L-Rhamnose Dehydrogenase LraA of Aspergillus niger Shows High Substrate Specificity Matching Its Expression Profile
Abstract
:1. Introduction
2. Materials and Methods
2.1. Phylogenetic Analysis
2.2. Transcriptome Analysis
2.3. Growth Profile Analysis
2.4. Construction of an Expression Plasmid
2.5. Recombinant Protein Production and Purification
2.6. Enzyme Assays
3. Results and Discussion
3.1. The lraA Gene Is Expressed Exclusively on L-Rhamnose Unlike Its Close Homolog of PF00106
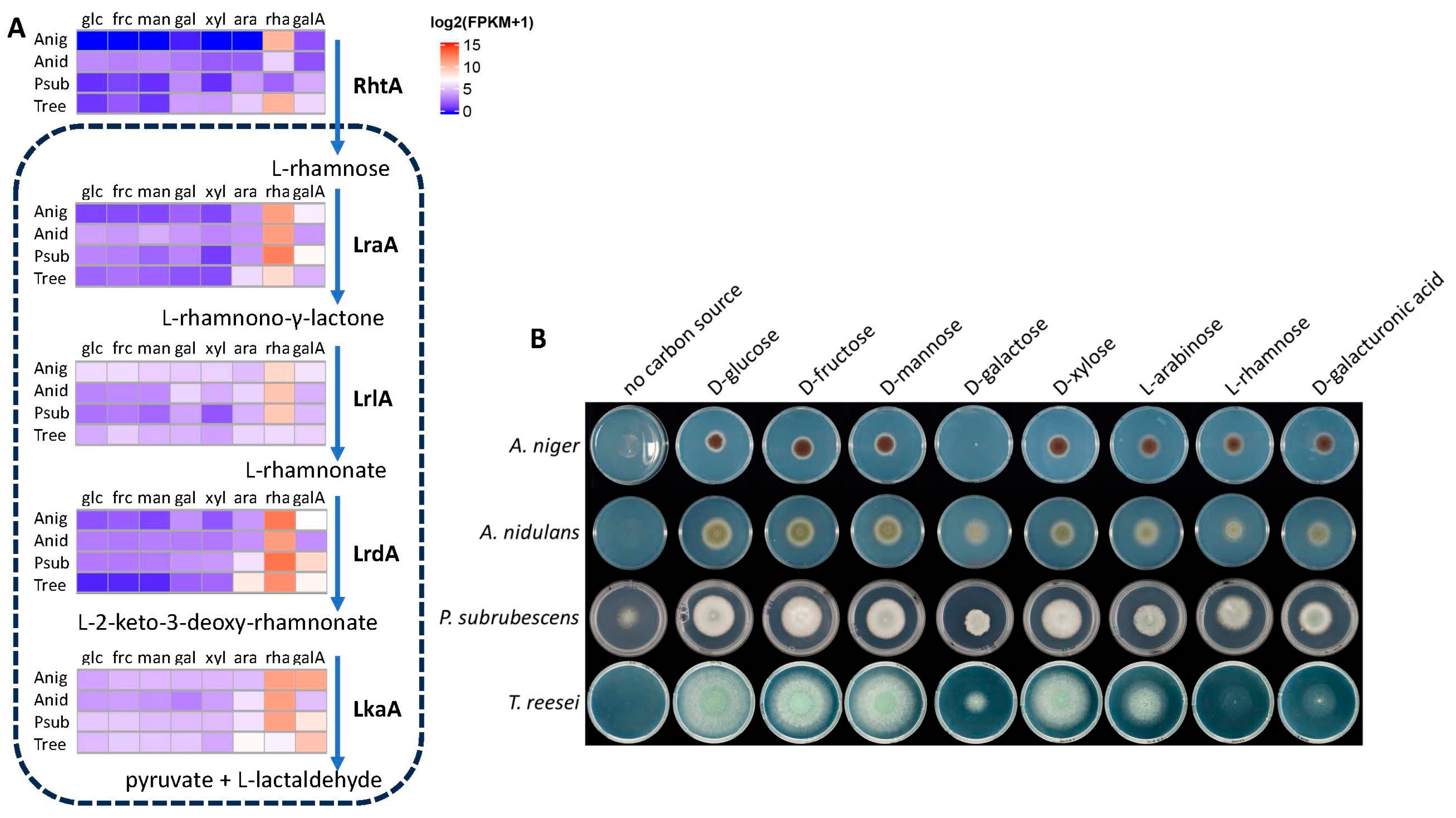
3.2. Expression of L-Rhamnose Catabolic Pathway Genes in Four Fungi Correlates with Their Growth on L-Rhamnose
3.3. LraA Is Highly Specific for L-Rhamnose
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Albersheim, P.; Darvill, A.G.; O’Neill, M.A.; Schols, H.A.; Voragen, A.G.J. An hypothesis: The same six polysaccharides are components of the primary cell walls of all higher plants. In Pectins and Pectinases; Visser, J., Voragen, A.G.J., Eds.; Elsevier Science: Amsterdam, The Netherlands, 1996; Volume 14, pp. 47–55. [Google Scholar]
- Kaczmarska, A.; Pieczywek, P.M.; Cybulska, J.; Zdunek, A. Structure and functionality of Rhamnogalacturonan I in the cell wall and in solution: A review. Carbohydr. Polym. 2022, 278, 118909. [Google Scholar] [CrossRef] [PubMed]
- Mikshina, P.V.; Petrova, A.A.; Faizullin, D.A.; Zuev, Y.F.; Gorshkova, T.A. Tissue-specific rhamnogalacturonan I forms the gel with hyperelastic properties. Biochemistry 2015, 80, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, C.; Kun, R.S.; Visser, J.; Aguilar-Pontes, M.V.; de Vries, R.P.; Battaglia, E. In vivo functional analysis of L-rhamnose metabolic pathway in Aspergillus niger: A tool to identify the potential inducer of RhaR. BMC Microbiol. 2017, 17, 214. [Google Scholar] [CrossRef]
- Rodionova, I.A.; Li, X.; Thiel, V.; Stolyar, S.; Stanton, K.; Fredrickson, J.K.; Bryant, D.A.; Osterman, A.L.; Best, A.A.; Rodionov, D.A. Comparative genomics and functional analysis of rhamnose catabolic pathways and regulons in bacteria. Front. Microbiol. 2013, 4, 407. [Google Scholar] [CrossRef]
- Sloothaak, J.; Odoni, D.I.; Martins dos Santos, V.A.P.; Schaap, P.J.; Tamayo-Ramos, J.A. Identification of a novel L-rhamnose uptake transporter in the filamentous fungus Aspergillus niger. PLoS Genet. 2016, 12, e1006468. [Google Scholar] [CrossRef]
- Wilson, D.M.; Ajl, S. Metabolism of L-rhamnose by Escherichia coli. I. L-rhamnose isomerase. J. Bacteriol. 1957, 73, 410–414. [Google Scholar] [CrossRef]
- Akhy, M.T.; Brown, C.M.; Old, D.C. L-Rhamnose utilisation in Salmonella typhimurium. J. Appl. Bacteriol. 1984, 56, 269–274. [Google Scholar] [CrossRef]
- Badia, J.; Baldoma, L.; Aguilar, J.; Boronat, A. Identification of the rhaA, rhaB and rhaD gene products from Escherichia coli K-12. FEMS Microbiol. Lett. 1989, 53, 253–257. [Google Scholar] [CrossRef]
- Schwartz, N.B.; Abram, D.; Feingold, D.S. L-Rhamnulose 1-phosphate aldolase of Escherichia coli. The role of metal in enzyme structure. Biochemistry 1974, 13, 1726–1730. [Google Scholar] [CrossRef]
- MacCabe, A.P.; Ninou, E.I.; Pardo, E.; Orejas, M. Catabolism of L-rhamnose in A. nidulans proceeds via the non-phosphorylated pathway and is glucose repressed by a CreA-independent mechanism. Microb. Cell Factories 2020, 19, 188. [Google Scholar] [CrossRef]
- Watanabe, S.; Saimura, M.; Makino, K. Eukaryotic and bacterial gene clusters related to an alternative pathway of nonphosphorylated L-rhamnose metabolism. J. Biol. Chem. 2008, 283, 20372–20382. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Makino, K. Novel modified version of nonphosphorylated sugar metabolism-an alternative L-rhamnose pathway of Sphingomonas sp. FEBS J. 2009, 276, 1554–1567. [Google Scholar] [CrossRef]
- Rigo, L.U.; Marechal, L.R.; Vieira, M.M.; Veiga, L.A. Oxidative pathway for L-rhamnose degradation in Pullularia pullulans. Can. J. Microbiol. 1985, 31, 817–822. [Google Scholar] [CrossRef]
- Koivistoinen, O.M.; Arvas, M.; Headman, J.R.; Andberg, M.; Penttilä, M.; Jeffries, T.W.; Richard, P. Characterisation of the gene cluster for L-rhamnose catabolism in the yeast Scheffersomyces (Pichia) stipitis. Gene 2012, 492, 177–185. [Google Scholar] [CrossRef]
- Riley, R.; Haridas, S.; Wolfe, K.H.; Lopes, M.R.; Hittinger, C.T.; Goker, M.; Salamov, A.A.; Wisecaver, J.H.; Long, T.M.; Calvey, C.H.; et al. Comparative genomics of biotechnologically important yeasts. Proc. Natl. Acad. Sci. USA 2016, 113, 9882–9887. [Google Scholar] [CrossRef] [PubMed]
- Chroumpi, T.; Aguilar-Pontes, M.V.; Peng, M.; Wang, M.; Lipzen, A.; Ng, V.; Grigoriev, I.V.; Mäkelä, M.R.; de Vries, R.P. Identification of a gene encoding the last step of the L-rhamnose catabolic pathway in Aspergillus niger revealed the inducer of the pathway regulator. Microbiol. Res. 2020, 234, 126426. [Google Scholar] [CrossRef]
- Koivistoinen, O.M.; Hilditch, S.; Voutilainen, S.P.; Boer, H.; Penttilä, M.; Richard, P. Identification in the yeast Pichia stipitis of the first l-rhamnose-1-dehydrogenase gene. FEBS J. 2008, 275, 2482–2488. [Google Scholar] [CrossRef]
- Li, J.; Chroumpi, T.; Garrigues, S.; Kun, R.S.; Meng, J.; Salazar-Cerezo, S.; Aguilar-Pontes, M.V.; Zhang, Y.; Tejomurthula, S.; Lipzen, A.; et al. The sugar metabolic model of Aspergillus niger can only be reliably transferred to fungi of its phylum. J. Fungi 2022, 8, 1315. [Google Scholar] [CrossRef]
- Chroumpi, T.; Peng, M.; Aguilar-Pontes, M.V.; Muller, A.; Wang, M.; Yan, J.; Lipzen, A.; Ng, V.; Grigoriev, I.V.; Mäkelä, M.R.; et al. Revisiting a ‘simple’ fungal metabolic pathway reveals redundancy, complexity and diversity. Microb. Biotechnol. 2021, 14, 2525–2537. [Google Scholar] [CrossRef]
- Mojzita, D.; Vuoristo, K.; Koivistoinen, O.M.; Penttilä, M.; Richard, P. The ‘true’ L-xylulose reductase of filamentous fungi identified in Aspergillus niger. FEBS Lett. 2010, 584, 3540–3544. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Asimenos, G.; Toh, H. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 2009, 537, 39–64. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Xu, L.; Li, J.; Gonzalez Ramos, V.M.; Lyra, C.; Wiebenga, A.; Grigoriev, I.V.; de Vries, R.P.; Mäkelä, M.R.; Peng, M. Genome-wide prediction and transcriptome analysis of sugar transporters in four ascomycete fungi. Bioresour. Technol. 2024, 391, 130006. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Bjellqvist, B.; Hughes, G.J.; Pasquali, C.; Paquet, N.; Ravier, F.; Sanchez, J.C.; Frutiger, S.; Hochstrasser, D. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 1993, 14, 1023–1031. [Google Scholar] [CrossRef]
- Kim, S.M.; Paek, K.H.; Lee, S.B. Characterization of NADP+-specific L-rhamnose dehydrogenase from the thermoacidophilic Archaeon Thermoplasma acidophilum. Extremophiles 2012, 16, 447–454. [Google Scholar] [CrossRef]
- Yoshiwara, K.; Watanabe, S.; Watanabe, Y. Crystal structure of l-rhamnose 1-dehydrogenase involved in the nonphosphorylative pathway of l-rhamnose metabolism in bacteria. FEBS Lett. 2021, 595, 637–646. [Google Scholar] [CrossRef]
- Fekete, E.; de Vries, R.P.; Seiboth, B.; vanKuyk, P.A.; Sandor, E.; Metz, B.; Kubicek, C.P.; Karaffa, L. D-Galactose uptake is nonfunctional in the conidiospores of Aspergillus niger. FEMS Microbiol. Lett. 2012, 329, 198–203. [Google Scholar] [CrossRef]
- Twerdochlib, A.L.; Pedrosa, F.O.; Funayama, S.; Rigo, L.U. L-rhamnose metabolism in Pichia stipitis and Debaryomyces polymorphus. Can. J. Microbiol. 1994, 40, 896–902. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Pletnev, V.Z.; Weeks, C.M.; Duax, W.L. Rational proteomics II: Electrostatic nature of cofactor preference in the short-chain oxidoreductase (SCOR) enzyme family. Proteins 2004, 57, 294–301. [Google Scholar] [CrossRef]
- Tanaka, N.; Nonaka, T.; Nakanishi, M.; Deyashiki, Y.; Hara, A.; Mitsui, Y. Crystal structure of the ternary complex of mouse lung carbonyl reductase at 1.8 A resolution: The structural origin of coenzyme specificity in the short-chain dehydrogenase/reductase family. Structure 1996, 4, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Kallberg, Y.; Oppermann, U.; Persson, B. Classification of the short-chain dehydrogenase/reductase superfamily using hidden Markov models. FEBS J. 2010, 277, 2375–2386. [Google Scholar] [CrossRef]
- Shah, B.S.; Tetu, S.G.; Harrop, S.J.; Paulsen, I.T.; Mabbutt, B.C. Structure of a short-chain dehydrogenase/reductase (SDR) within a genomic island from a clinical strain of Acinetobacter baumannii. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 1318–1323. [Google Scholar] [CrossRef] [PubMed]
- Elshafei, A.M.; Abdel-Fatah, O.M. Evidence for a non-phosphorylated route of galactose breakdown in cell-free extracts of Aspergillus niger. Enzym. Microb. Technol. 2001, 29, 76–83. [Google Scholar] [CrossRef]
- Frey, P.A. The Leloir pathway: A mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 1996, 10, 461–470. [Google Scholar] [CrossRef]
- Mojzita, D.; Herold, S.; Metz, B.; Seiboth, B.; Richard, P. L-xylo-3-hexulose reductase is the missing link in the oxidoreductive pathway for D-galactose catabolism in filamentous fungi. J. Biol. Chem. 2012, 287, 26010–26018. [Google Scholar] [CrossRef]
- Mäkelä, M.R.; Aguilar-Pontes, M.V.; van Rossen-Uffink, D.; Peng, M.; de Vries, R.P. The fungus Aspergillus niger consumes sugars in a sequential manner that is not mediated by the carbon catabolite repressor CreA. Sci. Rep. 2018, 8, 6655. [Google Scholar] [CrossRef]




| Organism | Enzyme | Km [mM] | kcat [min−1] | kcat/Km [mM−1 min−1] | Reference |
|---|---|---|---|---|---|
| Aspergillus niger | LraA | 2.4 ± 0.9 | 2149.2 ± 183.4 | 904.8 ± 30.7 | This study |
| Scheffersomyces stipitis | PsLRA1 | 1.7 ± 0.0 | 1510.0 ± 20.0 | 885.0 ± 8.0 | [13] |
| S. stipitis | Rha1 | 1.5 ± 0.0 | NM | NM | [19] |
| Debaryomyces hansenii | DhLRA1 | 9.4 ± 1.1 | 2860.0 ± 230.0 | 307.0 ± 11.0 | [13] |
| Azotobacter vinelandii | AvLRA1 | 2.2 ± 0.1 | 5010.0 ± 178.0 | 2250.0 ± 51.0 | [13,30] |
| 2.6 ± 0.1 * | 2230.0 ± 43.0 * | 856.0 ± 28.0 * | |||
| Thermoplasma acidophilum | - | 0.5 * | 1341.3 * | 2915.9 * | [29] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terebieniec, A.; Xu, L.; Peng, M.; Mäkelä, M.R.; Vries, R.P.d. L-Rhamnose Dehydrogenase LraA of Aspergillus niger Shows High Substrate Specificity Matching Its Expression Profile. J. Fungi 2025, 11, 301. https://doi.org/10.3390/jof11040301
Terebieniec A, Xu L, Peng M, Mäkelä MR, Vries RPd. L-Rhamnose Dehydrogenase LraA of Aspergillus niger Shows High Substrate Specificity Matching Its Expression Profile. Journal of Fungi. 2025; 11(4):301. https://doi.org/10.3390/jof11040301
Chicago/Turabian StyleTerebieniec, Agata, Li Xu, Mao Peng, Miia R. Mäkelä, and Ronald P. de Vries. 2025. "L-Rhamnose Dehydrogenase LraA of Aspergillus niger Shows High Substrate Specificity Matching Its Expression Profile" Journal of Fungi 11, no. 4: 301. https://doi.org/10.3390/jof11040301
APA StyleTerebieniec, A., Xu, L., Peng, M., Mäkelä, M. R., & Vries, R. P. d. (2025). L-Rhamnose Dehydrogenase LraA of Aspergillus niger Shows High Substrate Specificity Matching Its Expression Profile. Journal of Fungi, 11(4), 301. https://doi.org/10.3390/jof11040301





