Long-Chain Acyl-CoA Synthetase is Associated with the Growth of Malassezia spp.
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains, Media, and Growth Conditions
2.2. Plasmids
2.3. Transformation of S. cerevisiae
2.4. Triacsin C Minimum Inhibitory Concentration (MIC) Analysis
2.5. Isolation of M. pachydermatis Total RNA
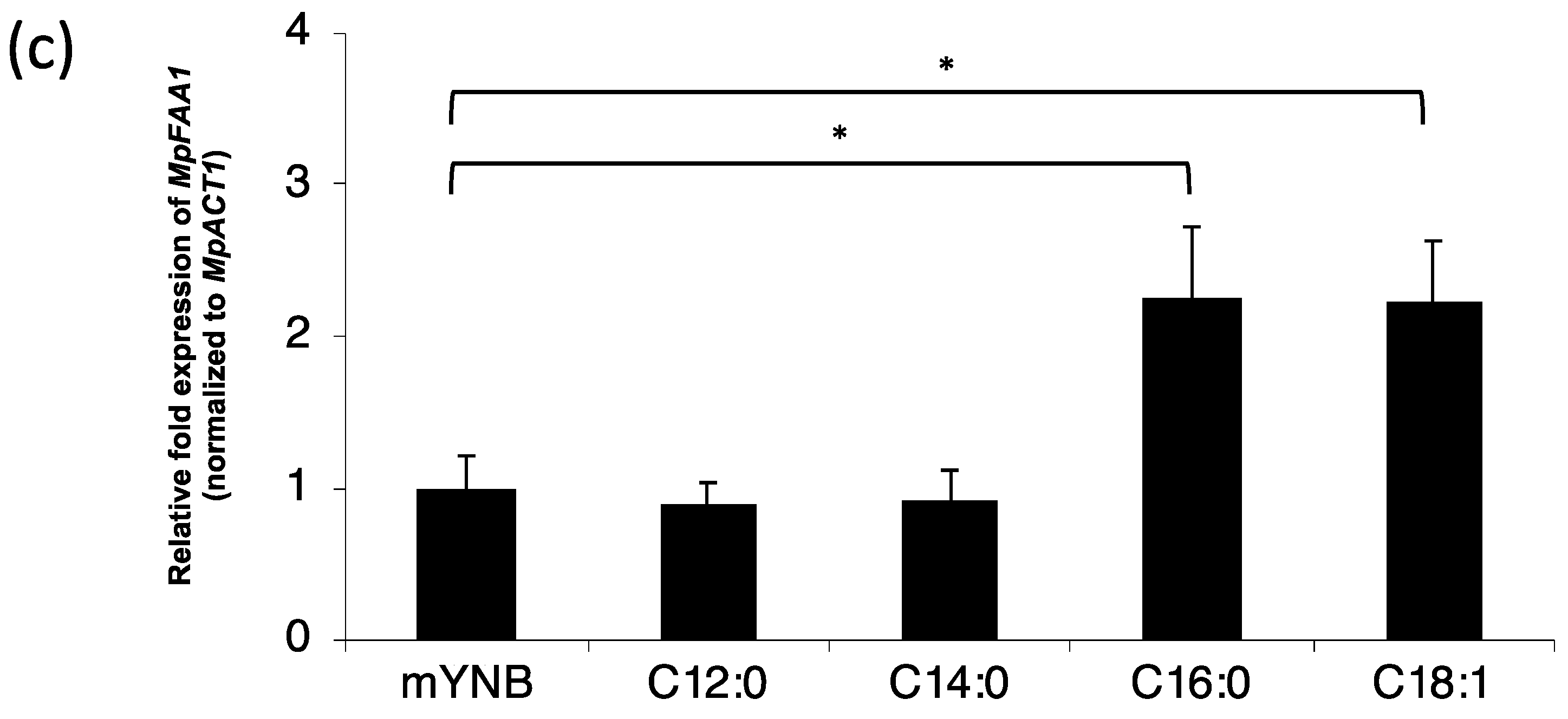
2.6. cDNA Synthesis and Transcription Level Analysis
3. Results
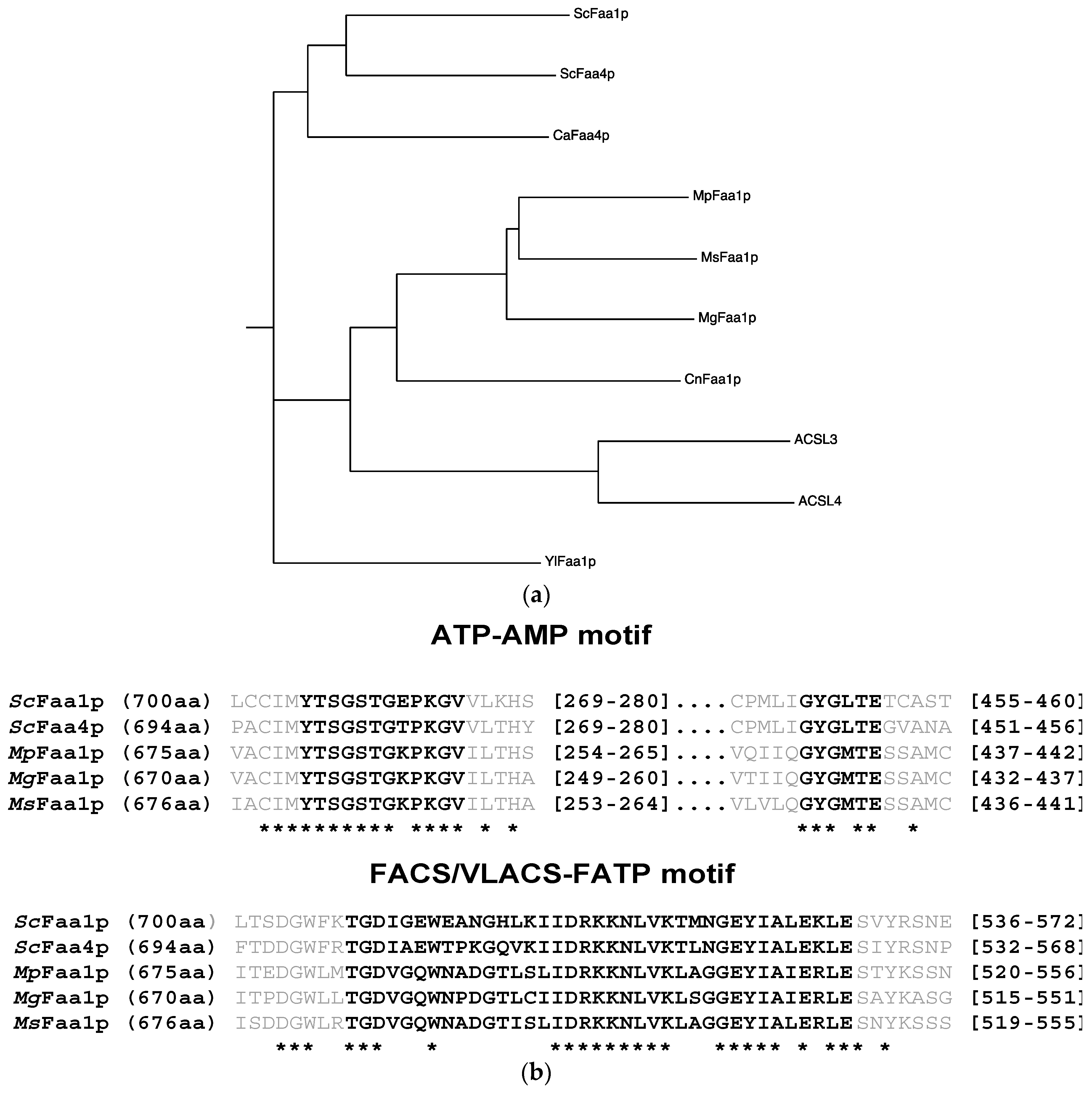
3.1. Identification of Malassezia orthologs of S. cerevisiae FAA1 Genes
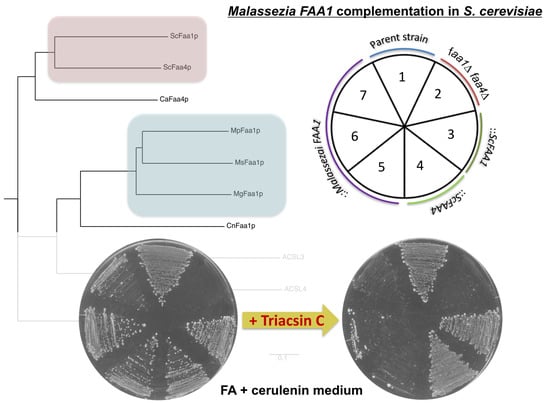
3.2. Malassezia Faa1 Proteins Complement the Function of S. cerevisiae Faa1p and Faa4p
3.3. Inhibition of Malassezia Faa1p by Triacsin C
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sugita, T.; Suto, H.; Unno, T. Molecular analysis of Malassezia microflora on the skin of atopic dermatitis patients and healthy subjects. J. Clin. Microbiol. 2001, 39, 3486–3490. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Saunders, C.W.; Hu, P.; Grant, R.A.; Boekhout, T.; Kuramae, E.E.; Kronstad, J.W.; DeAngelis, Y.M.; Reeder, N.L.; Johnstone, K.R.; et al. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc. Natl. Acad. Sci. USA 2007, 104, 18730–18735. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Saito, M.; Sugita, T.; Tsuboi, R. Malassezia species and their associated skin diseases. J. Dermatol. 2015, 42, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Yurayart, C.; Chindamporn, A.; Suradhat, S.; Tummaruk, P.; Kajiwara, S.; Prapasarakul, N. Comparative analysis of the frequency, distribution and population sizes of yeasts associated with canine seborrheic dermatitis and healthy skin. Vet. Microbiol. 2011, 148, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Triana, S.; Ohm, R.A.; De Cock, H.; Restrepo, S.; Celis, A. Draft Genome Sequence of the Animal and Human Pathogen Malassezia pachydermatis Strain CBS 1879. Genome Announc. 2015, 3, e01197-15. [Google Scholar] [CrossRef] [PubMed]
- Lorch, J.M.; Palmer, J.M.; Vanderwolf, K.J.; Schmidt, K.Z.; Verant, M.L.; Weller, T.J.; Blehert, D.S. A new cold-tolerant species of yeast isolated from bats. Persoonia 2018, 41, 56–70. [Google Scholar] [CrossRef]
- Wu, G.; Zhao, H.; Li, C.; Rajapakse, M.P.; Wong, W.C.; Xu, J.; Saunders, C.W.; Reeder, N.L.; Reilman, R.A.; Scheynius, A.; et al. Genus-Wide Comparative Genomics of Malassezia Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin. PLoS Genet. 2015, 11, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Juntachai, W.; Oura, T.; Murayama, S.Y.; Kajiwara, S. The lipolytic enzymes activities of Malassezia species. Med. Mycol. 2009, 47, 477–484. [Google Scholar] [CrossRef]
- Deangelis, Y.M.; Saunders, C.W.; Johnstone, K.R.; Reeder, N.L.; Coleman, C.G.; Kaczvinsky, J.R.; Gale, C.; Walter, R.; Mekel, M.; Lacey, M.P.; et al. Isolation and expression of a Malassezia globosa lipase gene, LIP1. J. Investig. Dermatol. 2007, 127, 2138–2146. [Google Scholar] [CrossRef]
- Sommer, B.; Overy, D.P.; Kerr, R.G. Identification and characterization of lipases from Malassezia restricta, a causative agent of dandruff. FEMS Yeast Res. 2015, 15, fov078. [Google Scholar] [CrossRef]
- Dawson, T.L. Malassezia globosa and restricta: Breakthrough understanding of the etiology and treatment of dandruff and seborrheic dermatitis through whole-genome analysis. J. Investig. Dermatol. Symp. Proc. 2007, 12, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Knoll, L.J.; Johnson, D.R.; Gordon, J.I. Biochemical Studies of Three Saccharomyces cerevisiae Acyl-CoA Synthetases, Faa1p, Faa2p, and Faa3p. J. Biol. Chem. 1994, 269, 16348–16356. [Google Scholar] [PubMed]
- Black, P.N.; Dirusso, C.C. Transmembrane Movement of Exogenous Long-Chain Fatty Acids: Proteins, Enzymes, and Vectorial Esterification Transmembrane Movement of Exogenous Long-Chain Fatty Acids: Proteins, Enzymes, and Vectorial Esterification. Microbiol. Mol. Biol. Rev. 2003, 67, 454–472. [Google Scholar] [CrossRef] [PubMed]
- Hettema, E.H.; van Roermund, C.W.; Distel, B.; van den Berg, M.; Vilela, C.; Rodrigues-Pousada, C.; Wanders, R.J.; Tabak, H.F. The ABC transporter proteins Pat1 and Pat2 are required for import of long-chain fatty acids into peroxisomes of Saccharomyces cerevisiae. EMBO J. 1996, 15, 3813–3822. [Google Scholar] [CrossRef] [PubMed]
- Van Roermund, C.W.T.; Ijlst, L.; Majczak, W.; Waterham, H.R.; Folkerts, H.; Wanders, R.J.A.; Hellingwerf, K.J. Peroxisomal fatty acid uptake mechanism in saccharomyces cerevisiae. J. Biol. Chem. 2012, 287, 20144–20153. [Google Scholar] [CrossRef]
- Johnson, D.R.; Knoll, J.; Rowleyo, N.; Gordon, J.I. Genetic Analysis of the Role of Saccharomyces cerevisiae Acyl-CoA Synthetase Genes in Regulating Protein N-Myristoylation. J. Biol. Chem. 1994, 269, 18037–18046. [Google Scholar] [PubMed]
- Thomaz, D.Y.; de Almeida, J.N.; Lima, G.M.E.; Nunes, M.D.O.; Camargo, C.H.; Grenfell, R.D.C.; Benard, G.; Del Negro, G.M.B. An Azole-Resistant Candida parapsilosis Outbreak: Clonal Persistence in the Intensive Care Unit of a Brazilian Teaching Hospital. Front. Microbiol. 2018, 9, 2997. [Google Scholar] [CrossRef]
- Popp, C.; Ramírez-Zavala, B.; Schwanfelder, S.; Krüger, I.; Morschhäuser, J. Evolution of Fluconazole-Resistant Candida albicans Strains by Drug-Induced Mating Competence and Parasexual Recombination. mBio 2019, 10, e02740-18. [Google Scholar] [CrossRef]
- Smith, K.D.; Achan, B.; Hullsiek, K.H.; McDonald, T.R.; Okagaki, L.H.; Alhadab, A.A.; Akampurira, A.; Rhein, J.R.; Meya, D.B.; Boulware, D.R.; et al. Increased Antifungal Drug Resistance in Clinical Isolates of Cryptococcus neoformans in Uganda. Antimicrob. Agents Chemother. 2015, 59, 7197–7204. [Google Scholar] [CrossRef]
- Nijima, M.; Kano, R.; Nagata, M.; Hasegawa, A.; Kamata, H. An azole-resistant isolate of Malassezia pachydermatis. Vet. Microbiol. 2011, 149, 288–290. [Google Scholar] [CrossRef]
- Kano, R.; Yokoi, S.; Kariya, N.; Oshimo, K.; Kamata, H. Multi-azole-resistant strain of Malassezia pachydermatis isolated from a canine Malassezia dermatitis. Med. Mycol. 2018, 57, 346–350. [Google Scholar] [CrossRef]
- Omura, S.; Tomoda, H.; Xu, Q.M.; Takahashi, Y.; Iwai, Y. Triacsins, New Inhibitore of Acyl-CoA Synthetase produced by Streptomyces sp. J. Antibiot. 1986, 39, 1211–1218. [Google Scholar] [CrossRef]
- Tomoda, H.; Igarashi, K.; Omura, S. Inhibition of acyl-CoA synthetase by triacsins. Biochim. Biophys. Acta 1987, 921, 595–598. [Google Scholar]
- Igal, R.A.; Wang, P.; Coleman, R.A. Triacsin C blocks de novo synthesis of glycerolipids and cholesterol esters but not recycling of fatty acid into phospholipid: Evidence for functionally separate pools of acyl-CoA. Biochem. J. 1997, 324 Pt 2, 529–534. [Google Scholar] [CrossRef]
- Guo, F.; Zhang, H.; Fritzler, J.M.; Rider, S.D., Jr.; Xiang, L.; Mcnair, N.N.; Mead, J.R. Amelioration of Cryptosporidium parvum Infection In Vitro and In Vivo by Targeting Parasite Fatty Acyl-Coenzyme A Synthetases. J. Infect. Dis. 2014, 209, 1279–1287. [Google Scholar] [CrossRef]
- Guo, F.; Ortega-Pierres, G.; Argüello-García, R.; Zhang, H.; Zhu, G. Giardia fatty acyl-CoA synthetases as potential drug targets. Front. Microbiol. 2015, 6, 753. [Google Scholar] [CrossRef]
- Vessey, D.A.; Kelley, M.; Warren, R.S. Characterization of Triacsin C Inhibition of Short-, Medium-, and Long-Chain Fatty Acid: CoA Ligases of Human Liver. J. Biochem. Mol. Toxicol. 2004, 18, 100–106. [Google Scholar] [CrossRef]
- Knoll, L.J.; Schall, O.F.; Suzuki, I.; Gokel, G.W.; Gordon, J.I. Comparison of the Reactivity of Tetradecanoic Acids, a Triacsin, and Unsaturated Oximes with Four Purified Saccharomyces cerevisiae Fatty Acid Activation Proteins. J. Biol. Chem. 1995, 270, 20090–20097. [Google Scholar] [CrossRef] [PubMed]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K.; Wiley, C.J.; Ausubel, F.M.; Brent, R.; et al. Current Protocols in Molecular Biology; John Wiley & Sons, Inc.: New York, NY, USA, 2003; ISBN 047150338X. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning. A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Boeke, J.D.; Lacroute, F.; Fink, G.R. A positive selection for mutants lacking orotidine-5’-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 1984, 197, 345–346. [Google Scholar] [CrossRef] [PubMed]
- Collart, M.A.; Oliviero, S. Preparation of Yeast RNA. In Current Protocols in Molecular Biology; John Wiley & Sons, Inc.: New York, USA, 2001; pp. 13.12.1–13.12.5. [Google Scholar]
- Tejima, K.; Ishiai, M.; Murayama, S.O.; Iwatani, S. Candida albicans fatty acyl-CoA synthetase, Ca Faa4p, is involved in the uptake of exogenous long-chain fatty acids and cell activity in the biofilm. Curr. Genet. 2017, 64, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Mashima, T.; Sato, S.; Okabe, S.; Miyata, S.; Matsuura, M.; Sugimoto, Y.; Tsuruo, T.; Seimiya, H. Acyl-CoA synthetase as a cancer survival factor: Its inhibition enhances the efficacy of etoposide. Cancer Sci. 2009, 100, 1561. [Google Scholar] [CrossRef] [PubMed]




| Gene Name | Accession No. | Malassezia Gene ID | Protein Size | Description | % Identity to | Query Cover to | ||
|---|---|---|---|---|---|---|---|---|
| ScFAA1 | ScFAA4 | ScFAA1 | ScFAA4 | |||||
| MpFAA1 | XP_017991190.1 | Malapachy_0054 | 675 aa | long-chain fatty acid ligase | 38 | 37 | 87 | 99 |
| MgFAA1 | XP_001729159.1 | MGL_3626 | 670 aa | hypothetical protein | 37 | 36 | 97 | 98 |
| MsFAA1 | SHO79486.1 | MSYG_3835 | 676 aa | similar to S. cerevisiae FAA4 | 37 | 39 | 99 | 98 |
| MsFAA1 | XP_018739945.1 | MSY001_1358 | 662 aa | hypothetical protein | 36 | 38 | 99 | 98 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tenagy; Tejima, K.; Chen, X.; Iwatani, S.; Kajiwara, S. Long-Chain Acyl-CoA Synthetase is Associated with the Growth of Malassezia spp. J. Fungi 2019, 5, 88. https://doi.org/10.3390/jof5040088
Tenagy, Tejima K, Chen X, Iwatani S, Kajiwara S. Long-Chain Acyl-CoA Synthetase is Associated with the Growth of Malassezia spp. Journal of Fungi. 2019; 5(4):88. https://doi.org/10.3390/jof5040088
Chicago/Turabian StyleTenagy, Kengo Tejima, Xinyue Chen, Shun Iwatani, and Susumu Kajiwara. 2019. "Long-Chain Acyl-CoA Synthetase is Associated with the Growth of Malassezia spp." Journal of Fungi 5, no. 4: 88. https://doi.org/10.3390/jof5040088
APA StyleTenagy, Tejima, K., Chen, X., Iwatani, S., & Kajiwara, S. (2019). Long-Chain Acyl-CoA Synthetase is Associated with the Growth of Malassezia spp. Journal of Fungi, 5(4), 88. https://doi.org/10.3390/jof5040088