The TOR Signaling Pathway Governs Fungal Development, Virulence and Ustiloxin Biosynthesis in Ustilaginoidea virens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Fungal Strains
2.2. Bioinformatics Analysis
2.3. Microscopic Examinations of Hyphae and Conidiation
2.4. Infection Assay of U. virens
2.5. Quantification of Ustilaginoidins by HPLC
2.6. Transcriptome Sequencing and Analysis
2.7. qRT-PCR Assay
2.8. Statistical Analysis
3. Results
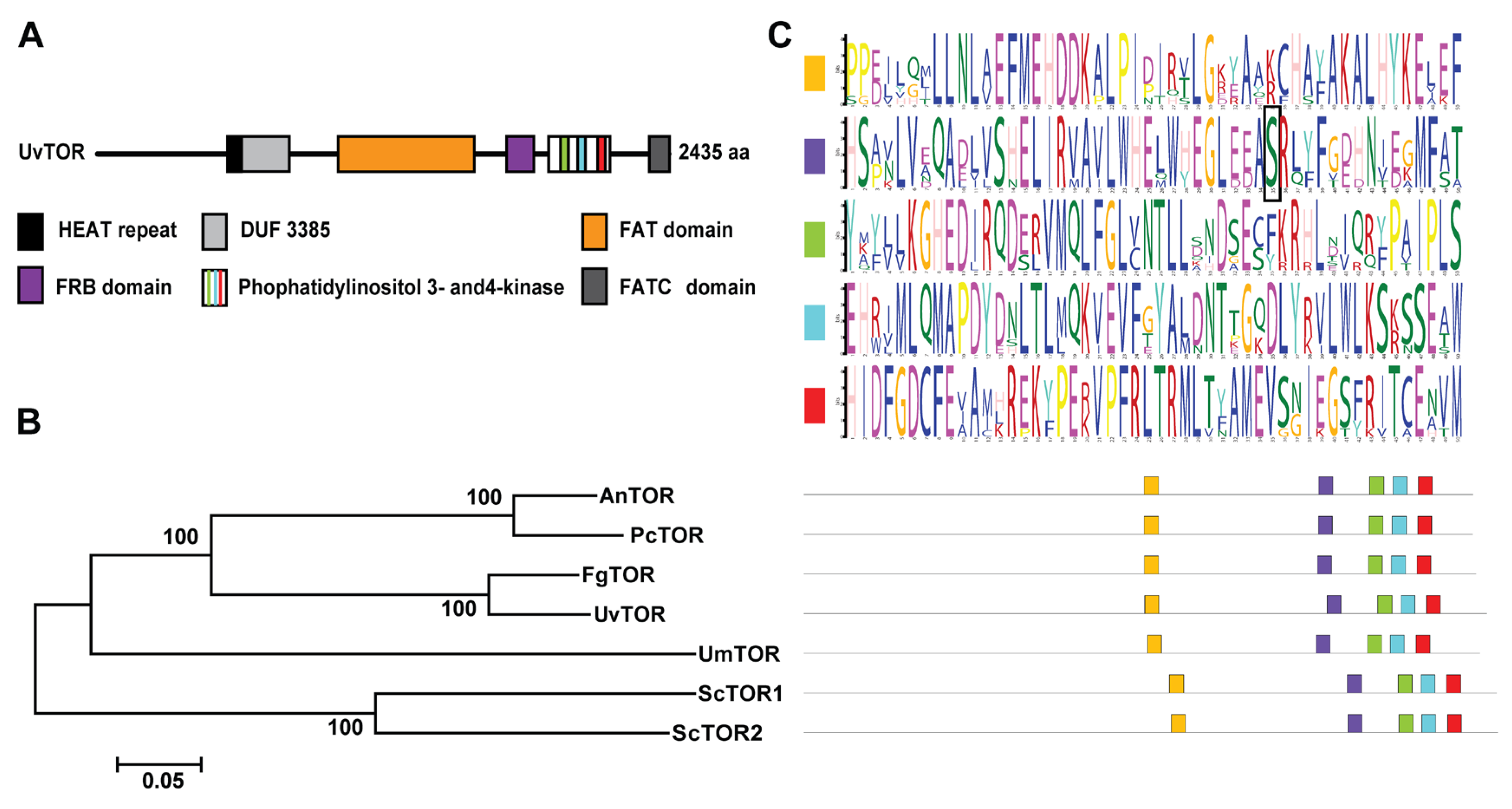
3.1. Molecular Components of the TOR Protein in U. virens
3.2. The TOR Signaling Pathway Is Involved in the Regulation of Hyphal Growth and Conidiation
3.3. The TOR Signaling Pathway Is Associated with Pathogenicity
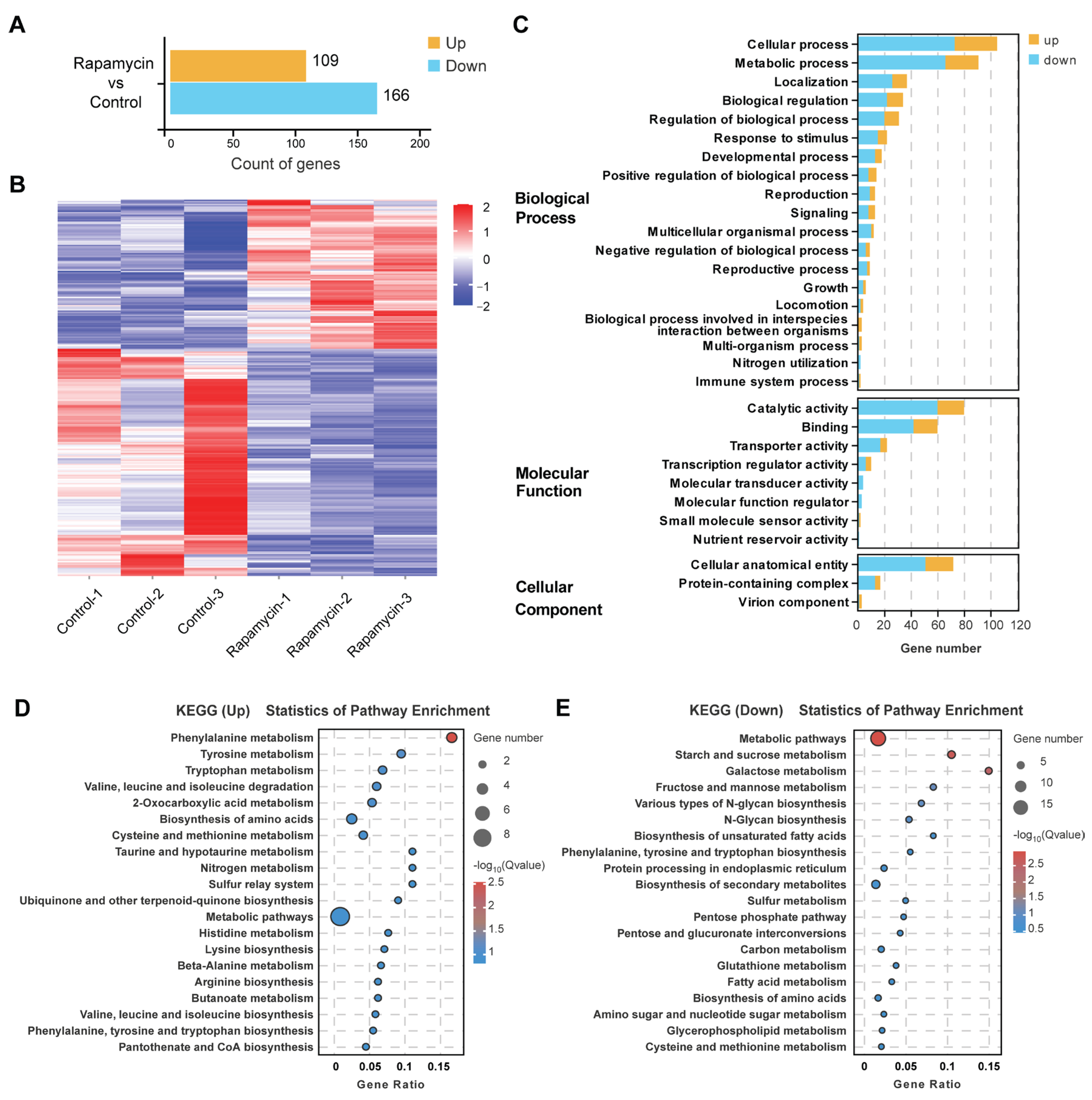
3.4. The TOR Signaling Pathway Regulates Related Gene Expression to Control Growth and Metabolism
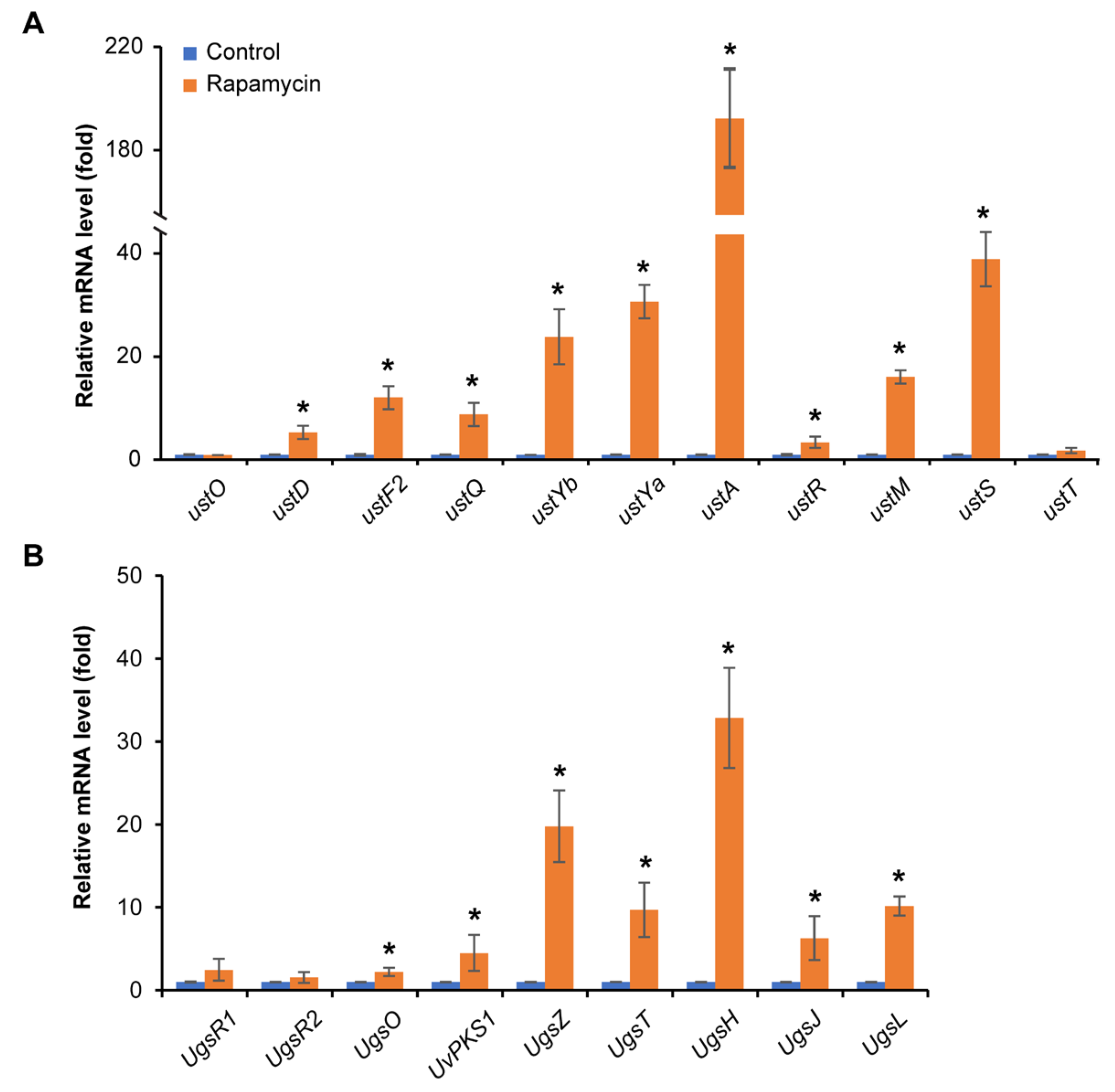
3.5. Rapamycin Exhibits a Strong Induction of Mycotoxin Biosynthesis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, W.; Fan, J.; Fang, A.; Li, Y.; Tariqjaveed, M.; Li, D.; Hu, D.; Wang, W.M. Ustilaginoidea virens: Insights into an emerging rice pathogen. Annu. Rev. Phytopathol. 2020, 58, 363–385. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Liu, J.; Gong, Z.Y.; Xu, P.Z.; Hu, X.H.; Wu, J.L.; Li, G.B.; Yang, J.; Wang, Y.Q.; Zhou, Y.F.; et al. The false smut pathogen Ustilaginoidea virens requires rice stamens for false smut ball formation. Environ. Microbiol. 2020, 22, 646–659. [Google Scholar] [PubMed]
- Van Dingenen, J. A virulence effector resolved: How a fungal phosphatase effector promotes rice false smut. Plant Cell 2022, 34, 2831–2832. [Google Scholar] [PubMed]
- Li, Y.; Wang, M.; Liu, Z.; Zhang, K.; Cui, F.; Sun, W. Towards understanding the biosynthetic pathway for ustilaginoidin mycotoxins in Ustilaginoidea virens. Environ. Microbiol. 2019, 21, 2629–2643. [Google Scholar]
- Wang, X.; Fu, X.; Lin, F.; Sun, W.; Meng, J.; Wang, A.; Lai, D.; Zhou, L.; Liu, Y. The contents of ustiloxins A and B along with their distribution in rice false smut balls. Toxins 2016, 8, 262. [Google Scholar] [CrossRef]
- Ye, Y.; Minami, A.; Igarashi, Y.; Izumikawa, M.; Umemura, M.; Nagano, N.; Machida, M.; Kawahara, T.; Shin-Ya, K.; Gomi, K.; et al. Unveiling the biosynthetic pathway of the ribosomally synthesized and post-translationally modified peptide ustiloxin B in filamentous fungi. Angew. Chem. Int. Ed. Engl. 2016, 55, 8072–8075. [Google Scholar]
- Tsukui, T.; Nagano, N.; Umemura, M.; Kumagai, T.; Terai, G.; Machida, M.; Asai, K. Ustiloxins, fungal cyclic peptides, are ribosomally synthesized in Ustilaginoidea virens. Bioinformatics 2015, 31, 981–985. [Google Scholar]
- Wang, X.; Wang, J.; Lai, D.; Wang, W.; Dai, J.; Zhou, L.; Liu, Y. Ustiloxin G, a new cyclopeptide mycotoxin from rice false smut balls. Toxins 2017, 9, 54. [Google Scholar] [CrossRef]
- Nakamura, K.; Izumiyama, N.; Ohtsubo, K.; Koiso, Y.; Iwasaki, S.; Sonoda, R.; Fujita, Y.; Yaegashi, H.; Sato, Z. “Lupinosis”-like lesions in mice caused by ustiloxin, produced by Ustilaginoieda virens: A morphological study. Nat. Toxins 1994, 2, 22–28. [Google Scholar] [CrossRef]
- Hu, Z.; Zheng, L.; Huang, J.; Zhou, L.; Liu, C.; Liu, H. Ustiloxin A is produced early in experimental Ustilaginoidea virens infection and affects transcription in rice. Curr. Microbiol. 2020, 77, 2766–2774. [Google Scholar]
- Sun, Q.; Liu, H.; Zhang, Y.; Yi, X.; Kong, R.; Cheng, S.; Man, J.; Zheng, L.; Huang, J.; Su, G.; et al. Global distribution of ustiloxins in rice and their male-biased hepatotoxicity. Environ. Pollut. 2022, 15, 118992. [Google Scholar]
- Cheng, S.; Liu, H.; Sun, Q.; Kong, R.; Letcher, R.J.; Liu, C. Occurrence of the fungus mycotoxin, ustiloxin A, in surface waters of paddy fields in Enshi, Hubei, China, and toxicity in Tetrahymena thermophila. Environ. Pollut. 2019, 251, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Guo, X.Y.; Li, L.; Huang, F.; Sun, W.X.; Li, Y.; Huang, Y.Y.; Xu, Y.J.; Shi, J.; Lei, Y.; et al. Infection of Ustilaginoidea virens intercepts rice seed formation but activates grain-filling-related genes. J. Integr. Plant Biol. 2015, 57, 577–590. [Google Scholar]
- Song, J.H.; Wei, W.; Lv, B.; Lin, Y.; Yin, W.X.; Peng, Y.L.; Schnabel, G.; Huang, J.B.; Jiang, D.H.; Luo, C.X. Rice false smut fungus hijacks the rice nutrients supply by blocking and mimicking the fertilization of rice ovary. Environ. Microbiol. 2016, 18, 3840–3849. [Google Scholar]
- Wang, Y.; Engel, T.; Teng, X. Post-translational regulation of the mTORC1 pathway: A switch that regulates metabolism-related gene expression. Biochim. Biophys. Acta Gene Regul. Mech. 2024, 1867, 195005. [Google Scholar]
- Baretić, D.; Williams, R.L. The structural basis for mTOR function. Semin. Cell Dev. Biol. 2014, 36, 91–101. [Google Scholar]
- Sauer, E.; Imseng, S.; Maier, T.; Hall, M.N. Conserved sequence motifs and the structure of the mTOR kinase domain. Biochem. Soc. Trans. 2013, 41, 889–895. [Google Scholar]
- Yu, F.; Gu, Q.; Yun, Y.; Yin, Y.; Xu, J.R.; Shim, W.B.; Ma, Z. The TOR signaling pathway regulates vegetative development and virulence in Fusarium graminearum. New Phytol. 2014, 203, 219–232. [Google Scholar]
- Li, G.; Cao, X.; Tumukunde, E.; Zeng, Q.; Wang, S. The target of rapamycin signaling pathway regulates vegetative development, aflatoxin biosynthesis, and pathogenicity in Aspergillus flavus. eLife 2024, 12, RP89478. [Google Scholar]
- Sun, G.; Qi, X.; Wilson, R.A. A feed-forward subnetwork emerging from integrated TOR- and cAMP/PKA-signaling architecture reinforces Magnaporthe oryzae appressorium morphogenesis. Mol. Plant Microbe Interact. 2019, 32, 593–607. [Google Scholar]
- Brunkard, J.O. Exaptive evolution of target of rapamycin signaling in multicellular eukaryotes. Dev. Cell 2020, 54, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR signaling in growth and metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Baldin, C.; Valiante, V.; Krüger, T.; Schafferer, L.; Haas, H.; Kniemeyer, O.; Brakhage, A.A. Comparative proteomics of a tor inducible Aspergillus fumigatus mutant reveals involvement of the Tor kinase in iron regulation. Proteomics 2015, 15, 2230–2243. [Google Scholar] [CrossRef]
- Xiong, F.; Liu, M.; Zhuo, F.; Yin, H.; Deng, K.; Feng, S.; Liu, Y.; Luo, X.; Feng, L.; Zhang, S.; et al. Host-induced gene silencing of BcTOR in Botrytis cinerea enhances plant resistance to grey mould. Mol. Plant Pathol. 2019, 20, 1722–1739. [Google Scholar] [CrossRef]
- Gu, Q.; Zhang, C.; Yu, F.; Yin, Y.; Shim, W.B.; Ma, Z. Protein kinase FgSch9 serves as a mediator of the target of rapamycin and high osmolarity glycerol pathways and regulates multiple stress responses and secondary metabolism in Fusarium graminearum. Environ. Microbiol. 2015, 17, 2661–2676. [Google Scholar] [CrossRef]
- Liu, L.; Wang, B.; Duan, G.; Wang, J.; Pan, Z.; Ou, M.; Bai, X.; Wang, P.; Zhao, D.; Nan, N.; et al. Histone deacetylase UvHST2 Is a global regulator of secondary metabolism in Ustilaginoidea virens. J. Agric. Food Chem. 2023, 71, 13124–13136. [Google Scholar] [CrossRef]
- Terensan, S.; Fernando, H.N.S.; Silva, J.N.; Perera, S.A.C.N.; Kottearachchi, N.S.; Weerasena, O.V.D.S.J. Morphological and molecular analysis of fungal species associated with blast and brown spot diseases of Oryza sativa. Plant Dis. 2022, 106, 1617–1625. [Google Scholar] [CrossRef]
- Sharma, P.; Khadka, R.B.; Baidya, S. Evaluation of fungicides to manage rice false smut (Ustilaginoidea virens) in the hills of Nepal. Heliyon 2024, 10, e34151. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Huang, J.; Liu, S.; Hu, W.; Li, P.; Xiao, Z.; Zhang, P.; Liu, R.; Li, C.; Yi, Y. Physiological biochemistry and multi-omics reveal the underlying mechanism of citral in controlling Colletotrichum scovillei in postharvest chili. LWT 2024, 214, 117123. [Google Scholar]
- Liu, Y.; Wu, Y.; Lv, X.; Li, K.; Xiong, J.; Liu, X.; Li, J.; Liu, L.; Du, G.; Chen, J.; et al. Improving cellular protein content of Saccharomyces cerevisiae based on adaptive evolution and flow cytometry-aided high throughput screening. J. Agric. Food Chem. 2025, 73, 706–717. [Google Scholar] [PubMed]
- Battaglioni, S.; Benjamin, D.; Wälchli, M.; Maier, T.; Hall, M.N. mTOR substrate phosphorylation in growth control. Cell 2022, 185, 1814–1836. [Google Scholar] [PubMed]
- Song, Y.; Wang, Y.; Zhang, H.; Saddique, M.A.B.; Luo, X.; Ren, M. The TOR signalling pathway in fungal phytopathogens: A target for plant disease control. Mol. Plant Pathol. 2024, 25, e70024. [Google Scholar]
- Seong, K.; Hou, Z.; Tracy, M.; Kistler, H.C.; Xu, J.R. Random insertional mutagenesis identifies genes associated with virulence in the wheat scab fungus Fusarium graminearum. Phytopathology 2005, 95, 744–750. [Google Scholar]
- Foster, A.J.; Jenkinson, J.M.; Talbot, N.J. Trehalose synthesis and metabolism are required at different stages of plant infection by Magnaporthe grisea. EMBO J. 2003, 22, 225–235. [Google Scholar]
- Fu, R.; Wang, J.; Chen, C.; Liu, Y.; Zhao, L.; Lu, D. Transcriptomic and metabolomic analyses provide insights into the pathogenic mechanism of the rice false smut pathogen Ustilaginoidea virens. Int. J. Mol. Sci. 2023, 24, 10805. [Google Scholar] [CrossRef]
- Liu, N.; Yun, Y.; Yin, Y.; Hahn, M.; Ma, Z.; Chen, Y. Lipid droplet biogenesis regulated by the FgNem1/Spo7-FgPah1 phosphatase cascade plays critical roles in fungal development and virulence in Fusarium graminearum. New Phytol. 2019, 223, 412–429. [Google Scholar]
- Wilfling, F.; Haas, J.T.; Walther, T.C.; Farese, R.V., Jr. Lipid droplet biogenesis. Curr. Opin. Cell Biol. 2014, 29, 39–45. [Google Scholar]
- Zhang, T.; Li, Y.N.; Li, X.; Gu, W.; Moeketsi, E.K.; Zhou, R.; Zheng, X.; Zhang, Z.; Zhang, H. The peroxisomal-CoA synthetase MoPcs60 is important for fatty acid metabolism and infectious growth of the rice blast fungus. Front. Plant Sci. 2022, 12, 811041. [Google Scholar]
- Teichert, S.; Wottawa, M.; Schönig, B.; Tudzynski, B. Role of the Fusarium fujikuroi TOR kinase in nitrogen regulation and secondary metabolism. Eukaryot. Cell 2006, 5, 1807–1819. [Google Scholar] [PubMed]
- Segreto, R.; Bazafkan, H.; Millinger, J.; Schenk, M.; Atanasova, L.; Doppler, M.; Büschl, C.; Boeckstaens, M.; Soto Diaz, S.; Schreiner, U.; et al. The TOR kinase pathway is relevant for nitrogen signaling and antagonism of the mycoparasite Trichoderma atroviride. PLoS ONE 2021, 16, e0262180. [Google Scholar]
- Umemura, M.; Kuriiwa, K.; Tamano, K.; Kawarabayasi, Y. Ustiloxin biosynthetic machinery is not compatible between Aspergillus flavus and Ustilaginoidea virens. Fungal Genet. Biol. 2020, 143, 103434. [Google Scholar]

| Name | Gene ID in U. virens | Homologue in S. cerevisiae | Blast E Value | |
|---|---|---|---|---|
| TORC1 | UvTOR | UV8b_00876 | ScTor1 | 0 |
| UvKog1 | UV8b_04344 | ScKog1 | 4 × 10−134 | |
| UvLst8 | UV8b_04658 | ScLst8 | 1 × 10−153 | |
| - | - | ScTco89 | - | |
| TORC2 | - | - | ScTor2 | - |
| UvAvo1 | UV8b_06451 | ScAvo1 | 1 × 10−18 | |
| - | - | ScAvo2 | - | |
| - | - | ScAvo3 | - | |
| - | - | ScBit61 | - | |
| - | - | ScBit2 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Sun, S.; Li, G.; Yang, Z.; Xing, Y.; Wang, R.; Xuan, Y.; Yang, X. The TOR Signaling Pathway Governs Fungal Development, Virulence and Ustiloxin Biosynthesis in Ustilaginoidea virens. J. Fungi 2025, 11, 239. https://doi.org/10.3390/jof11040239
Li Y, Sun S, Li G, Yang Z, Xing Y, Wang R, Xuan Y, Yang X. The TOR Signaling Pathway Governs Fungal Development, Virulence and Ustiloxin Biosynthesis in Ustilaginoidea virens. Journal of Fungi. 2025; 11(4):239. https://doi.org/10.3390/jof11040239
Chicago/Turabian StyleLi, Yuejiao, Shuqin Sun, Guangsheng Li, Zezhong Yang, Yuqi Xing, Ruixiang Wang, Yuanhu Xuan, and Xiurong Yang. 2025. "The TOR Signaling Pathway Governs Fungal Development, Virulence and Ustiloxin Biosynthesis in Ustilaginoidea virens" Journal of Fungi 11, no. 4: 239. https://doi.org/10.3390/jof11040239
APA StyleLi, Y., Sun, S., Li, G., Yang, Z., Xing, Y., Wang, R., Xuan, Y., & Yang, X. (2025). The TOR Signaling Pathway Governs Fungal Development, Virulence and Ustiloxin Biosynthesis in Ustilaginoidea virens. Journal of Fungi, 11(4), 239. https://doi.org/10.3390/jof11040239






