Five New Species of Wood-Decaying Brown-Rot Fungi within Postiaceae (Polyporales, Basidiomycota) from Xinjiang, Northwest China
Abstract
1. Introduction
2. Materials and Methods
2.1. Morphological Studies
2.2. DNA Extraction, PCR, and Sequencing
2.3. Phylogenetic Analyses
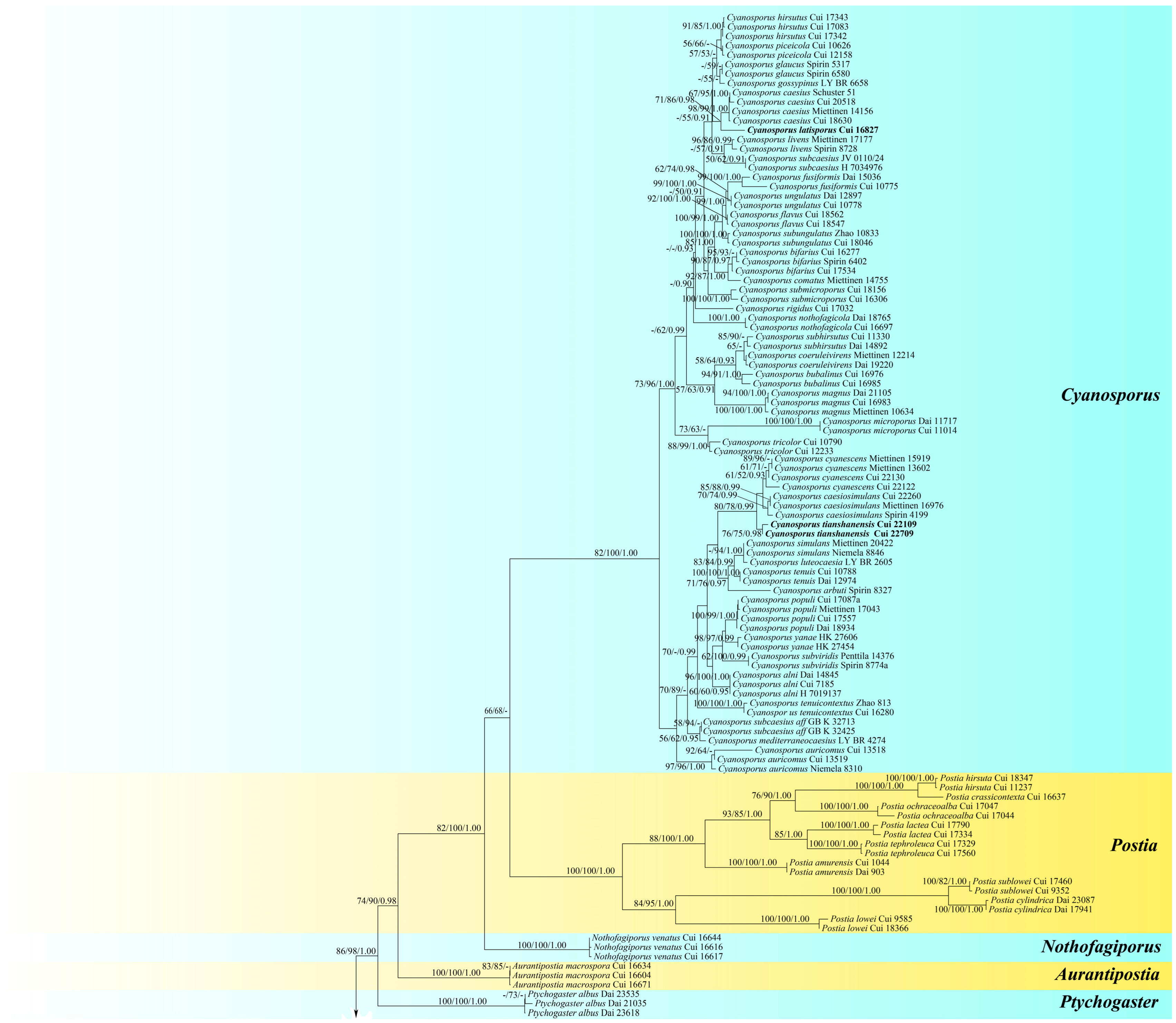
3. Results
3.1. Molecular Phylogeny
3.2. Taxonomy
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Atiwesh, G.; Parrish, C.C.; Banoub, J.; Le, T.A.T. Lignin degradation by microorganisms: A review. Biotechnol. Prog. 2022, 38, e3226. [Google Scholar] [CrossRef] [PubMed]
- Krah, F.S.; Bässler, C.; Heibl, C.; Heibl, C.; Soghigian, J.; Schaefer, H.; Hibbett, D.S. Evolutionary dynamics of host specialization in wood-decay fungi. BMC Evol. Biol. 2018, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hatakka, A.; Hammel, K.E. Fungal biodegradation of lignocelluloses. In The Mycota: Industrial Applications, 2nd ed.; Hofrichter, M., Ed.; Springer: Berlin, Germany, 2010; Volume 10, pp. 319–340. [Google Scholar]
- Chen, Y.Y.; Cui, B.K. Phylogenetic analysis and taxonomy of the Antrodia heteromorpha complex in China. Mycoscience 2016, 57, 1–10. [Google Scholar] [CrossRef]
- Cui, B.K.; Vlasák, J.; Dai, Y.C. The phylogenetic position of Osteina obducta (Polyporales, Basidiomycota) based on samples from northern hemisphere. Chiang Mai J. Sci. 2014, 41, 838–845. [Google Scholar]
- Han, M.L.; Chen, Y.Y.; Shen, L.L.; Song, J.; Vlasák, J.; Dai, Y.C.; Cui, B.K. Taxonomy and phylogeny of the brown-rot fungi: Fomitopsis and its related genera. Fungal Divers. 2016, 80, 343–373. [Google Scholar] [CrossRef]
- Liu, S.; Chen, Y.Y.; Sun, Y.F.; He, X.L.; Song, C.G.; Si, J.; Liu, D.M.; Gates, G.; Cui, B.K. Systematic classification and phylogenetic relationships of the brown-rot fungi within the Polyporales. Fungal Divers. 2023, 118, 1–94. [Google Scholar] [CrossRef]
- Liu, S.; Han, M.L.; Xu, T.M.; Wang, Y.; Wu, D.M.; Cui, B.K. Taxonomy and phylogeny of the Fomitopsis pinicola complex with descriptions of six new species from East Asia. Front. Microbiol. 2021, 12, 644979. [Google Scholar] [CrossRef]
- Liu, S.; Shen, L.L.; Wang, Y.; Xu, T.M.; Gates, G.; Cui, B.K. Species diversity and molecular phylogeny of Cyanosporus (Polyporales, Basidiomycota). Front. Microbiol. 2021, 12, e631166. [Google Scholar] [CrossRef]
- Liu, S.; Shen, L.L.; Xu, T.M.; Song, C.G.; Gao, N.; Wu, D.M.; Sun, Y.F.; Cui, B.K. Global diversity, molecular phylogeny and di-vergence times of the brown-rot fungi within the Polyporales. Mycosphere 2023, 14, 1564–1664. [Google Scholar] [CrossRef]
- Liu, S.; Xu, T.M.; Song, C.G.; Zhao, C.L.; Wu, D.M.; Cui, B.K. Species diversity, molecular phylogeny and ecological habits of Cyanosporus (Polyporales, Basidiomycota) with an emphasis on Chinese collections. MycoKeys 2022, 86, 19–46. [Google Scholar] [CrossRef]
- Shen, L.L.; Liu, H.X.; Cui, B.K. Morphological characters and molecular data reveal two new species of Postia (Basidiomycota) from China. Mycol. Prog. 2015, 14, 7. [Google Scholar] [CrossRef]
- Shen, L.L.; Wang, M.; Zhou, J.L.; Xing, J.H.; Cui, B.K.; Dai, Y.C. Taxonomy and phylogeny of Postia. Multi-gene phylogeny and taxonomy of the brown-rot fungi: Postia and its related genera. Persoonia 2019, 42, 101–126. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Chen, Y.Y.; Cui, B.K.; Liu, H.G.; Wang, Y.Z. Morphological and molecular evidence for two new species of Laetiporus (Basidiomycota, Polyporales) from southwestern China. Mycologia 2014, 106, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, C.G.; Wu, Y.D.; Liu, S.; Yuan, Y. Two new brown-rot polypores from tropical China. MycoKeys 2021, 82, 173–197. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Shen, L.L. Morphological characters and molecular data reveal a new species of Rhodonia (Polyporales, Basidiomycota) from China. Phytotaxa 2017, 328, 175–182. [Google Scholar] [CrossRef]
- Hu, W.; Yao, J.; He, Q.; Chen, J. Changes in precipitation amounts and extremes across Xinjiang (northwest China) and their connection to climate indices. PeerJ 2021, 9, e10792. [Google Scholar] [CrossRef] [PubMed]
- Li, L.P.; Zhang, C.Y.; Lughadha, E.N.; Leao, T.C.C.; Hardwick, K.T.; Zheng, Y.M.; Wan, H.W.; Ma, M.; Abudusalih, N.; Hai, Y.; et al. Geographic range size patterns across plants and animals of Xinjiang, China. Sci. Cold Arid Reg. 2022, 14, 54–67. [Google Scholar] [CrossRef]
- Yao, J.Q.; Li, M.Y.; Dilinuer, T.; Chen, J.; Mao, W.Y. The assessment on “warming-wetting” trend in Xinjiang at multi-scale during 1961-2019. Arid. Zone Res. 2022, 39, 333–346. [Google Scholar]
- Yang, C.Y. Sylva Xinjiangensis; China Forestry Publishing House: Beijing, China, 2010; pp. 9–16. [Google Scholar]
- Wang, J.Y.; Ayinuer. Macrofungi of Hanasi Lake Rigion from MT. Altal. J. Xinjiang Univ. (Nat. Sci. Ed.) 2004, 21, 88–91. [Google Scholar]
- Dai, Y.C.; Wei, Y.L.; Yuan, H.S.; Huang, M.Y.; Penzina, T. Polypores from Altay and Tian Mts. in Xinjiang, northwest China. Cryptogam. Mycol. 2007, 28, 269–279. [Google Scholar]
- Bau, T.; Hu, J.W.; Zhou, Z.B.; Xu, B. The new distribution of macrofungi in Xinjiang. J. Tarim Univ. 2008, 20, 4. [Google Scholar]
- Guli, A.; Feng, L.; Qin, X.Z.; Chen, J.; Marhaba; Yang, X.P.; Liu, A.M.; Wang, Z.H. Investigation on wild fungi resources in Habahe Plain of Altai Prefecture (I). Xinjiang Agric. Sci. 2015, 52, 1707–1714. [Google Scholar]
- Zhao, Z.X. Diversity of Macrofungi in Tomur National Nature Reserve and Molecular Systematics of Important Taxa. Master’s Thesis, Tarim University, Alar, China, 2022. [Google Scholar]
- McGinty, N.J. A new genus, Cyanosporus. Mycol. Notes 1909, 33, 436. [Google Scholar]
- Donk, M.A. Osteina, a new genus of Polyporaceae. Schweiz. Z. Pilzkd. 1966, 44, 83–87. [Google Scholar]
- Cui, B.K.; Li, H.J.; Ji, X.; Zhou, J.L.; Song, J.; Si, J.; Yang, Z.L.; Dai, Y.C. Species diversity, taxonomy and phylogeny of Polyporaceae (Basidiomycota) in China. Fungal Divers. 2019, 97, 137–392. [Google Scholar] [CrossRef]
- Sun, Y.F.; Xing, J.H.; He, X.L.; Wu, D.M.; Song, C.G.; Liu, S.; Vlasák, J.; Gates, G.; Gibertoni, T.B.; Cui, B.K. Species diversity, systematic revision and molecular phylogeny of Ganodermataceae (Polyporales, Basidiomycota) with an emphasis on Chinese collections. Stud. Mycol. 2022, 101, 287–415. [Google Scholar] [CrossRef]
- Petersen, J.H. Farvekort. In The Danish Mycological Society’s Color-Chart; Foreningen til Svampekundskabens Fremme: Hornbæk, Denmark, 1996. [Google Scholar]
- Sun, Y.F.; Costa-Rezende, D.H.; Xing, J.H.; Zhou, J.L.; Zhang, B.; Gibertoni, T.B.; Gates, G.; Glen, M.; Dai, Y.C.; Cui, B.K. Multi-gene phylogeny and taxonomy of Amauroderma s. lat. (Ganodermataceae). Persoonia 2020, 44, 206–239. [Google Scholar] [CrossRef]
- Ji, X.; Zhou, J.L.; Song, C.G.; Xu, T.M.; Wu, D.M.; Cui, B.K. Taxonomy, phylogeny and divergence times of Polyporus (Basidiomycota) and related genera. Mycosphere 2022, 13, 1–52. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Matheny, P.B.; Liu, Y.J.; Ammirati, J.F.; Hall, B.D. Using RPB1 sequences to improve phylogenetic inference among mush-rooms (Inocybe, Agaricales). Am. J. Bot. 2002, 89, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Matheny, P.B. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe, Agaricales). Mol. Phylogenet. Evol. 2005, 35, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Santana, B.; Lindner, D.L.; Miettinen, O.; Justo, A.; Hibbett, D.S. A phylogenetic overview of the antrodia clade (Basidiomycota, Polyporales). Mycologia 2013, 105, 1391–1411. [Google Scholar] [CrossRef] [PubMed]
- Pildain, M.B.; Rajchenberg, M. The phylogenetic position of Postia s.l. (Polyporales, Basidiomycota) from Patagonia, Argentina. Mycologia 2013, 105, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, O.; Vlasák, J.; Rivoire, B.; Spirin, V. Postia caesia complex (Polyporales, Basidiomycota) in temperate Northern Hemisphere. Fungal Syst. Evol. 2018, 1, 101–129. [Google Scholar] [CrossRef][Green Version]
- Vampola, P.; Ordynets, A.; Vlasák, J. The identity of Postia lowei (Basidiomycota, Polyporales) and notes on related or similar species. Czech Mycol. 2014, 66, 39–52. [Google Scholar] [CrossRef]
- Spirin, V.; Vlasák, J.; MiLakovsky, B.; Miettinen, O. Searching for indicator species of old-growth spruce forests: Studies in the genus Jahnoporus (Polyporales, Basidiomycota). Cryptogamie Mycol. 2015, 36, 409–417. [Google Scholar] [CrossRef]
- Lutzoni, F.; Kauff, F.; Cox, C.J.; McLaughlin, D.; Celio, G.; Dentinger, B.; Padamsee, M.; Hibbett, D.; James, T.Y.; Baloch, E.; et al. Assembling the fungal tree of life: Progress, classifcation, and evolution of subcellular traits. Am. J. Bot. 2004, 91, 1446–1480. [Google Scholar] [CrossRef]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.2. 2017. Available online: http://mesquiteproject.org (accessed on 22 July 2024).
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b10; Sinauer Associates: Sunderland, MA, USA, 2002; Volume 56, pp. 1776–1788. Available online: http://paup.csit.fsu.edu/ (accessed on 22 July 2024).
- Felsenstein, J. Confidence intervals on phylogenetics: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Page, R.D.M. TreeView: Application to display phylogenetic trees on personal computers. Bioinformatics 1996, 12, 357–358. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hőhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Gilbertson, R.L.; Ryvarden, L. North American Polypores. Syn. Fung. 1987, 2, 1–885. [Google Scholar]
- Spirin, W.A.; Zmitrovich, I.V.; Wasser, S.P. Oligoporus balsameus: Rare Eurasian species plus notes on some related taxa. Mycotaxon 2006, 97, 73–82. [Google Scholar]
- Wei, Y.L.; Dai, Y.C. Three new species of Postia (Aphyllophorales, Basidiomycota) from China. Fungal Divers. 2006, 23, 391–402. [Google Scholar]











| Species | Sample No. | Locality | GenBank Accessions | References | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS | nLSU | mtSSU | nuSSU | RPB1 | RPB2 | TEF1 | ||||
| Amaropostia altaiensis | Cui 18983 | Xinjiang, China | PP917921 | PP917955 | PP917972 | PP917938 | – | – | PP944601 | Present study |
| A. altaiensis | Cui 19000 (holotype) | Xinjiang, China | PP917922 | PP917956 | PP917973 | PP917939 | – | PP918933 | PP944589 | Present study |
| A. hainanensis | Cui 13739 (holotype) | Hainan, China | KX900909 | KX900979 | KX901051 | KX901123 | KX901171 | KX901223 | – | [13] |
| A. hainanensis | Cui 5367 | Hainan, China | KX900910 | KX900980 | KX901052 | KX901124 | KX901172 | KX901224 | – | [13] |
| A. hainanensis | Dai 15208 | Hainan, China | KX900911 | KX900981 | KX901053 | KX901125 | – | KX901225 | – | [13] |
| A. stiptica | Cui 10043 | Jilin, China | KX900906 | KX900976 | KX901046 | KX901119 | KX901167 | KX901219 | – | [13] |
| A. stiptica | Cui 10981 | Shandong, China | KX900907 | KX900977 | KX901047 | KX901120 | KX901168 | KX901220 | – | [13] |
| A. stiptica | Cui 18013 | Yunnan, China | OM039270 | OM039170 | OM039205 | OM039236 | OM037742 | OM037768 | OM037792 | [7] |
| A. stiptica | Cui 17037 | Yunnan, China | OK045504 | OK045510 | OK045498 | OK045492 | OK076902 | OK076930 | OK076958 | [7] |
| A. stiptica | Cui 9268 | Xizang, China | KF727431 | KX900978 | KX901048 | – | – | – | – | [12] |
| A. tianshanensis | Cui 22197 | Xinjiang, China | PP917919 | PP917953 | PP917970 | PP917936 | – | – | PP944599 | Present study |
| A. tianshanensis | Cui 22201 (holotype) | Xinjiang, China | PP917920 | PP917954 | PP917971 | PP917937 | – | – | PP944600 | Present study |
| Amylocystis lapponica | FP-105131 | Colorado, United States | KY948805 | KY948879 | – | – | KY948973 | – | – | [38] |
| A. lapponica | HHB 13400 | Alaska, United States | KC585237 | KC585059 | AF518667 | AF518570 | – | – | – | [38] |
| A. lapponica | OKM 4118 | Montana, United States | KC585238 | KC585060 | – | – | – | – | – | [38] |
| Antrodia serpens | Dai 7465 | Luxembourg | KR605813 | KR605752 | KR606013 | KR605913 | ON424666 | KR610832 | KR610742 | [7] |
| Aurantipostia macrospora | Cui 16604 (holotype) | Tasmania, Australia | MW377258 | MW377339 | – | MW377417 | MW337157 | MW337026 | MW337089 | [7] |
| A. macrospora | Cui 16634 | Tasmania, Australia | MW377259 | MW377340 | – | MW377418 | MW337158 | MW337027 | MW337090 | [7] |
| A macrospora | Cui 16671 | Tasmania, Australia | MW377260 | MW377341 | – | MW377419 | MW337159 | MW337028 | MW337091 | [7] |
| Austropostia brunnea | Dai 18591A | Victoria, Australia | MW377272 | MW377352 | – | MW377430 | MW337169 | MW337038 | MW337101 | [7] |
| A. brunnea | NLB 1135 | Australia | MT536995 | MT524530 | – | – | – | – | – | Unpublished |
| A. hirsuta | Cui 16660 (holotype) | Tasmania, Australia | MW377267 | MW377347 | MW382055 | MW377425 | MW337164 | MW337033 | MW337096 | [7] |
| A. hirsuta | Cui 16661 | Tasmania, Australia | MW377268 | MW377348 | MW382056 | MW377426 | MW337165 | MW337034 | MW337097 | [7] |
| A. hirsuta | Cui 16662 | Tasmania, Australia | MW377269 | MW377349 | MW382057 | MW377427 | MW337166 | MW337035 | MW337098 | [7] |
| A. pelliculosa | MR 10592 | Chubut, Argentina | JX090102 | JX090124 | – | – | – | – | – | [39] |
| A. pelliculosa | MR 10671 | Neuquén, Argentina | JX090101 | JX090123 | – | – | – | – | – | [39] |
| A. plumbea | Cui 16550 (holotype) | Victoria, Australia | MW377270 | MW377350 | MW382058 | MW377428 | MW337167 | MW337036 | MW337099 | [7] |
| A. plumbea | Cui 16639 | Tasmania, Australia | MW377271 | MW377351 | MW382059 | MW377429 | MW337168 | MW337037 | MW337100 | [7] |
| A. punctata | MR 11100 | Neuquén, Argentina | JX090112 | JX090128 | – | – | – | – | – | [7] |
| A. punctata | MR 12398 | Región X, Chile | JX090111 | JX090127 | – | – | – | – | – | [7] |
| A. subpunctata | Cui 16675 (holotype) | Tasmania, Australia | MW377273 | MW377353 | MW382060 | MW377431 | MW337170 | MW337039 | MW337102 | [7] |
| A. subpunctata | Cui 16685 | Tasmania, Australia | MW377274 | MW377354 | MW382061 | MW377432 | MW337171 | MW337040 | MW337103 | [7] |
| A. subpunctata | Cui 16686 | Tasmania, Australia | MW377275 | MW377355 | MW382062 | MW377433 | MW337172 | MW337041 | MW337104 | [7] |
| Calcipostia guttulata | Cui 10018 | Jilin, China | KF727432 | KJ684978 | KX901065 | KX901138 | KX901181 | KX901236 | KX901276 | [13] |
| C. guttulata | Cui 10028 | Jilin, China | KF727433 | KJ684979 | KX901066 | KX901139 | KX901182 | KX901237 | KX901277 | [13] |
| C. guttulata | Cui 16274 | Yunnan, China | OM039274 | OM039174 | OM039209 | OM039240 | OM037746 | OM037772 | OM037796 | [7] |
| C. guttulata | Cui 16281 | Yunnan, China | OM039275 | OM039175 | OM039210 | OM039241 | OM037747 | OM037773 | OM037797 | [7] |
| Cyanosporus alni | Cui 7185 | Hebei, China | KX900879 | KX900949 | KX901017 | KX901092 | KX901155 | KX901202 | KX901254 | [13] |
| C. alni | Dai 14845 | Poland | KX900880 | KX900950 | KX901018 | KX901093 | KX901156 | KX901203 | KX901255 | [13] |
| C. alni | H 7019137 | Bratislava, Slovakia | MG137026 | – | – | – | – | – | – | [40] |
| C. arbuti | Spirin 8327 (holotype) | Washington, United States | MG137039 | – | – | – | – | – | MG137132 | [40] |
| C. auricomus | Cui 13518 | Inner Mongolia, China | KX900887 | KX900957 | KX901025 | KX901100 | – | KX901209 | – | [13] |
| C. auricomus | Cui 13519 | Inner Mongolia, China | KX900888 | KX900958 | KX901026 | KX901101 | – | – | – | [13] |
| C. auricomus | Niemela 8310 (holotype) | Pohjois-Savo, Finland | MG137040 | – | – | – | – | – | – | [40] |
| C. bifarius | Cui 16277 | Yunnan, China | OL423599 | OL423609 | OL437196 | OL423621 | OL444986 | OL447000 | OL444995 | [11] |
| C. bifarius | Cui 17534 | Sichuan, China | OL423598 | OL423608 | OL437195 | OL423620 | OL444985 | OL446999 | OL444994 | [11] |
| C. bifarius | Spirin 6402 (holotype) | Primorie, Russia | MG137043 | – | – | – | – | – | MG137133 | [40] |
| C. bubalinus | Cui 16976 | Yunnan, China | MW182172 | MW182225 | MW182208 | MW182189 | MW191547 | MW191563 | MW191530 | [9] |
| C. bubalinus | Cui 16985 (holotype) | Yunnan, China | MW182173 | MW182226 | MW182209 | MW182190 | MW191548 | MW191564 | MW191531 | [9] |
| C. caesiosimulans | Miettinen 16976 | New York, United States | MG137054 | – | – | – | – | – | MG137137 | [40] |
| C. caesiosimulans | Spirin 4199 | Khabarovsk, Russia | MG137061 | – | – | – | – | – | MG137140 | [40] |
| C. caesiosimulans | Cui 22260 | Xinjiang, China | PP917932 | PP917966 | PP917983 | PP917949 | – | – | – | Present study |
| C. caesius | Cui 18630 | Nancy, France | OL423600 | OL423610 | OL437197 | OL423622 | – | – | OL444996 | [11] |
| C. caesius | Schuster 51 | Niedersachsen, Germany | MG137045 | – | – | – | – | – | – | [40] |
| C. caesius | Miettinen 14156 | Uusimaa, Finland | MG137048 | – | – | – | – | – | MG137134 | [40] |
| C. caesius | Cui 20518 | Xinjiang, China | PP917933 | PP917967 | PP917984 | PP917950 | – | – | – | Present study |
| C. caesius aff GB | K 32425 | United Kingdom | AY599575 | – | – | – | – | – | – | [40] |
| C. caesius aff GB | K 32713 | United Kingdom | AY599576 | – | – | – | – | – | – | [40] |
| C. coeruleivirens | Dai 19220 | Hunan, China | MW182174 | MW182227 | MW182210 | MW182191 | MW191549 | MW191532 | [9] | |
| C. coeruleivirens | Miettinen 12214 | Bali, Indonesia | MG137063 | – | – | – | – | – | – | [40] |
| C. comatus | Miettinen 14755 (holotype) | Massachusetts, United States | MG137066 | – | – | – | – | – | – | [40] |
| C. cyanescens | Miettinen 13602 (holotype) | Uusimaa, Finland | MG137067 | – | – | – | – | – | MG137142 | [40] |
| C. cyanescens | Miettinen 15919 | Huesca, Spain | MG137071 | – | – | – | – | – | MG137144 | [40] |
| C. cyanescens | Cui 22122 | Xinjiang, China | PP917934 | PP917968 | PP917985 | PP917951 | – | – | PP944597 | Present study |
| C. cyanescens | Cui 22130 | Xinjiang, China | PP917935 | PP917969 | PP917986 | PP917952 | – | – | PP944598 | Present study |
| C. flavus | Cui 18547 (holotype) | Sichuan, China | MW448564 | MW448561 | – | MW448557 | MW452596 | MW452599 | MW452601 | [11] |
| C. flavus | Cui 18562 | Sichuan, China | MW448565 | MW448562 | – | MW448558 | MW452597 | MW452600 | MW452602 | [11] |
| C. fusiformis | Cui 10775 | Sichuan, China | KX900868 | KX900938 | KX901006 | KX901081 | – | KX901191 | KX901245 | [13] |
| C. fusiformis | Dai 15036 (holotype) | Guizhou, China | KX900867 | KX900937 | KX901005 | KX901080 | – | KX901190 | KX901244 | [13] |
| C. glaucus | Spirin 5317 (holotype) | Khabarovsk, Russia | MG137078 | – | – | – | – | – | – | [40] |
| C. glaucus | Spirin 6580 | Khabarovsk, Russia | MG137081 | – | – | – | – | – | MG137145 | [40] |
| C. gossypinus | LY BR 6658 | Vaucluse, France | – | – | – | – | – | – | MG137146 | [40] |
| C. hirsutus | Cui 17083 (holotype) | Yunnan, China | MW182179 | MW182233 | MW182214 | MW182197 | MW191554 | MW191568 | MW191538 | [9] |
| C. hirsutus | Cui 17342 | Sichuan, China | OL423602 | OL423612 | OL437199 | OL423624 | OL444988 | OL447002 | OL444998 | [11] |
| C. hirsutus | Cui 17343 | Sichuan, China | OL423601 | OL423611 | OL437198 | OL423623 | OL444987 | OL447001 | OL444997 | [11] |
| C. latisporus | Cui 16827 (holotype) | Xinjiang, China | PP917925 | PP917959 | PP917976 | PP917942 | PP918931 | PP918932 | PP944588 | Present study |
| C. livens | Miettinen 17177 (holotype) | New York, United States | MG137082 | – | – | – | – | – | MG137147 | [40] |
| C. livens | Spirin 8728 | Washington, United States | MG137090 | – | – | – | – | – | MG137150 | [40] |
| C. luteocaesia | LY BR 2605 | France | MG137091 | – | – | – | – | – | – | [40] |
| C. magnus | Cui 16983 | Yunnan, China | MW182180 | MW182234 | MW182215 | MW182198 | MW191555 | MW191569 | MW191539 | [9] |
| C. magnus | Dai 21105 | Chongqin, China | OL423603 | OL423613 | OL437200 | OL423625 | OL444989 | OL447003 | OL444999 | [11] |
| C. magnus | Miettinen 10634 (holotype) | Jilin, China | KC595944 | KC595944 | – | – | – | – | MG137151 | [40] |
| C. mediterraneocaesius | LY BR 4274 | Bonnieux, France | KX900886 | – | KX901024 | KX901099 | – | – | – | [13] |
| C. microporus | Cui 11014 (holotype) | Yunnan, China | KX900878 | KX900948 | KX901016 | KX901091 | – | KX901201 | – | [13] |
| C. microporus | Dai 11717 | Yunnan, China | KX900877 | KX900947 | KX901015 | KX901090 | – | KX901200 | – | [13] |
| C. nothofagicola | Cui 16697 (holotype) | Tasmania, Australia | MW182181 | MW182235 | MW182216 | MW182199 | MW191556 | MW191570 | MW191540 | [9] |
| C. nothofagicola | Dai 18765 | Tasmania, Australia | MW182182 | MW182236 | MW182217 | MW182200 | MW191557 | MW191541 | [9] | |
| C. piceicola | Cui 10626 (holotype) | Sichuan, China | KX900862 | KX900932 | KX901001 | KX901075 | KX901185 | [13] | ||
| C. piceicola | Cui 12158 | Xizang, China | KX900866 | KX900936 | KX901004 | KX901079 | KX901153 | KX901189 | KX901243 | [13] |
| C. populi | Cui 17087a | Yunnan, China | MW182183 | MW182237 | MW182218 | MW182201 | MW191558 | MW191571 | MW191542 | [9] |
| C. populi | Cui 17557 | Sichuan, China | OL423605 | OL423615 | OL437202 | OL423627 | OL444991 | OL447005 | OL445001 | [11] |
| C. populi | Dai 18934 | Qinghai, China | OL423604 | OL423614 | OL437201 | OL423626 | OL444990 | OL447004 | OL445000 | [11] |
| C. populi | Miettinen 17043 (holotype) | NewYork, United States | MG137092 | – | – | – | – | – | MG137153 | [40] |
| C. rigidus | Cui 17032 (holotype) | Yunnan, China | OL423606 | OL423617 | OL437204 | OL423629 | OL444993 | – | OL445003 | [11] |
| C. simulans | Miettinen 20422 | Satakunta, Finland | MG137110 | – | – | – | – | – | MG137160 | [40] |
| C. simulans | Niemela 8846 | Satakunta, Finland | MG137103 | – | – | – | – | – | – | [40] |
| C. subcaesius | H 7034976 | Isère, France | MG137116 | – | – | – | – | – | – | [40] |
| C. subcaesius | JV 0110/24 | Jihomoravský, Czech Republic | MG137117 | – | – | – | – | MG137164 | [40] | |
| C. subhirsutus | Cui 11330 | Fujian, China | KX900873 | KX900943 | KX901011 | KX901086 | – | KX901196 | KX901250 | [13] |
| C. subhirsutus | Dai 14892 (holotype) | Guizhou, China | KX900871 | KX900941 | KX901009 | KX901084 | – | KX901194 | KX901248 | [13] |
| C. tianshanensis | Cui 22109 | Xinjiang, China | PP917923 | PP917957 | PP917974 | PP917940 | – | – | PP944596 | Present study |
| C. tianshanensis | Cui 22709 (holotype) | Xinjiang, China | PP917924 | PP917958 | PP917975 | PP917941 | – | – | – | Present study |
| C. submicroporus | Cui 16306 | Yunnan, China | MW182184 | MW182239 | MW182220 | MW182203 | MW191560 | MW191573 | MW191544 | [9] |
| C. submicroporus | Cui 18156 (holotype) | Yunnan, China | MW182186 | MW182241 | MW182222 | MW182205 | – | MW191574 | – | [9] |
| C. subungulatus | Cui 18046 (holotype) | Yunnan, China | MW448566 | MW448563 | MW448560 | MW448559 | MW452598 | – | MW452603 | [11] |
| C. subungulatus | Zhao 10833 | Yunnan, China | MW742586 | OL423616 | OL437203 | OL423628 | OL444992 | – | OL445002 | [11] |
| C. subviridis | Penttila 14376 | Pohjois-Karjala, Finland | – | – | – | – | – | – | MG137165 | [40] |
| C. subviridis | Spirin 8774a | Washington, United States | MG137120 | – | – | – | – | – | MG137166 | [40] |
| C. tenuicontextus | Cui 16280 (holotype) | Yunnan, China | OL423607 | OL423618 | OL437205 | OL423630 | – | – | OL445004 | [11] |
| C. tenuicontextus | Zhao 813 | Yunnan, China | MG231802 | OL423619 | OL437206 | OL423631 | – | – | OL445005 | [11] |
| C. tenuis | Cui 10788 (holotype) | Sichuan, China | KX900885 | KX900955 | KX901023 | KX901098 | KX901161 | KX901208 | – | [13] |
| C. tenuis | Dai 12974 | Sichuan, China | KX900884 | KX900954 | KX901022 | KX901097 | KX901160 | KX901207 | KX901258 | [13] |
| C. tricolor | Cui 10790 | Sichuan, China | KX900875 | KX900945 | KX901013 | KX901088 | – | KX901198 | KX901252 | [13] |
| C. tricolor | Cui 12233 (holotype) | Xizang, China | KX900876 | KX900946 | KX901014 | KX901089 | – | KX901199 | KX901253 | [13] |
| C. ungulatus | Cui 10778 | Sichuan, China | KX900870 | KX900940 | KX901008 | KX901083 | – | KX901193 | KX901247 | [13] |
| C. ungulatus | Dai 12897 (holotype) | Sichuan, China | KX900869 | KX900939 | KX901007 | KX901082 | KX901154 | KX901192 | KX901246 | [13] |
| C. yanae | HK 27454 (holotype) | Sakha, Russia | MG137121 | – | – | – | – | – | MG137167 | [40] |
| C. yanae | HK 27606 | Sakha, Russia | MG137122 | – | – | – | – | – | MG137168 | [40] |
| Cystidiopostia hibernica | Cui 17624 | Sichuan, China | MW377277 | MW377357 | MW382064 | MW377435 | MW337173 | – | MW337105 | [7] |
| C. hibernica | Cui 2658 | Zhejiang, China | KX900905 | KX900975 | KX901045 | KX901118 | – | KX901218 | – | [13] |
| C. inocybe | LY BR 3703 | France | KX900903 | KX900973 | KX901044 | KX901116 | – | – | KX901267 | [13] |
| C. pileata | Cui 10034 | Jilin, China | KX900908 | KX900956 | KX901050 | KX901122 | KX901170 | KX901222 | KX901269 | [13] |
| C. pileata | Cui 5721 | Liaoning, China | KF699127 | KX900960 | KX901049 | KX901121 | KX901169 | KX901221 | KX901268 | [13] |
| C. subhibernica | Cui 17095 (holotype) | Yunnan, China | MW377278 | MW377358 | MW382065 | MW377436 | MW337174 | MW337042 | MW337106 | [7] |
| C. subhibernica | Dai 17621 | Sichuan, China | OM039276 | OM039176 | OM039211 | OM039242 | OM037749 | OM037774 | OM037798 | [7] |
| Fomitopsis betulina | Cui 17121 | Yunnan, China | OL621853 | OL621242 | OL621753 | OL621779 | ON424683 | OL588969 | OL588982 | [11] |
| Fuscopostia duplicata | Cui 10366 | Yunnan, China | KF699124 | KJ684975 | KR606026 | KR605927 | KX901173 | KR610844 | KR610755 | [6] |
| F. duplicata | Dai 13411 (holotype) | Zhejiang, China | KF699125 | KJ684976 | KR606027 | KR605928 | KX901174 | KR610845 | KR610756 | [6] |
| F. fragilis | Cui 10020 | Jilin, China | KX900912 | KX900982 | KX901054 | KX901126 | ON424693 | KX901226 | KX901270 | [13] |
| F. fragilis | Cui 10088 | Jilin, China | KF699120 | KJ684977 | KT893749 | KX901127 | ON424692 | KT893745 | KT893747 | [6] |
| F. lateritia | Dai 2652 | Helsinki, Finland | KX900913 | KX900983 | – | – | – | – | – | [13] |
| F. lateritia | KUO 0211531 | Khabarovsk, Russia | JF950567 | – | – | – | – | – | – | [41] |
| F. leucomallella | Cui 9577 | Xizang, China | KF699122 | KJ684982 | KX901055 | KX901128 | KX901175 | KX901227 | KX901271 | [13] |
| F. leucomallella | Cui 9599 | Xizang, China | KF699123 | KJ684983 | KX901056 | KX901129 | KX901176 | KX901228 | KX901272 | [13] |
| F. subfragilis | Cui 16282 | Yunnan, China | MW377296 | MW377375 | MW382082 | MW377454 | MW337189 | MW337057 | MW337123 | [7] |
| F. subfragilis | Cui 16302 (holotype) | Yunnan, China | MW377297 | MW377376 | MW382083 | MW377455 | MW337190 | MW337058 | MW337124 | [7] |
| Jahnoporus brachiatus | X 3232 (holotype) | Khabarovsk, Russia | KU165781 | – | – | – | – | – | – | [42] |
| J. hirtus | AFTOL ID 1687 | Washington, United States | DQ911605 | DQ911606 | – | DQ911607 | – | DQ911608 | – | [43] |
| J. hirtus | Spinosa 10X2014 | Washington, United States | KU165784 | – | – | – | KY949044 | – | – | [42] |
| J. oreinus | X 3241 (holotype) | Khabarovsk, Russia | KU165785 | – | – | – | – | – | – | [42] |
| Nothofagiporus venatus | Cui 16616 | Tasmania, Australia | MW377310 | MW377388 | MW382091 | MW377467 | MW337196 | MW337067 | MW337133 | [7] |
| N. venatus | Cui 16617 | Tasmania, Australia | MW377311 | MW377389 | MW382092 | MW377468 | MW337197 | MW337068 | MW337134 | [7] |
| N. venatus | Cui 16644 | Tasmania, Australia | ON417170 | ON417220 | ON417084 | ON417034 | – | ON424786 | ON424848 | [7] |
| Oligoporus podocarpi | Dai 22042 (holotype) | Hainan, China | MW937877 | MW937884 | MW937891 | MW937870 | MZ005579 | MZ082976 | MZ082982 | [15] |
| O. podocarpi | Dai 22043 | Hainan, China | MW937878 | MW937885 | MW937892 | MW937871 | MZ005580 | MZ082977 | MZ082983 | [7] |
| O. rennyi | Cui 17054 | Yunnan, China | OK045508 | OK045514 | OK045502 | OK045496 | OK076906 | OK076934 | OK076962 | [7] |
| O. rennyi | Dai 21016 | Belarus | ON417173 | ON417223 | ON417085 | ON417037 | ON424713 | ON424789 | ON424851 | [7] |
| O. romellii | Dai 21034 | Belarus | MW377312 | MW377390 | MW382093 | MW377469 | MW337198 | ON424790 | MW337135 | [7] |
| O. romellii | Dai 23576 | Xizang, China | ON417174 | ON417224 | ON417086 | ON417038 | ON424714 | ON424791 | ON424852 | [7] |
| O. sericeomollis | Cui 9560 | Xizang, China | KX900919 | KX900989 | KX901067 | KX901140 | KX901183 | ON424792 | ON424853 | [13] |
| O. sericeomollis | Dai 23473 | Xizang, China | ON417175 | ON417225 | ON417087 | ON417039 | ON424715 | ON424793 | ON424854 | [7] |
| Osteina obducta | Cui 10074 | Jilin, China | KX900924 | KX900994 | KX901071 | KX901144 | – | KX901240 | – | [13] |
| O. obducta | Cui 9832 | Heilongjiang, China | KX900925 | KX900995 | – | – | – | – | – | [13] |
| O. obducta | Cui 9959 | Jilin, China | KX900923 | KX900993 | KX901070 | KX901143 | – | KX901239 | – | [13] |
| O. undosa | Cui 16651 | Tasmania, Australia | MW377313 | MW377391 | MW382094 | MW377470 | MW337199 | MW337069 | MW337136 | [7] |
| O. undosa | Dai 6942 | Jilin, China | KX900922 | KX900992 | – | – | – | – | – | [13] |
| O. undosa | Dai 7105 | Jilin, China | KX900921 | KX900991 | KX901069 | KX901142 | – | KX901238 | – | [13] |
| O. undosa | L 10830 | North Carolina, United States | KC585396 | KC585229 | – | – | – | – | – | [38] |
| O. undosa | L 6646 | Colorado, United States | KC585399 | KC585232 | – | – | – | – | – | [38] |
| O. altaiensis | Cui 20555 | Xinjiang, China | PP917926 | PP917960 | PP917977 | PP917943 | – | – | PP944590 | Present study |
| O. altaiensis | Cui 20920 (holotype) | Xinjiang, China | PP917927 | PP917961 | PP917978 | PP917944 | – | – | PP944591 | Present study |
| O. altaiensis | Cui 20963 | Xinjiang, China | PP917928 | PP917962 | PP917979 | PP917945 | – | – | PP944592 | Present study |
| O. altaiensis | Cui 20964 | Xinjiang, China | PP917929 | PP917963 | PP917980 | PP917946 | – | – | PP944593 | Present study |
| O. altaiensis | Cui 20970 | Xinjiang, China | PP917930 | PP917964 | PP917981 | PP917947 | – | – | PP944594 | Present study |
| O. altaiensis | Cui 20972 | Xinjiang, China | PP917931 | PP917965 | PP917982 | PP917948 | – | – | PP944595 | Present study |
| Postia amurensis | Cui 1044 | Liaoning, China | KX900902 | KX900972 | KX901043 | – | – | – | – | [13] |
| P. amurensis | Dai 903 (holotype) | Jilin, China | KX900901 | KX900971 | KX901042 | – | – | – | – | [13] |
| P. crassicontexta | Cui 16637 (holotype) | Tasmania, Australia | MW377315 | MW377393 | MW382096 | MW377472 | MW337200 | MW337071 | MW337138 | [7] |
| P. cylindrica | Dai 17941 | Hubei, China | ON417183 | ON417233 | ON417091 | ON417047 | – | – | ON424862 | [7] |
| P. cylindrica | Dai 23087 | Yunnan, China | ON417182 | ON417232 | ON417090 | ON417046 | – | – | ON424861 | [7] |
| P. hirsuta | Cui 11237 (holotype) | Shanxi, China | KJ684970 | KJ684984 | KX901038 | KX901113 | – | – | KX901266 | [13] |
| P. hirsuta | Cui 18347 | Hunan, China | OM039286 | OM039186 | OM039221 | OM039253 | – | ON424800 | OM037809 | [7] |
| P. lactea | Cui 17334 | Sichuan, China | OM039287 | OM039187 | OM039222 | OM039254 | OM037753 | OM037782 | OM037810 | [7] |
| P. lactea | Cui 17790 | Sichuan, China | OM039288 | OM039188 | OM039223 | OM039255 | OM037754 | OM037783 | OM037811 | [7] |
| P. lowei | Cui 18366 | Sichuan, China | OM039289 | OM039189 | OM039224 | OM039256 | – | ON424801 | ON424863 | [7] |
| P. lowei | Cui 9585 | Xizang, China | KX900898 | KX900968 | KX901035 | KX901110 | – | – | – | [13] |
| P. ochraceoalba | Cui 17044 | Yunnan, China | OM039290 | OM039190 | OM039225 | OM039257 | OM037755 | OM037784 | OM037812 | [7] |
| P. ochraceoalba | Cui 17047 | Yunnan, China | OM039291 | OM039191 | OM039226 | OM039258 | OM037756 | OM037785 | OM037813 | [7] |
| P. sublowei | Cui 17460 | Sichuan, China | OM039294 | OM039194 | OM039229 | OM039261 | OM037759 | ON424802 | ON424864 | [7] |
| P. sublowei | Cui 9352 | Xizang, China | KX900899 | KX900969 | KX901036 | KX901111 | ON424723 | – | KX901264 | [7] |
| P. tephroleuca | Cui 17329 | Sichuan, China | OK045509 | OK045515 | OK045503 | OK045497 | OK076907 | OK076935 | OK076963 | [7] |
| P. tephroleuca | Cui 17560 | Sichuan, China | OM039295 | OM039195 | OM039230 | OM039262 | OM037760 | OM037788 | OM037816 | [7] |
| Ptychogaster albus | Dai 21035 | Belarus | OM039293 | OM039193 | OM039228 | OM039260 | OM037758 | OM037787 | OM037815 | [7] |
| P. albus | Dai 23535 | Xizang, China | ON417184 | ON417235 | ON417092 | ON417048 | ON424724 | ON424804 | ON424866 | [7] |
| P. albus | Dai 23618 | Xizang, China | OM039292 | OM039192 | OM039227 | OM039259 | OM037757 | OM037786 | OM037814 | [7] |
| Spongiporus balsameus | Cui 9835 | Heilongjiang, China | KX900916 | KX900986 | KX901061 | KX901134 | – | KX901233 | – | [13] |
| S. balsameus | Dai 22714 | Yunnan, China | ON417194 | ON417246 | ON417101 | ON417058 | – | ON424814 | ON424880 | [7] |
| S. floriformis | Cui 10292 | Yunnan, China | KM107899 | KM107904 | KX901058 | KX901131 | KX901178 | KX901230 | KX901274 | [13] |
| S. floriformis | Cui 17066 | Yunnan, China | OM039300 | OM039200 | OM039231 | OM039265 | OM037762 | ON424815 | OM037818 | [7] |
| S. floriformis | Dai 13887 | Yunnan, China | KX900914 | KX900984 | KX901057 | KX901130 | KX901177 | KX901229 | KX901273 | [13] |
| S. gloeoporus | Cui 10401 | Yunnan, China | KX900915 | KX900985 | KX901060 | KX901133 | ON424742 | KX901232 | ON424881 | [13] |
| S. gloeoporus | Cui 17813 | Singapore | OM039301 | OM039201 | OM039232 | OM039266 | OM037763 | ON424816 | OM037819 | [7] |
| S. zebra | Cui 9973 | Jilin, China | KX900917 | KX900987 | KX901062 | KX901135 | KX901179 | KX901234 | – | [13] |
| S. zebra | Dai 7131 (holotype) | Jilin, China | KF727430 | KM190902 | KX901063 | KX901136 | KX901180 | KX901235 | – | [13] |
| Tenuipostia dissecta | Cui 16555 | Victoria, Australia | MW377330 | MW377406 | MW382106 | MW377487 | MW337207 | ON424818 | MW337149 | [7] |
| T. dissecta | Cui 16560 | Victoria, Australia | MW377331 | MW377407 | MW382107 | MW377488 | MW337208 | ON424819 | MW337150 | [7] |
| T. dissecta | Cui 16653 | Tasmania, Australia | OM039302 | OM039202 | OM039233 | OM039267 | OM037764 | OM037789 | OM037820 | [7] |
| T. dissecta | Dai 18747 | Tasmania, Australia | OM039303 | OM039203 | OM039234 | OM039268 | OM037765 | OM037790 | OM037821 | [7] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, T.-M.; Wu, D.-M.; Gao, N.; Zeng, L.; Xu, Y.-H.; Fan, X.-P.; Sun, Y.-F.; Cui, B.-K. Five New Species of Wood-Decaying Brown-Rot Fungi within Postiaceae (Polyporales, Basidiomycota) from Xinjiang, Northwest China. J. Fungi 2024, 10, 655. https://doi.org/10.3390/jof10090655
Xu T-M, Wu D-M, Gao N, Zeng L, Xu Y-H, Fan X-P, Sun Y-F, Cui B-K. Five New Species of Wood-Decaying Brown-Rot Fungi within Postiaceae (Polyporales, Basidiomycota) from Xinjiang, Northwest China. Journal of Fungi. 2024; 10(9):655. https://doi.org/10.3390/jof10090655
Chicago/Turabian StyleXu, Tai-Min, Dong-Mei Wu, Neng Gao, Long Zeng, Yi-Hua Xu, Xiang-Ping Fan, Yi-Fei Sun, and Bao-Kai Cui. 2024. "Five New Species of Wood-Decaying Brown-Rot Fungi within Postiaceae (Polyporales, Basidiomycota) from Xinjiang, Northwest China" Journal of Fungi 10, no. 9: 655. https://doi.org/10.3390/jof10090655
APA StyleXu, T.-M., Wu, D.-M., Gao, N., Zeng, L., Xu, Y.-H., Fan, X.-P., Sun, Y.-F., & Cui, B.-K. (2024). Five New Species of Wood-Decaying Brown-Rot Fungi within Postiaceae (Polyporales, Basidiomycota) from Xinjiang, Northwest China. Journal of Fungi, 10(9), 655. https://doi.org/10.3390/jof10090655







