartite ssDNA mycovirus from a plant pathogenic fungus is infectious as cloned DNA and purified virions. Sci. Adv. 2020, 6, eaay9634. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Battersby, J.L.; Stevens, D.A.; Coutts, R.H.A.; Havlíček, V.; Hsu, J.L.; Sass, G.; Kotta-Loizou, I. The Expanding Mycovirome of Aspergilli. J. Fungi 2024, 10, 585. https://doi.org/10.3390/jof10080585
Battersby JL, Stevens DA, Coutts RHA, Havlíček V, Hsu JL, Sass G, Kotta-Loizou I. The Expanding Mycovirome of Aspergilli. Journal of Fungi. 2024; 10(8):585. https://doi.org/10.3390/jof10080585
Chicago/Turabian StyleBattersby, Josephine L., David A. Stevens, Robert H. A. Coutts, Vladimír Havlíček, Joe L. Hsu, Gabriele Sass, and Ioly Kotta-Loizou. 2024. "The Expanding Mycovirome of Aspergilli" Journal of Fungi 10, no. 8: 585. https://doi.org/10.3390/jof10080585
APA StyleBattersby, J. L., Stevens, D. A., Coutts, R. H. A., Havlíček, V., Hsu, J. L., Sass, G., & Kotta-Loizou, I. (2024). The Expanding Mycovirome of Aspergilli. Journal of Fungi, 10(8), 585. https://doi.org/10.3390/jof10080585







