The Role of Secretory Pathways in Candida albicans Pathogenesis
Abstract
1. Introduction
Genomic Overview and Comparison to S. cerevisiae and Other Fungi
2. Translocation and ER Transport
2.1. Translocation
2.2. Co-Translational Translocation
2.3. Post-Translational Translocation
2.4. Protein Folding and Maturation
3. ER-Golgi Secretion
ER–Golgi Anterograde Transport
4. Intra-Golgi Transport
5. Pre-Vacuolar Secretion
6. The Role of the Vacuole in Secretion and Virulence
7. Post-Golgi Secretion
8. Extracellular Vesicles
9. Endocytosis and Endocytic Pathways
The Endosomal Network
10. Polarized Secretion in Candida albicans Hyphae
11. Non-Classical Secretion
12. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Neville, B.W. Oral and Maxillofacial Pathology; Saunders/Elsevier: St. Louis, MO, USA, 2009; ISBN 978-1-4377-2197-3. [Google Scholar]
- Schaller, M.; Borelli, C.; Korting, H.C.; Hube, B. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses 2005, 48, 365–377. [Google Scholar] [CrossRef]
- Sundstrom, P. Adhesion in Candida spp. Cell. Microbiol. 2002, 4, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L.; Green, C.B.; Oh, S.-H.; Zhao, X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—A sticky pursuit. Med. Mycol. 2008, 46, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chaffin, W.L.; López-Ribot, J.L.; Casanova, M.; Gozalbo, D.; Martínez, J.P. Cell Wall and Secreted Proteins of Candida albicans: Identification, Function, and Expression. Microbiol. Mol. Biol. Rev. 1998, 62, 130–180. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, C.A.; Vinces, M.D. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence: Candida albicans morphogenesis and virulence. Cell. Microbiol. 2005, 7, 1546–1554. [Google Scholar] [CrossRef]
- Soll, D.R. Mating-type locus homozygosis, phenotypic switching and mating: A unique sequence of dependencies in Candida albicans. Bioessays 2004, 26, 10–20. [Google Scholar] [CrossRef]
- Blankenship, J.R.; Mitchell, A.P. How to build a biofilm: A fungalperspective. Curr. Opin. Microbiol. 2006, 9, 588–594. [Google Scholar] [CrossRef]
- Nett, J.; Andes, D. Candida albicans biofilm development, modeling a host–pathogen interaction. Curr. Opin. Microbiol. 2006, 9, 340–345. [Google Scholar] [CrossRef]
- De La Rosa, J.M.; Ruiz, T.; Fonzi, W.A.; Rodríguez, L. Analysis of heterologous expression of Candida albicans SEC61 gene reveals differences in Sec61p homologues related to species-specific functionality. Fungal Genet. Biol. 2004, 41, 941–953. [Google Scholar] [CrossRef]
- Lee, M.C.S.; Miller, E.A.; Goldberg, J.; Orci, L.; Schekman, R. BI-DIRECTIONAL PROTEIN TRANSPORT BETWEEN THE ER AND GOLGI. Annu. Rev. Cell Dev. Biol. 2004, 20, 87–123. [Google Scholar] [CrossRef]
- Von Heijne, G. Membrane-protein topology. Nat.Rev. Mol. Cell Biol. 2006, 7, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Payne, W.E.; Kaiser, C.A.; Bevis, B.J.; Soderholm, J.; Fu, D.; Sears, I.B.; Glick, B.S. Isolation of Pichia pastoris genes involved in ER-to-Golgi transport. Yeast 2000, 16, 979–993. [Google Scholar] [PubMed]
- Guo, W.; Sacher, M.; Barrowman, J.; Ferro-Novick, S.; Novick, P. Protein complexes in transport vesicle targeting. Trends Cell Biol. 2000, 10, 251–255. [Google Scholar] [CrossRef]
- Harsay, E.; Bretscher, A. Parallel secretory pathways to the cell surface in yeast. J. Cell Biol. 1995, 131, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.D.; Brent Heath, I. Predicting the distribution, conservation, and functions of SNAREs and related proteins in fungi. Fungal Genet. Biol. 2002, 36, 1–21. [Google Scholar] [CrossRef]
- Bonifacino, J.S.; Glick, B.S. The Mechanisms of Vesicle Budding and Fusion. Cell 2004, 116, 153–166. [Google Scholar] [CrossRef]
- Sato, K.; Nakano, A. Mechanisms of COPII vesicle formation and protein sorting. FEBS Lett. 2007, 581, 2076–2082. [Google Scholar] [CrossRef]
- Schmoranzer, J.; Simon, S.M. Role of Microtubules in Fusion of Post-Golgi Vesicles to the Plasma Membrane. Mol. Biol. Cell 2003, 14, 12. [Google Scholar]
- Harsay, E.; Schekman, R. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J. Cell Biol. 2002, 156, 271–286. [Google Scholar] [CrossRef]
- Lee, S.A.; Jones, J.; Khalique, Z.; Kot, J.; Alba, M.; Bernardo, S.; Seghal, A.; Wong, B. A functional analysis of the Candida albicans homolog of Saccharomyces cerevisiae VPS4. FEMS Yeast Res. 2007, 7, 973–985. [Google Scholar] [CrossRef][Green Version]
- Juan, T. Biogenesis and function of ESCRT-dependent extracellular vesicles. Dev. Biol. 2018, 74, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-R.; Lee, R.T.-H.; Wang, Y.-M.; Zheng, X.-D.; Wang, Y. Candida albicans hyphal morphogenesis occurs in Sec3p-independent and Sec3p-dependent phases separated by septin ring formation. J. Cell Sci. 2007, 120, 1898–1907. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Schorey, J.S.; Cheng, Y.; Singh, P.P.; Smith, V.L. Exosomes and other extracellular vesicles in host–pathogen interactions. EMBO Rep. 2015, 16, 24–43. [Google Scholar] [CrossRef] [PubMed]
- Podinovskaia, M.; Spang, A. The Endosomal Network: Mediators and Regulators of Endosome Maturation. In Endocytosis and Signaling; Lamaze, C., Prior, I., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 57, pp. 1–38. ISBN 978-3-319-96703-5. [Google Scholar]
- Delic, M.; Valli, M.; Graf, A.B.; Pfeffer, M.; Mattanovich, D.; Gasser, B. The secretory pathway: Exploring yeast diversity. FEMS Microbiol.Rev. 2013, 37, 872–914. [Google Scholar] [CrossRef] [PubMed]
- Fonzi, W.A. The protein secretory pathway of Candida albicans. Mycoses 2009, 52, 291–303. [Google Scholar] [CrossRef]
- Kabir, M.A.; Hussain, M.A. Human fungal pathogen Candida albicans in the postgenomic era: An overview. Expert Rev. Anti-Infect. Ther. 2009, 7, 121–134. [Google Scholar] [CrossRef]
- Braun, B.R.; Van Het Hoog, M.; D’Enfert, C.; Martchenko, M.; Dungan, J.; Kuo, A.; Inglis, D.O.; Uhl, M.A.; Hogues, H.; Berriman, M.; et al. A human-curated annotation of the Candida albicans genome. PLoS Genet. 2005, 1, 36–57. [Google Scholar] [CrossRef]
- Mitrovich, Q.M.; Tuch, B.B.; Guthrie, C.; Johnson, A.D. Computational and experimental approaches double the number of known introns in the pathogenic yeast Candida albicans. Genome Res. 2007, 17, 492–502. [Google Scholar] [CrossRef]
- Swennen, D.; Beckerich, J.-M. Yarrowia lipolytica vesicle-mediated protein transport pathways. BMC Evol. Biol. 2007, 7, 219. [Google Scholar] [CrossRef]
- Sorgo, A.G.; Heilmann, C.J.; Dekker, H.L.; Brul, S.; De Koster, C.G.; Klis, F.M. Mass spectrometric analysis of the secretome of Candida albicans. Yeast 2010, 27, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Nagano, M.; Toshima, J.Y.; Toshima, J. Rab GTPases Networks in Membrane Traffic in Saccharomyces cerevisiae. YAKUGAKU ZASSHI 2015, 135, 483–492. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mao, Y.; Zhang, Z.; Lee, S.A.; Wong, B. Overexpression of a dominant-negative allele of YPT1 inhibits growth and aspartyl protease secretion in Candida albicans. Microbiology 2001, 147, 1961–1970. [Google Scholar]
- Cle, M. Isolation and characterization of the Candida albicans SEC4 Gene. Yeast 1998, 14, 675–680. [Google Scholar]
- Johnston, D.A.; Tapia, A.L.; Eberle, K.E.; Palmer, G.E. Three Prevacuolar Compartment Rab GTPases Impact Candida albicans Hyphal Growth. Eukaryot. Cell 2013, 12, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E.; Lepak, A.J.; Marchillo, K.; Andes, D.R. Time Course Global Gene Expression Analysis of an In Vivo Candida Biofilm. J. Infect. Dis. 2009, 200, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Johnston, D.A.; Eberle, K.E.; Sturtevant, J.E.; Palmer, G.E. Role for Endosomal and Vacuolar GTPases in Candida albicans Pathogenesis. Infect. Immun. 2009, 77, 2343–2355. [Google Scholar] [CrossRef]
- Mora-Montes, H.M.; Bates, S.; Netea, M.G.; Díaz-Jiménez, D.F.; López-Romero, E.; Zinker, S.; Ponce-Noyola, P.; Kullberg, B.J.; Brown, A.J.P.; Odds, F.C.; et al. Endoplasmic Reticulum α-Glycosidases of Candida albicans Are Required for N Glycosylation, Cell Wall Integrity, and Normal Host-Fungus Interaction. Eukaryot. Cell 2007, 6, 2184–2193. [Google Scholar] [CrossRef]
- Egea, P.F.; Stroud, R.M.; Walter, P. Targeting proteins to membranes: Structure of the signal recognition particle. Curr. Opin. Struct. Biol. 2005, 15, 213–220. [Google Scholar] [CrossRef]
- Jones, T.; Federspiel, N.A.; Chibana, H.; Dungan, J.; Kalman, S.; Magee, B.B.; Newport, G.; Thorstenson, Y.R.; Agabian, N.; Magee, P.T.; et al. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 2004, 101, 7329–7334. [Google Scholar] [CrossRef]
- Ogg, S.C.; Barz, W.P.; Walter, P. A Functional GTPase Domain, but not its Transmembrane Domain, is Required for Function of the SRP Receptor beta-subunit. J. Cell Biol. 1998, 142, 14. [Google Scholar] [CrossRef] [PubMed]
- Maglott, D.; Ostell, J.; Pruitt, K.D.; Tatusova, T. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res. 2007, 35, D26–D31. [Google Scholar] [CrossRef] [PubMed]
- Rosenblad, M.A.; Zwieb, C.; Samuelsson, T. Identification and comparative analysis of components from the signal recognition particle in protozoa and fungi. BMC Genom. 2004, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Rosenblad, M.A. SRPDB: Signal Recognition Particle Database. Nucleic Acids Res. 2003, 31, 363–364. [Google Scholar] [CrossRef] [PubMed]
- Helmers, J.; Schmidt, D.; Glavy, J.S.; Blobel, G.; Schwartz, T. The β-Subunit of the Protein-conducting Channel of the Endoplasmic Reticulum Functions as the Guanine Nucleotide Exchange Factor for the β-Subunit of the Signal Recognition Particle Receptor. J. Biol. Chem. 2003, 278, 23686–23690. [Google Scholar] [CrossRef] [PubMed]
- Ogg, S.C.; Poritz, M.A.; Walter, P. Signal recognition particle receptor is important for cell growth and protein secretion in Saccharomyces cerevisiae. MBoC 1992, 3, 895–911. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pfeffer, S.; Burbaum, L.; Unverdorben, P.; Pech, M.; Chen, Y.; Zimmermann, R.; Beckmann, R.; Förster, F. Structure of the native Sec61 protein-conducting channel. Nat. Commun. 2015, 6, 8403. [Google Scholar] [CrossRef]
- Regnacq, M.; Hewitt, E.; Allen, J.; Rosamond, J.; Stirling, C.J. Deletion analysis of yeast Sec65p reveals a central domain that is sufficient for function in vivo. Mol. Microbiol. 1998, 29, 753–762. [Google Scholar] [CrossRef]
- Schlenker, O.; Hendricks, A.; Sinning, I.; Wild, K. The Structure of the Mammalian Signal Recognition Particle (SRP) Receptor as Prototype for the Interaction of Small GTPases with Longin Domains. J. Biol. Chem. 2006, 281, 8898–8906. [Google Scholar] [CrossRef]
- Brodsky, J.L.; Goeckeler, J.; Schekman, R. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 1995, 92, 9643–9646. [Google Scholar] [CrossRef]
- Corsi, A.K.; Schekman, R. Mechanism of Polypeptide Translocation into the Endoplasmic Reticulum. J. Biol. Chem. 1996, 271, 30299–30302. [Google Scholar] [CrossRef] [PubMed]
- De la Rosa, J.M.; González, J.M.; Gutiérrez, F.; Ruíz, T.; Rodríguez, L. Characterization ofCandida albicans orthologue of theSaccharomyces cerevisiae signal-peptidase-subunit encoding geneSPC3. Yeast 2004, 21, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Chaillot, J.; Tebbji, F.; Remmal, A.; Boone, C.; Brown, G.W.; Bellaoui, M.; Sellam, A. The Monoterpene Carvacrol Generates Endoplasmic Reticulum Stress in the Pathogenic Fungus Candida albicans. Antimicrob. Agents Chemother. 2015, 59, 4584–4592. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Martín, E.; Ongay-Larios, L.; Kawasaki, L.; Vincent, O.; Coello, G.; Coria, R.; Escalante, R. IreA controls endoplasmic reticulum stress-induced autophagy and survival through homeostasis recovery. Mol. Cell. Biol. 2018, 38, MCB.00054-18. [Google Scholar] [CrossRef] [PubMed]
- Schlenstedt, G.; Gudmundsson, G.; Boman, H.G.; Zimmermann, R. A large presecretory protein translocates both cotranslationally, using signal recognition particle and ribosome, and post-translationally, without these ribonucleoparticles, when synthesized in the presence of mammalian microsomes. J. Boil. Chem. 1990, 265, 13960–13968. [Google Scholar]
- Fewell, S.W.; Brodsky, J.L. Entry into the Endoplasmic Reticulum: Protein Translocation, Folding and Quality Control. In Trafficking Inside Cells; Springer: New York, NY, USA, 2009; pp. 119–142. ISBN 978-0-387-93876-9. [Google Scholar]
- Morrow, M.W.; Janke, M.R.; Lund, K.; Morrison, E.P.; Paulson, B.A. The Candida albicans Kar2 protein is essential and functions during the translocation of proteins into the endoplasmic reticulum. Curr.Genet. 2011, 57, 25–37. [Google Scholar] [CrossRef]
- Gong, Y.; Li, T.; Yu, C.; Sun, S. Candida albicans Heat Shock Proteins and Hsps-Associated Signaling Pathways as Potential Antifungal Targets. Front. Cell. Infect. Microbiol. 2017, 7, 520. [Google Scholar] [CrossRef]
- López-Ribot, J.L.; Alloush, H.M.; Masten, B.J.; Chaffin, W.L. Evidence for presence in the cell wall of Candida albicans of a protein related to the hsp70 family. Infect. Immun. 1996, 64, 3333–3340. [Google Scholar] [CrossRef]
- Sharma, D.; Martineau, C.N.; Le Dall, M.-T.; Reidy, M.; Masison, D.C.; Kabani, M. Function of SSA Subfamily of Hsp70 Within and Across Species Varies Widely in Complementing Saccharomyces cerevisiae Cell Growth and Prion Propagation. PLoS ONE 2009, 4, e6644. [Google Scholar] [CrossRef]
- Becker, J.; Walter, W.; Yan, W.; Craig, E.A. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 1996, 16, 4378–4386. [Google Scholar] [CrossRef]
- Verghese, J.; Abrams, J.; Wang, Y.; Morano, K.A. Biology of the Heat Shock Response and Protein Chaperones: Budding Yeast (Saccharomyces cerevisiae) as a Model System. Microbiol. Mol. Biol. Rev. 2012, 76, 115–158. [Google Scholar] [CrossRef] [PubMed]
- Li, X.S.; Sun, J.N.; Okamoto-Shibayama, K.; Edgerton, M. Candida albicans Cell Wall Ssa Proteins Bind and Facilitate Import of Salivary Histatin 5 Required for Toxicity. J. Biol. Chem. 2006, 281, 22453–22463. [Google Scholar] [CrossRef] [PubMed]
- Maneu, V.; Cervera, A.M.; Martinez, J.P.; Gozalbo, D. Molecular Cloning of a Candida albicans Gene (SSB1) Coding for a Protein Related to the Hsp70 Family. Yeast 1997, 13, 677–681. [Google Scholar] [CrossRef]
- Nimrichter, L.; De Souza, M.M.; Del Poeta, M.; Nosanchuk, J.D.; Joffe, L.; Tavares, P.D.M.; Rodrigues, M.L. Extracellular Vesicle-Associated Transitory Cell Wall Components and Their Impact on the Interaction of Fungi with Host Cells. Front. Microbiol. 2016, 7, 1034. [Google Scholar] [CrossRef]
- Pitarch, A.; Pardo, M.; Jiménez, A.; Pla, J.; Gil, C.; Sánchez, M.; Nombela, C. Two-dimensional gel electrophoresis as analytical tool for identifying Candida albicans immunogenic proteins. Electrophoresis 1999, 20, 1001–1010. [Google Scholar] [CrossRef]
- Pitarch, A.; Jiménez, A.; Nombela, C.; Gil, C. Decoding Serological Response to Candida Cell Wall Immunome into Novel Diagnostic, Prognostic, and Therapeutic Candidates for Systemic Candidiasis by Proteomic and Bioinformatic Analyses. Mol. Cell.Proteom. 2006, 5, 79–96. [Google Scholar] [CrossRef]
- Luo, T.; Krüger, T.; Knüpfer, U.; Kasper, L.; Wielsch, N.; Hube, B.; Kortgen, A.; Bauer, M.; Giamarellos-Bourboulis, E.J.; Dimopoulos, G.; et al. Immunoproteomic Analysis of Antibody Responses to Extracellular Proteins of Candida albicans Revealing the Importance of Glycosylation for Antigen Recognition. J. Proteome Res. 2016, 15, 2394–2406. [Google Scholar] [CrossRef]
- Bromuro, C.; Torosantucci, A.; Gomez, M.J.; Urbani, F.; Cassone, A. Differential release of an immunodominant 65 kDa mannoprotein antigen from yeast and mycelial forms of Candida albicans. J. Med Vet. Mycol. 1994, 32, 447–459. [Google Scholar] [CrossRef]
- Nagao, J.; Cho, T.; Uno, J.; Ueno, K.; Imayoshi, R.; Nakayama, H.; Chibana, H.; Kaminishi, H. Candida albicans Msi3p, a homolog of the Saccharomyces cerevisiae Sse1p of the Hsp70 family, is involved in cell growth and fluconazole tolerance. FEMS Yeast Res. 2012, 12, 728–737. [Google Scholar] [CrossRef]
- Deshaies, R.J.; Sanders, S.L.; Feldheim, D.A.; Schekman, R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature 1991, 349, 806–808. [Google Scholar] [CrossRef]
- Johnson, N.; Powis, K.; High, S. Post-translational translocation into the endoplasmic reticulum. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2013, 1833, 2403–2409. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.-J.; Kim, J.E.H.; Reithinger, J.H.; Kim, H. The Sec62-Sec63 translocon facilitates translocation of the C-terminus of membrane proteins. J. Cell Sci. 2014, 127, 4270–4278. [Google Scholar] [CrossRef] [PubMed]
- Matlack, K.E.S.; Misselwitz, B.; Plath, K.; Rapoport, T.A. BiP Acts as a Molecular Ratchet during Posttranslational Transport of Prepro-alphaFactor across the ER Membrane. Cell 1999, 97, 553–564. [Google Scholar] [CrossRef]
- Johnson, A.E.; Van Waes, M.A. The Translocon: A Dynamic Gateway at the ER Membrane. Annu. Rev. Cell Dev. Biol. 1999, 15, 799–842. [Google Scholar] [CrossRef] [PubMed]
- Reyna-Beltrán, E.; Isaac Bazán Méndez, C.; Iranzo, M.; Mormeneo, S.; Pedro Luna-Arias, J. The Cell Wall of Candida albicans: A Proteomics View. In Candida Albicans; Sandai, D., Ed.; IntechOpen: London, UK, 2019; ISBN 978-1-83880-159-5. [Google Scholar]
- Tiwari, S.; Thakur, R.; Shankar, J. Role of Heat-Shock Proteins in Cellular Function and in the Biology of Fungi. Biotechnol. Res. Int. 2015, 2015, 132635. [Google Scholar] [CrossRef]
- Neckers, L.; Tatu, U. Molecular Chaperones in Pathogen Virulence: Emerging New Targets for Therapy. Cell Host Microbe 2008, 4, 519–527. [Google Scholar] [CrossRef]
- Bukau, B.; Deuerling, E.; Pfund, C.; Craig, E.A. Getting Newly Synthesized Proteins into Shape. Cell 2000, 101, 119–122. [Google Scholar] [CrossRef]
- Ciechanover, A.; Kwon, Y.T. Protein Quality Control by Molecular Chaperones in Neurodegeneration. Front. Neurosci. 2017, 11, 185. [Google Scholar] [CrossRef]
- Hernández, M.P.; Sullivan, W.P.; Toft, D.O. The Assembly and Intermolecular Properties of the hsp70-Hop-hsp90 Molecular Chaperone Complex. J. Biol. Chem. 2002, 277, 38294–38304. [Google Scholar] [CrossRef]
- Fan, C.-Y.; Lee, S.; Cyr, D.M. Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress 2003, 8, 309–316. [Google Scholar] [CrossRef]
- Xie, J.L.; Bohovych, I.; Wong, E.O.Y.; Lambert, J.-P.; Gingras, A.-C.; Khalimonchuk, O.; Cowen, L.E.; Leach, M.D. Ydj1 governs fungal morphogenesis and stress response, and facilitates mitochondrial protein import via Mas1 and Mas2. Microb. Cell 2017, 4, 342–361. [Google Scholar] [CrossRef] [PubMed]
- Greene, M.K.; Maskos, K.; Landry, S.J. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl. Acad. Sci. USA 1998, 95, 6108–6113. [Google Scholar] [CrossRef] [PubMed]
- Sahi, C.; Craig, E.A. Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. USA 2007, 104, 7163–7168. [Google Scholar] [CrossRef] [PubMed]
- Makio, T.; Nishikawa, S.; Nakayama, T.; Nagai, H.; Endo, T. Identification and characterization of a Jem1p ortholog of Candida albicans: Dissection of Jem1p functions in karyogamy and protein quality control in Saccharomyces cerevisiae. Genes Cells 2008, 13, 1015–1026. [Google Scholar] [CrossRef]
- Hernández-Elvira, M.; Torres-Quiroz, F.; Escamilla-Ayala, A.; Domínguez-Martin, E.; Escalante, R.; Kawasaki, L.; Ongay-Larios, L.; Coria, R. The Unfolded Protein Response Pathway in the Yeast Kluyveromyces lactis. A Comparative View among Yeast Species. Cells 2018, 7, 106. [Google Scholar] [CrossRef]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat.Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef]
- Miyazaki, T.; Nakayama, H.; Nagayoshi, Y.; Kakeya, H.; Kohno, S. Dissection of Ire1 Functions Reveals Stress Response Mechanisms Uniquely Evolved in Candida glabrata. PLoS Pathog. 2013, 9, e1003160. [Google Scholar] [CrossRef]
- Kastora, S.L.; Herrero-de-Dios, C.; Avelar, G.M.; Munro, C.A.; Brown, A.J.P. Sfp1 and Rtg3 reciprocally modulate carbon source-conditional stress adaptation in the pathogenic yeast Candida albicans. Mol. Microbiol. 2017, 105, 620–636. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Chen, H.-F.; Yeh, Y.-C.; Xue, Y.-P.; Lan, C.-Y. The Transcription Factor Sfp1 Regulates the Oxidative Stress Response in Candida albicans. Microorganisms 2019, 7, 131. [Google Scholar] [CrossRef]
- Zhang, X.; De Micheli, M.; Coleman, S.T.; Sanglard, D.; Moye-Rowley, W.S. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p: Regulation of C. albicans Cap1p. Mol. Microbiol. 2002, 36, 618–629. [Google Scholar] [CrossRef]
- Yu, Q.; Zhang, B.; Li, J.; Zhang, B.; Wang, H.; Li, M. Endoplasmic reticulum-derived reactive oxygen species (ROS) is involved in toxicity of cell wall stress to Candida albicans. Free Radic. Biol. Med. 2016, 99, 572–583. [Google Scholar] [CrossRef]
- Yu, Q.; Ding, X.; Zhang, B.; Xu, N.; Cheng, X.; Qian, K.; Zhang, B.; Xing, L.; Li, M. The P-type ATPase Spf1 is required for endoplasmic reticulum functions and cell wall integrity in Candida albicans. Int. J. Med Microbiol. 2013, 303, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Ma, T.; Ma, C.; Zhang, B.; Li, M. Multifunction of the ER P-Type Calcium Pump Spf1 during Hyphal Development in Candida albicans. Mycopathologia 2019, 184, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Wang, H.; Xu, N.; Cheng, X.; Wang, Y.; Zhang, B.; Xing, L.; Li, M. Spf1 strongly influences calcium homeostasis, hyphal development, biofilm formation and virulence in Candida albicans. Microbiology 2012, 158, 2272–2282. [Google Scholar] [CrossRef] [PubMed]
- Feige, M.J.; Hendershot, L.M. Disulfide bonds in ER protein folding and homeostasis. Curr. Opin. Cell Biol. 2011, 23, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Sideris, D.P.; Sevier, C.S.; Kaiser, C.A. Balanced Ero1 activation and inactivation establishes ER redox homeostasis. J. Cell Biol. 2012, 196, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.L.; Plaine, A. Comprehensive Analysis of Glycosylphosphatidylinositol-Anchored Proteins in Candida albicans. Eukaryot. Cell 2007, 6, 119–133. [Google Scholar] [CrossRef]
- Jain, P.; Sethi, S.C.; Pratyusha, V.A.; Garai, P.; Naqvi, N.; Singh, S.; Pawar, K.; Puri, N.; Komath, S.S. Ras signaling activates glycosylphosphatidylinositol (GPI) anchor biosynthesis via the GPI– N -acetylglucosaminyltransferase (GPI–GnT) in Candida albicans. J. Biol. Chem. 2018, 293, 12222–12238. [Google Scholar] [CrossRef]
- Plaine, A.; Walker, L.; Da Costa, G.; Mora-Montes, H.M.; McKinnon, A.; Gow, N.A.R.; Gaillardin, C.; Munro, C.A.; Richard, M.L. Functional analysis of Candida albicans GPI-anchored proteins: Roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. 2008, 45, 1404–1414. [Google Scholar] [CrossRef]
- Martínez-Duncker, I.; Díaz-Jímenez, D.F.; Mora-Montes, H.M. Comparative Analysis of Protein Glycosylation Pathways in Humans and the Fungal Pathogen Candida albicans. Int. J. Microbiol. 2014, 2014, 267497. [Google Scholar] [CrossRef]
- Lehle, L.; Strahl, S.; Tanner, W. Protein Glycosylation, Conserved from Yeast to Man: A Model Organism Helps Elucidate Congenital Human Diseases. Angew. Chem. Int. Ed. 2006, 45, 6802–6818. [Google Scholar] [CrossRef] [PubMed]
- Shahana, S.; Mora-Montes, H.M.; Castillo, L.; Bohovych, I.; Sheth, C.C.; Odds, F.C.; Gow, N.A.R.; Brown, A.J.P. Reporters for the analysis of N-glycosylation in Candida albicans. Fungal Genet. Biol. 2013, 56, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Juchimiuk, M.; Kruszewska, J.; Palamarczyk, G. Dolichol phosphate mannose synthase from the pathogenic yeast Candida albicans is a multimeric enzyme. Biochim. Biophys. Acta (BBA) Gen. Subj. 2015, 1850, 2265–2275. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.A.; Bates, S.; Lenardon, M.D.; MacCallum, D.M.; Wagener, J.; Lowman, D.W.; Kruppa, M.D.; Williams, D.L.; Odds, F.C.; Brown, A.J.P.; et al. The Mnn2 Mannosyltransferase Family Modulates Mannoprotein Fibril Length, Immune Recognition and Virulence of Candida albicans. PLoS Pathog. 2013, 9, e1003276. [Google Scholar] [CrossRef]
- Ferreira, C.; Silva, S.; Faria-Oliveira, F.; Pinho, E.; Henriques, M.; Lucas, C. Candida albicans virulence and drug-resistance requires the O-acyltransferase Gup1p. BMC Microbiol. 2010, 10, 238. [Google Scholar] [CrossRef]
- Prill, S.K.-H.; Klinkert, B.; Timpel, C.; Gale, C.A.; Schröppel, K.; Ernst, J.F. PMT family of Candida albicans: Five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance: Pmt family. Mol. Microbiol. 2004, 55, 546–560. [Google Scholar] [CrossRef]
- Rouabhia, M.; Schaller, M.; Corbucci, C.; Vecchiarelli, A.; Prill, S.K.-H.; Giasson, L.; Ernst, J.F. Virulence of the Fungal Pathogen Candida albicans Requires the Five Isoforms of Protein Mannosyltransferases. Infect. Immun. 2005, 73, 4571–4580. [Google Scholar] [CrossRef]
- Bates, S.; Hughes, H.B.; Munro, C.A.; Thomas, W.P.H.; MacCallum, D.M.; Bertram, G.; Atrih, A.; Ferguson, M.A.J.; Brown, A.J.P.; Odds, F.C.; et al. Outer Chain N -Glycans Are Required for Cell Wall Integrity and Virulence of Candida albicans. J. Biol. Chem. 2006, 281, 90–98. [Google Scholar] [CrossRef]
- Netea, M.G. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J. Clin. Investig. 2006, 116, 1642–1650. [Google Scholar] [CrossRef]
- Murciano, C.; Moyes, D.L.; Runglall, M.; Islam, A.; Mille, C.; Fradin, C.; Poulain, D.; Gow, N.A.R.; Naglik, J.R. Candida albicans Cell Wall Glycosylation May Be Indirectly Required for Activation of Epithelial Cell Proinflammatory Responses. Infect. Immun. 2011, 79, 4902–4911. [Google Scholar] [CrossRef]
- Dennehy, K.M.; Ferwerda, G.; Faro-Trindade, I.; Pyż, E.; Willment, J.A.; Taylor, P.R.; Kerrigan, A.; Tsoni, S.V.; Gordon, S.; Meyer-Wentrup, F.; et al. Syk kinase is required for collaborative cytokine production induced through Dectin-1 and Toll-like receptors. Eur. J. Immunol. 2008, 38, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Gow, N.A.R.; Joosten, L.A.B.; Verschueren, I.; Van Der Meer, J.W.M.; Kullberg, B.J. Variable recognition of Candida albicans strains by TLR4 and lectin recognition receptors. Med. Mycol. 2010, 48, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Cota, E.; Hoyer, L.L. The Candida albicans agglutinin-like sequence family of adhesins: Functional insights gained from structural analysis. Future Microbiol. 2015, 10, 1635-1548. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.C.S.; Orci, L.; Hamamoto, S.; Futai, E.; Ravazzola, M.; Schekman, R. Sar1p N-Terminal Helix Initiates Membrane Curvature and Completes the Fission of a COPII Vesicle. Cell 2005, 122, 605–617. [Google Scholar] [CrossRef]
- Miller, E.A.; Beilharz, T.H.; Malkus, P.N.; Lee, M.C.S.; Hamamoto, S.; Orci, L.; Schekman, R. Multiple Cargo Binding Sites on the COPII Subunit Sec24p Ensure Capture of Diverse Membrane Proteins into Transport Vesicles. Cell 2003, 114, 497–509. [Google Scholar] [CrossRef]
- Bevis, B.J.; Hammond, A.T.; Reinke, C.A.; Glick, B.S. De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat. Cell Biol. 2002, 4, 750–756. [Google Scholar] [CrossRef]
- Shindiapina, P.; Barlowe, C. Requirements for Transitional Endoplasmic Reticulum Site Structure and Function in Saccharomyces cerevisiae. MBoC 2010, 21, 1530–1545. [Google Scholar] [CrossRef]
- Bharucha, N.; Liu, Y.; Papanikou, E.; McMahon, C.; Esaki, M.; Jeffrey, P.D.; Hughson, F.M.; Glick, B.S. Sec16 influences transitional ER sites by regulating rather than organizing COPII. MBoC 2013, 24, 3406–3419. [Google Scholar] [CrossRef]
- Budnik, A.; Stephens, D.J. ER exit sites—Localization and control of COPII vesicle formation. FEBS Lett. 2009, 583, 3796–3803. [Google Scholar] [CrossRef]
- Nantel, A.; Dignard, D.; Bachewich, C.; Harcus, D.; Marcil, A.; Bouin, A.-P.; Sensen, C.W. Transcription Profiling of Candida albicans Cells Undergoing the Yeast-to-Hyphal Transition□D. Mol. Biol. Cell 2002, 13, 14. [Google Scholar] [CrossRef]
- Matsuoka, K.; Schekman, R.; Orci, L.; Heuser, J.E. Surface structure of the COPII-coated vesicle. Proc. Natl. Acad. Sci. USA 2001, 98, 13705–13709. [Google Scholar] [CrossRef] [PubMed]
- Lederkremer, G.Z.; Cheng, Y.; Petre, B.M.; Vogan, E.; Springer, S.; Schekman, R.; Walz, T.; Kirchhausen, T. Structure of the Sec23p/24p and Sec13p/31p complexes of COPII. Proc. Natl. Acad. Sci. USA 2001, 98, 10704–10709. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Lee, C.-M.; Shen, S.-H. Functional characterization of theCandida albicans homologue of secretion-associated and Ras-related (Sar1) protein. Yeast 2002, 19, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Monteoliva, L.; López Matas, M.; Gil, C.; Nombela, C.; Pla, J. Large-Scale Identification of Putative Exported Proteins in Candida albicans by Genetic Selection. Eukaryot. Cell 2002, 1, 514–525. [Google Scholar] [CrossRef][Green Version]
- Barlowe, C.; d’Enfert, C.; Schekman, R. Purification and characterization of SAR1p, a small GTP-binding protein required for transport vesicle formation from the endoplasmic reticulum. J. Biol. Chem. 1993, 268, 873–879. [Google Scholar]
- Bao, J.; Huang, M.; Petranovic, D.; Nielsen, J. Moderate Expression of SEC16 Increases Protein Secretion by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2017, 83, e03400–e03416. [Google Scholar] [CrossRef]
- Supek, F.; Madden, D.T.; Hamamoto, S.; Orci, L.; Schekman, R. Sec16p potentiates the action of COPII proteins to bud transport vesicles. J. Cell Biol. 2002, 158, 1029–1038. [Google Scholar] [CrossRef]
- Watson, P.; Townley, A.K.; Koka, P.; Palmer, K.J.; Stephens, D.J. Sec16 Defines Endoplasmic Reticulum Exit Sites and is Required for Secretory Cargo Export in Mammalian Cells. Traffic 2006, 7, 1678–1687. [Google Scholar] [CrossRef]
- Antonny, B.; Madden, D.; Hamamoto, S.; Orci, L.; Schekman, R. Dynamics of the COPII coat with GTP and stable analogues. Nat. Cell Biol. 2001, 3, 531–537. [Google Scholar] [CrossRef]
- Bi, X.; Corpina, R.A.; Goldberg, J. Structure of the Sec23/24–Sar1 pre-budding complex of the COPII vesicle coat. Nature 2002, 419, 271–277. [Google Scholar] [CrossRef]
- Antonny, B.; Beraud-Dufour, S.; Chardin, P.; Chabre, M. N-Terminal Hydrophobic Residues of the G-Protein ADP-Ribosylation Factor-1 Insert into Membrane Phospholipids upon GDP to GTP Exchange †. Biochemistry 1997, 36, 4675–4684. [Google Scholar] [CrossRef] [PubMed]
- Gimeno, R.E.; Espenshade, P.; Kaisert, C.A. COPII Coat Subunit Interactions: Sec24p and Sec23p Bind to Adjacent Regions of Secl6p. Mol. Biol. Cell 1996, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Willger, S.D.; Liu, Z.; Olarte, R.A.; Adamo, M.E.; Stajich, J.E.; Myers, L.C.; Kettenbach, A.N.; Hogan, D.A. Analysis of the Candida albicans Phosphoproteome. Eukaryot. Cell 2015, 14, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Lan, C.-Y.; Rodarte, G.; Murillo, L.A.; Jones, T.; Davis, R.W.; Dungan, J.; Newport, G.; Agabian, N. Regulatory networks affected by iron availability in Candida albicans: Iron regulation in C. albicans. Mol. Microbiol. 2004, 53, 1451–1469. [Google Scholar] [CrossRef]
- Matsuoka, K.; Orci, L.; Amherdt, M.; Bednarek, S.Y.; Hamamoto, S.; Schekman, R.; Yeung, T. COPII-Coated Vesicle Formation Reconstituted with Purified Coat Proteins and Chemically Defined Liposomes. Cell 1998, 93, 263–275. [Google Scholar] [CrossRef]
- Fath, S.; Mancias, J.D.; Bi, X.; Goldberg, J. Structure and Organization of Coat Proteins in the COPII Cage. Cell 2007, 129, 1325–1336. [Google Scholar] [CrossRef]
- Kung, L.F.; Pagant, S.; Futai, E.; D’Arcangelo, J.G.; Buchanan, R.; Dittmar, J.C.; Reid, R.J.D.; Rothstein, R.; Hamamoto, S.; Snapp, E.L.; et al. Sec24p and Sec16p cooperate to regulate the GTP cycle of the COPII coat: Sec24p and Sec16p regulate the COPII GTP cycle. EMBO J. 2012, 31, 1014–1027. [Google Scholar] [CrossRef]
- Lee, S.A.; Khalique, Z.; Gale, C.A.; Wong, B. Intracellular trafficking of fluorescently tagged proteins associated with pathogenesis in Candida albicans. Med. Mycol. 2005, 43, 423–430. [Google Scholar] [CrossRef]
- Thomas, L.L.; Joiner, A.M.N.; Fromme, J.C. The TRAPPIII complex activates the GTPase Ypt1 (Rab1) in the secretory pathway. J. Cell Biol. 2018, 217, 283–298. [Google Scholar] [CrossRef]
- Sacher, M.; Kim, Y.-G.; Lavie, A.; Oh, B.-H.; Segev, N. The TRAPP Complex: Insights into its Architecture and Function. Traffic 2008, 9, 2032–2042. [Google Scholar] [CrossRef]
- Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat.Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Liu, Y.; Zhang, X.Q.; Chen, Y.; Ye, M.; Zhu, X.; Yang, S.; Lipatova, Z.; Liang, Y.; Segev, N. Modular TRAPP Complexes Regulate Intracellular Protein Trafficking Through Multiple Ypt/Rab GTPases in Saccharomyces cerevisiae. Genetics 2012, 191, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Yu, S.; Menon, S.; Cai, Y.; Lazarova, D.; Fu, C.; Reinisch, K.; Hay, J.C.; Ferro-Novick, S. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature 2007, 445, 941–944. [Google Scholar] [CrossRef] [PubMed]
- Brunet, S.; Noueihed, B.; Shahrzad, N.; Saint-Dic, D.; Hasaj, B.; Guan, T.L.; Moores, A.; Barlowe, C.; Sacher, M. The SMS domain of Trs23p is responsible for the in vitro appearance of the TRAPP I complex in Saccharomyces cerevisiae. Cell. Logist. 2012, 2, 28–42. [Google Scholar] [CrossRef][Green Version]
- Lynch-Day, M.A.; Bhandari, D.; Menon, S.; Huang, J.; Cai, H.; Bartholomew, C.R.; Brumell, J.H.; Ferro-Novick, S.; Klionsky, D.J. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA 2010, 107, 7811–7816. [Google Scholar] [CrossRef]
- Oh, J.; Fung, E.; Schlecht, U.; Davis, R.W.; Giaever, G.; St. Onge, R.P.; Deutschbauer, A.; Nislow, C. Gene Annotation and Drug Target Discovery in Candida albicans with a Tagged Transposon Mutant Collection. PLoS Pathog. 2010, 6, e1001140. [Google Scholar] [CrossRef]
- Chen, Y.; Mallick, J.; Maqnas, A.; Sun, Y.; Choudhury, B.I.; Cote, P.; Yan, L.; Ni, T.-J.-H.; Li, Y.; Zhang, D.; et al. Chemogenomic Profiling of the Fungal Pathogen Candida albicans. Antimicrob. Agents Chemother. 2018, 62, e02365-17. [Google Scholar] [CrossRef]
- Cao, X. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998, 17, 2156–2165. [Google Scholar] [CrossRef]
- Sapperstein, S.K.; Lupashin, V.V.; Schmitt, H.D.; Waters, M.G. Assembly of the ER to Golgi SNARE Complex Requires Usolp. J. Cell Biol. 1996, 132, 13. [Google Scholar] [CrossRef]
- Romo, J.A.; Zhang, H.; Cai, H.; Kadosh, D.; Koehler, J.R.; Saville, S.P.; Wang, Y.; Lopez-Ribot, J.L. Global Transcriptomic Analysis of the Candida albicans Response to Treatment with a Novel Inhibitor of Filamentation. mSphere 2019, 4, e00620-19. [Google Scholar] [CrossRef]
- Cavalieri, D.; Di Paola, M.; Rizzetto, L.; Tocci, N.; De Filippo, C.; Lionetti, P.; Ardizzoni, A.; Colombari, B.; Paulone, S.; Gut, I.G.; et al. Genomic and Phenotypic Variation in Morphogenetic Networks of Two Candida albicans Isolates Subtends Their Different Pathogenic Potential. Front. Immunol. 2018, 8, 1997. [Google Scholar] [CrossRef] [PubMed]
- Veses, V.; Richards, A.; Gow, N.A.R. Vacuole inheritance regulates cell size and branching frequency of Candida albicans hyphae. Mol. Microbiol. 2009, 71, 505–519. [Google Scholar] [CrossRef] [PubMed]
- Herrero, A.B.; Uccelletti, D.; Hirschberg, C.B.; Dominguez, A.; Abeijon, C. The Golgi GDPase of the Fungal Pathogen Candida albicans Affects Morphogenesis, Glycosylation, and Cell Wall Properties. Eukaryot. Cell 2002, 1, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Munro, C.A.; Bates, S.; Buurman, E.T.; Hughes, H.B.; MacCallum, D.M.; Bertram, G.; Atrih, A.; Ferguson, M.A.J.; Bain, J.M.; Brand, A.; et al. Mnt1p and Mnt2p of Candida albicans Are Partially Redundant α-1,2-Mannosyltransferases That Participate in O -Linked Mannosylation and Are Required for Adhesion and Virulence. J. Biol. Chem. 2005, 280, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.; MacCallum, D.M.; Bertram, G.; Munro, C.A.; Hughes, H.B.; Buurman, E.T.; Brown, A.J.P.; Odds, F.C.; Gow, N.A.R. Candida albicans Pmr1p, a Secretory Pathway P-type Ca 2+ /Mn 2+ -ATPase, Is Required for Glycosylation and Virulence. J. Biol. Chem. 2005, 280, 23408–23415. [Google Scholar] [CrossRef]
- Carvalho-Pereira, J.; Vaz, C.; Carneiro, C.; Pais, C.; Sampaio, P. Genetic Variability of Candida albicans Sap8 Propeptide in Isolates from Different Types of Infection. BioMed Res. Int. 2015, 2015, 148343. [Google Scholar] [CrossRef]
- Newport, G.; Agabian, N. KEX2 Influences Candida albicans Proteinase Secretion and Hyphal Formation. J. Biol. Chem. 1997, 272, 28954–28961. [Google Scholar] [CrossRef]
- Glick, B.S.; Nakano, A. Membrane Traffic within the Golgi Apparatus. Annu. Rev. Cell Dev. Biol. 2009, 25, 113–132. [Google Scholar] [CrossRef]
- Zhang, B.; Yu, Q.; Huo, D.; Li, J.; Liang, C.; Li, H.; Yi, X.; Xiao, C.; Zhang, D.; Li, M. Arf1 regulates the ER –mitochondria encounter structure (ERMES) in a reactive oxygen species-dependent manner. FEBS J. 2018, 285, 2004–2018. [Google Scholar] [CrossRef]
- Labbaoui, H.; Bogliolo, S.; Ghugtyal, V.; Solis, N.V.; Filler, S.G.; Arkowitz, R.A.; Bassilana, M. Role of Arf GTPases in fungal morphogenesis and virulence. PLoS Pathog. 2017, 13, e1006205. [Google Scholar] [CrossRef]
- Wakade, R.; Labbaoui, H.; Stalder, D.; Arkowitz, R.A.; Bassilana, M. Overexpression of YPT6 restores invasive filamentous growth and secretory vesicle clustering in a Candida albicans arl1 mutant. Small GTPases 2017, 29, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Bannykh, S.I.; Plutner, H.; Matteson, J.; Balch, W.E. The Role of ARF1 and Rab GTPases in Polarization of the Golgi Stack: ARF1-Independent Golgi Sorting. Traffic 2005, 6, 803–819. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Solis, N.V.; Heilmann, C.J.; Phan, Q.T.; Mitchell, A.P.; Klis, F.M.; Filler, S.G. Role of Retrograde Trafficking in Stress Response, Host Cell Interactions, and Virulence of Candida albicans. Eukaryot. Cell 2014, 13, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhang, Y.; Pypaert, M.; Walker, L.; Ferro-Novick, S. Mutants in trs120 disrupt traffic from the early endosome to the late Golgi. J. Cell Biol. 2005, 171, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.; Davey, M.; Schluter, C.; Pandher, P.; Fang, Y.; Foster, L.J.; Conibear, E. Organization and Assembly of the TRAPPII Complex. Traffic 2011, 12, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Suvorova, E.S.; Duden, R.; Lupashin, V.V. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J. Cell Biol. 2002, 157, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Lupashin, V.; Sztul, E. Golgi tethering factors. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2005, 1744, 325–339. [Google Scholar] [CrossRef]
- Peng, R.; Gallwitz, D. Sly1 protein bound to Golgi syntaxin Sed5p allows assembly and contributes to specificity of SNARE fusion complexes. J. Cell Biol. 2002, 157, 645–655. [Google Scholar] [CrossRef]
- Klionsky, D.J. The Fungal Vacuole: Composition, Function, and Biogenesis. Microbiol. Rev. 1990, 54, 27. [Google Scholar] [CrossRef]
- Gerrard, S.R.; Bryant, N.J.; Stevens, T.H. VPS21 Controls Entry of Endocytosed and Biosynthetic Proteins into the Yeast Prevacuolar Compartment. MBoC 2000, 11, 613–626. [Google Scholar] [CrossRef]
- Palmer, G.E. Endosomal and AP-3-Dependent Vacuolar Trafficking Routes Make Additive Contributions to Candida albicans Hyphal Growth and Pathogenesis. Eukaryot. Cell 2010, 9, 1755–1765. [Google Scholar] [CrossRef] [PubMed]
- Palmer, G.E. Vacuolar trafficking and Candida albicans pathogenesis. Commun. Integr. Biol. 2011, 4, 240–242. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Williams, R.L.; Urbé, S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007, 8, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Horner, D.S.; Pasini, M.E.; Beltrame, M.; Mastrodonato, V.; Morelli, E.; Vaccari, T. ESCRT genes and regulation of developmental signaling. Semin. Cell Dev. Biol. 2018, 74, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Smith, F.J.; Subaran, R.; Mitchell, A.P. Multivesicular Body-ESCRT Components Function in pH Response Regulation in Saccharomyces cerevisiae and Candida albicans. MBoC 2004, 15, 5528–5537. [Google Scholar] [CrossRef] [PubMed]
- Gale, C.A. Linkage of Adhesion, Filamentous Growth, and Virulence in Candida albicans to a Single Gene, INT1. Science 1998, 279, 1355–1358. [Google Scholar] [CrossRef]
- Wolf, J.M.; Johnson, D.J.; Chmielewski, D.; Davis, D.A. The Candida albicans ESCRT Pathway Makes Rim101-Dependent and -Independent Contributions to Pathogenesis. Eukaryot. Cell 2010, 9, 1203–1215. [Google Scholar] [CrossRef]
- Babst, M. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997, 16, 1820–1831. [Google Scholar] [CrossRef]
- Babst, M. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998, 17, 2982–2993. [Google Scholar] [CrossRef]
- Katzmann, D.J.; Odorizzi, G.; Emr, S.D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002, 3, 893–905. [Google Scholar] [CrossRef]
- Russell, M.R.G.; Shideler, T.; Nickerson, D.P.; West, M.; Odorizzi, G. Class E compartments form in response to ESCRT dysfunction in yeast due to hyperactivity of the Vps21 Rab GTPase. J. Cell Sci. 2012, 125, 5208–5220. [Google Scholar] [CrossRef]
- Lee, S.A.; Jones, J.; Hardison, S.; Kot, J.; Khalique, Z.; Bernardo, S.M.; Lazzell, A.; Monteagudo, C.; Lopez-Ribot, J. Candida albicans VPS4 is Required for Secretion of Aspartyl Proteases and In Vivo Virulence. Mycopathologia 2009, 167, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.P.; Lopez-Ribot, J.L.; Lee, S.A. A proteomic analysis of secretory proteins of a pre-vacuolar mutant of Candida albicans. J. Proteom. 2009, 73, 342–351. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rane, H.S.; Hardison, S.; Botelho, C.; Bernardo, S.M.; Wormley, F.; Lee, S.A. Candida albicansVPS4 contributes differentially to epithelial and mucosal pathogenesis. Virulence 2014, 5, 810–818. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Weisman, L.S.; Emr, S.D.; Wickner, W.T. Mutants of Saccharomyces cerevisiae that block intervacuole vesicular traffic and vacuole division and segregation. Proc. Natl. Acad. Sci. USA 1990, 87, 1076–1080. [Google Scholar] [CrossRef]
- Weisman, L.S.; Wickner, W. Molecular characterization of VAC1, a gene required for vacuole inheritance and vacuole protein sorting. J. Biol. Chem. 1992, 267, 618–623. [Google Scholar]
- Franke, K. The vesicle transport protein Vac1p is required for virulence of Candida albicans. Microbiology 2006, 152, 3111–3121. [Google Scholar] [CrossRef]
- Bryant, N.J.; Stevens, T.H. Vacuole Biogenesis in Saccharomyces cerevisiae: Protein Transport Pathways to the Yeast Vacuole. Microbiol. Mol. Biol. Rev. 1998, 62, 18. [Google Scholar] [CrossRef]
- Bernardo, S.M.; Khalique, Z.; Kot, J.; Jones, J.K.; Lee, S.A. Candida albicans VPS1 contributes to protease secretion, filamentation, and biofilm formation. Fungal Genet. Biol. 2008, 45, 861–877. [Google Scholar] [CrossRef]
- Kane, P.M. The Where, When, and How of Organelle Acidification by the Yeast Vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 2006, 70, 15. [Google Scholar] [CrossRef]
- Rane, H.S.; Bernardo, S.M.; Raines, S.M.; Binder, J.L.; Parra, K.J.; Lee, S.A. Candida albicans VMA3 Is Necessary for V-ATPase Assembly and Function and Contributes to Secretion and Filamentation. Eukaryot. Cell 2013, 12, 1369–1382. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Umemoto, N.; Yoshihisa, T.; Hirata, R.; Anraku, Y. Roles of the VMA3 gene product, subunit c of the vacuolar membrane H(+)-ATPase on vacuolar acidification and protein transport. A study with VMA3-disrupted mutants of Saccharomyces cerevisiae. J. Biol. Chem. 1990, 265, 18447–18453. [Google Scholar] [PubMed]
- Parra, K.J. Vacuolar ATPase: A model proton pump for antifungal drug discovery. Antimicrob. Drug Discov. Emerg. Strateg. 2012, 89–100. [Google Scholar]
- Graham, L.A.; Flannery, A.R.; Stevens, T.H. Structure and Assembly of the Yeast V-ATPase. J. Bioenerg. Biomembr. 2003, 35, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki-Nishi, S.; Nishi, T.; Forgac, M. Yeast V-ATPase Complexes Containing Different Isoforms of the 100-kDa a-subunit Differ in Coupling Efficiency and in Vivo Dissociation. J. Biol. Chem. 2001, 276, 17941–17948. [Google Scholar] [CrossRef] [PubMed]
- Raines, S.M.; Rane, H.S.; Bernardo, S.M.; Binder, J.L.; Lee, S.A.; Parra, K.J. Deletion of Vacuolar Proton-translocating ATPase Voa Isoforms Clarifies the Role of Vacuolar pH as a Determinant of Virulence-associated Traits in Candida albicans. J. Biol. Chem. 2013, 288, 6190–6201. [Google Scholar] [CrossRef] [PubMed]
- Patenaude, C.; Zhang, Y.; Cormack, B.; Köhler, J.; Rao, R. Essential Role for Vacuolar Acidification in Candida albicans Virulence. J. Biol. Chem. 2013, 288, 26256–26264. [Google Scholar] [CrossRef]
- Hayek, S.R.; Lee, S.A.; Parra, K.J. Advances in targeting the vacuolar proton-translocating ATPase (V-ATPase) for anti-fungal therapy. Front. Pharmacol. 2014, 5, 4. [Google Scholar] [CrossRef]
- Anraku, Y.; Umemoto, N.; Hirata, R.; Ohya, Y. Genetic and cell biological aspects of the yeast vacuolar H+-ATPase. J. Bioenerg. Biomembr. 1992, 24, 395–405. [Google Scholar] [CrossRef]
- Leng, X.-H.; Manolson, M.F.; Liu, Q.; Forgac, M. Site-directed Mutagenesis of the 100-kDa Subunit (Vph1p) of the Yeast Vacuolar (H+)-ATPase. J. Biol. Chem. 1996, 271, 22487–22493. [Google Scholar] [CrossRef]
- Rane, H.S.; Bernardo, S.M.; Hayek, S.R.; Binder, J.L.; Parra, K.J.; Lee, S.A. The Contribution of Candida albicans Vacuolar ATPase Subunit V1 B, Encoded by VMA2, to Stress Response, Autophagy, and Virulence Is Independent of Environmental pH. Eukaryot. Cell 2014, 13, 1207–1221. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, T.; Vasilyeva, E.; Forgac, M. Subunit Interactions in the Clathrin-coated Vesicle Vacuolar (H+)-ATPase Complex. J. Biol. Chem. 1999, 274, 28909–28915. [Google Scholar] [CrossRef] [PubMed]
- Lindorfers, M.A.; Stevensq, T.H. Isolation of Vacuolar Membrane H+-ATPase-deficient YeasMt utants; the VMAS and VMA4 Genes Are Essential for Assembly and Activity of the Vacuolar H+-ATPase. J. Biol. Chem. 1993, 268, 221–227. [Google Scholar]
- Tabke, K.; Albertmelcher, A.; Vitavska, O.; Huss, M.; Schmitz, H.-P.; Wieczorek, H. Reversible disassembly of the yeast V-ATPase revisited under in vivo conditions. Biochem. J. 2014, 462, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Jia, C.; Yu, Q.; Xiao, C.; Dong, Y.; Zhang, M.; Zhang, D.; Zhao, Q.; Zhang, B.; Li, M. Contribution of VMA5 to vacuolar function, stress response, ion homeostasis and autophagy in Candida albicans. Future Microbiol. 2017, 12, 1147–1166. [Google Scholar] [CrossRef] [PubMed]
- Poltermann, S.; Nguyen, M.; Günther, J.; Wendland, J.; Härtl, A.; Künkel, W.; Zipfel, P.F.; Eck, R. The putative vacuolar ATPase subunit Vma7p of Candida albicans is involved in vacuole acidification, hyphal development and virulence. Microbiology 2005, 151, 1645–1655. [Google Scholar] [CrossRef]
- Tarsio, M.; Zheng, H.; Smardon, A.M.; Martínez-Muñoz, G.A.; Kane, P.M. Consequences of Loss of Vph1 Protein-containing Vacuolar ATPases (V-ATPases) for Overall Cellular pH Homeostasis. J. Biol. Chem. 2011, 286, 28089–28096. [Google Scholar] [CrossRef]
- Smardon, A.M.; Kane, P.M. Loss of Vacuolar H+-ATPase Activity in Organelles Signals Ubiquitination and Endocytosis of the Yeast Plasma Membrane Proton pump Pma1p. J. Biol. Chem. 2014, 289, 32316–32326. [Google Scholar] [CrossRef]
- Monk, B.C.; Kurtz, M.B.; Marrinan, J.A.; Perlin, D.S. Cloning and characterization of the plasma membrane H(+)-ATPase from Candida albicans. J. Bacteriol. 1991, 173, 6826–6836. [Google Scholar] [CrossRef]
- Rane, H.S.; Hayek, S.R.; Frye, J.E.; Abeyta, E.L.; Bernardo, S.M.; Parra, K.J.; Lee, S.A. Candida albicans Pma1p Contributes to Growth, pH Homeostasis, and Hyphal Formation. Front. Microbiol. 2019, 10, 1012. [Google Scholar] [CrossRef]
- Pfeffer, S.R. Transport-vesicle targeting: Tethers before SNAREs. Nat. Cell Biol. 1999, 1, E17–E22. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Styles, C.A.; Feng, Q.; Fink, G.R. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 2000, 97, 12158–12163. [Google Scholar] [CrossRef] [PubMed]
- Novick, P.; Guo, W. Ras family therapy: Rab, Rho and Ral talk to the exocyst. Trends Cell Biol. 2002, 12, 247–249. [Google Scholar] [CrossRef]
- Roemer, T.; Jiang, B.; Davison, J.; Ketela, T.; Veillette, K.; Breton, A.; Tandia, F.; Linteau, A.; Sillaots, S.; Marta, C.; et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery: C. albicans essential gene identification and antifungal drug discovery. Mol. Microbiol. 2003, 50, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, D.; Sen, A.; Boettner, D.R.; Fairn, G.D.; Schlam, D.; Bonilla Valentin, F.J.; McCaffery, J.M.; Hazbun, T.; Staiger, C.J.; Grinstein, S.; et al. Bem3, a Cdc42 GTPase-activating protein, traffics to an intracellular compartment and recruits the secretory Rab GTPase Sec4 to endomembranes. J. Cell Sci. 2013, 126, 4560–4571. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.G.; Pfeffert, S.R. Membrane tethering in intracellular transport. Curr. Opin. Cell Biol. 1999, 11, 453–459. [Google Scholar] [CrossRef]
- Guo, W.; Grant, A.; Novick, P. Exo84p Is an Exocyst Protein Essential for Secretion. J. Biol. Chem. 1999, 274, 23558–23564. [Google Scholar] [CrossRef]
- Matern, H.T.; Yeaman, C.; Nelson, W.J.; Scheller, R.H. The Sec6/8 complex in mammalian cells: Characterization of mammalian Sec3, subunit interactions, and expression of subunits in polarized cells. Proc. Natl. Acad. Sci. USA 2001, 98, 9648–9653. [Google Scholar] [CrossRef]
- Boyd, C.; Hughes, T.; Pypaert, M.; Novick, P. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J. Cell Biol. 2004, 167, 889–901. [Google Scholar] [CrossRef]
- He, B.; Xi, F.; Zhang, X.; Zhang, J.; Guo, W. Exo70 interacts with phospholipids and mediates the targeting of the exocyst to the plasma membrane. EMBO J. 2007, 26, 4053–4065. [Google Scholar] [CrossRef]
- Liu, J.; Zuo, X.; Yue, P.; Guo, W. Phosphatidylinositol 4,5-Bisphosphate Mediates the Targeting of the Exocyst to the Plasma Membrane for Exocytosis in Mammalian Cells. Mol. Biol. Cell 2007, 18, 4483–4492. [Google Scholar] [CrossRef]
- Zhang, X.; Orlando, K.; He, B.; Xi, F.; Zhang, J.; Zajac, A.; Guo, W. Membrane association and functional regulation of Sec3 by phospholipids and Cdc42. J. Cell Biol. 2008, 180, 145–158. [Google Scholar] [CrossRef]
- Shen, D.; Yuan, H.; Hutagalung, A.; Verma, A.; Kümmel, D.; Wu, X.; Reinisch, K.; McNew, J.A.; Novick, P. The synaptobrevin homologue Snc2p recruits the exocyst to secretory vesicles by binding to Sec6p. J. Cell Biol. 2013, 202, 509–526. [Google Scholar] [CrossRef]
- Finger, F.P.; Novick, P. Sec3p is involved in secretion and morphogenesis in Saccharomyces cerevisiae. MBoC 1997, 8, 647–662. [Google Scholar] [CrossRef]
- Finger, F.P.; Hughes, T.; Novick, P. Sec3p is a spatial landmark for polarized secretion in budding yeast. Cell 1998, 92, 559–571. [Google Scholar] [CrossRef]
- Wiederkehr, A.; Du, Y.; Pypaert, M.; Ferro-Novick, S.; Novick, P. Sec3p Is Needed for the Spatial Regulation of Secretion and for the Inheritance of the Cortical Endoplasmic Reticulum. Mol. Biol. Cell 2003, 14, 4770–4782. [Google Scholar] [CrossRef]
- Luo, G.; Zhang, J.; Guo, W. The role of Sec3p in secretory vesicle targeting and exocyst complex assembly. MBoC 2014, 25, 3813–3822. [Google Scholar] [CrossRef]
- Mei, K.; Yue, P.; Guo, W. Analysis of the Role of Sec3 in SNARE Assembly and Membrane Fusion. In SNAREs; Fratti, R., Ed.; Springer: New York, NY, USA, 2019; Volume 1860, pp. 175–189. ISBN 978-1-4939-8759-7. [Google Scholar]
- Jones, L.A.; Sudbery, P.E. Spitzenkörper, Exocyst, and Polarisome Components in Candida albicans Hyphae Show Different Patterns of Localization and Have Distinct Dynamic Properties. Eukaryot. Cell 2010, 9, 1455–1465. [Google Scholar] [CrossRef]
- Jahn, R.; Scheller, R.H. SNAREs—Engines for membrane fusion. Nat.Rev. Mol. Cell Biol. 2006, 7, 631–643. [Google Scholar] [CrossRef]
- Rothman, J.E.; Warren, G. Implications of the SNARE hypothesis for intracellular membrane topology and dynamics. Curr. Biol. 1994, 4, 220–233. [Google Scholar] [CrossRef]
- Marash, M. t-SNARE dephosphorylation promotes SNARE assembly and exocytosis in yeast. EMBO J. 2001, 20, 411–421. [Google Scholar] [CrossRef]
- Sutton, R.B.; Fasshauer, D.; Jahn, R.; Brunger, A.T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature 1998, 395, 347–353. [Google Scholar] [CrossRef]
- Hattendorf, D.A.; Andreeva, A.; Gangar, A.; Brennwald, P.J.; Weis, W.I. Structure of the yeast polarity protein Sro7 reveals a SNARE regulatory mechanism. Nature 2007, 446, 567–571. [Google Scholar] [CrossRef]
- Rossi, G.; Brennwald, P. Yeast homologues of lethal giant larvae and type V myosin cooperate in the regulation of Rab-dependent vesicle clustering and polarized exocytosis. MBoC 2011, 22, 842–857. [Google Scholar] [CrossRef]
- Bernardo, S.M.; Rane, H.S.; Chavez-Dozal, A.; Lee, S.A. Secretion and filamentation are mediated by the Candida albicans t-SNAREs Sso2p and Sec9p. FEMS Yeast Res. 2014, 14, 762–775. [Google Scholar] [CrossRef]
- Novick, P.; Field, C.; Schekman, R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 1980, 21, 205–215. [Google Scholar] [CrossRef]
- Novick, P.; Ferro, S.; Schekman, R. Order of events in the yeast secretory pathway. Cell 1981, 25, 461–469. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nimrichter, L.; Oliveira, D.L.; Frases, S.; Miranda, K.; Zaragoza, O.; Alvarez, M.; Nakouzi, A.; Feldmesser, M.; Casadevall, A. Vesicular Polysaccharide Export in Cryptococcus neoformans Is a Eukaryotic Solution to the Problem of Fungal Trans-Cell Wall Transport. Eukaryot. Cell 2007, 6, 48–59. [Google Scholar] [CrossRef]
- Ellen, A.F.; Zolghadr, B.; Driessen, A.J.M.; Albers, S.-V. Shaping the Archaeal Cell Envelope. Archaea 2010, 2010, 608243. [Google Scholar] [CrossRef]
- Marcilla, A.; Martin-Jaular, L.; Trelis, M.; De Menezes-Neto, A.; Osuna, A.; Bernal, D.; Fernandez-Becerra, C.; Almeida, I.C.; Del Portillo, H.A. Extracellular vesicles in parasitic diseases. J. Extracell. Vesicles 2014, 3, 25040. [Google Scholar] [CrossRef]
- Albuquerque, P.C.; Nakayasu, E.S.; Rodrigues, M.L.; Frases, S.; Casadevall, A.; Zancope-Oliveira, R.M.; Almeida, I.C.; Nosanchuk, J.D. Vesicular transport in Histoplasma capsulatum: An effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell. Microbiol. 2008, 10, 1695–1710. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.L.; Nakayasu, E.; Oliveira, D.L.; Nimrichter, L.; Nosanchuk, J.D.; Almeida, I.C.; Casadevall, A. Extracellular Vesicles Produced by Cryptococcus neoformans Contain Protein Components Associated with Virulence. Eukaryot. Cell 2007, 7, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Vargas, G.; Rocha, J.D.B.; Oliveira, D.L.; Albuquerque, P.C.; Frases, S.; Santos, S.S.; Nosanchuk, J.D.; Gomes, A.M.O.; Medeiros, L.C.A.S.; Miranda, K.; et al. Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans: Extracellular vesicles from Candida albicans. Cell. Microbiol. 2015, 17, 389–407. [Google Scholar] [CrossRef] [PubMed]
- Zarnowski, R.; Sanchez, H.; Covelli, A.S.; Dominguez, E.; Jaromin, A.; Bernhardt, J.; Mitchell, K.F.; Heiss, C.; Azadi, P.; Mitchell, A.; et al. Candida albicans biofilm–induced vesicles confer drug resistance through matrix biogenesis. PLoS Biol. 2018, 16, e2006872. [Google Scholar] [CrossRef] [PubMed]
- Escrevente, C.; Keller, S.; Altevogt, P.; Costa, J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 2011, 11, 108. [Google Scholar] [CrossRef]
- Gonzales, P.A.; Pisitkun, T.; Hoffert, J.D.; Tchapyjnikov, D.; Star, R.A.; Kleta, R.; Wang, N.S.; Knepper, M.A. Large-Scale Proteomics and Phosphoproteomics of Urinary Exosomes. JASN 2009, 20, 363–379. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Nawrocki, A.; Jensen, S.G.; Thorsen, K.; Whitehead, B.; Howard, K.A.; Dyrskjøt, L.; Ørntoft, T.F.; Larsen, M.R.; Ostenfeld, M.S. Quantitative proteomics of fractionated membrane and lumen exosome proteins from isogenic metastatic and nonmetastatic bladder cancer cells reveal differential expression of EMT factors. Proteomics 2014, 14, 699–712. [Google Scholar] [CrossRef]
- Østergaard, O.; Nielsen, C.T.; Iversen, L.V.; Jacobsen, S.; Tanassi, J.T.; Heegaard, N.H.H. Quantitative Proteome Profiling of Normal Human Circulating Microparticles. J. Proteome Res. 2012, 11, 2154–2163. [Google Scholar] [CrossRef]
- Gil-Bona, A.; Llama-Palacios, A.; Parra, C.M.; Vivanco, F.; Nombela, C.; Monteoliva, L.; Gil, C. Proteomics Unravels Extracellular Vesicles as Carriers of Classical Cytoplasmic Proteins in Candida albicans. J. Proteome Res. 2015, 14, 142–153. [Google Scholar] [CrossRef]
- Gil-Bona, A.; Monteoliva, L.; Gil, C. Global Proteomic Profiling of the Secretome of Candida albicans ecm33 Cell Wall Mutant Reveals the Involvement of Ecm33 in Sap2 Secretion. J. Proteome Res. 2015, 14, 4270–4281. [Google Scholar] [CrossRef]
- Vallejo, M.C.; Matsuo, A.L.; Ganiko, L.; Medeiros, L.C.S.; Miranda, K.; Silva, L.S.; Freymüller-Haapalainen, E.; Sinigaglia-Coimbra, R.; Almeida, I.C.; Puccia, R. The Pathogenic Fungus Paracoccidioides brasiliensis Exports Extracellular Vesicles Containing Highly Immunogenic α-Galactosyl Epitopes. Eukaryot. Cell 2011, 10, 343–351. [Google Scholar] [CrossRef]
- Konečná, K.; Klimentová, J.; Benada, O.; Němečková, I.; Janďourek, O.; Jílek, P.; Vejsová, M. A comparative analysis of protein virulence factors released via extracellular vesicles in two Candida albicans strains cultivated in a nutrient-limited medium. Microb. Pathog. 2019, 136, 103666. [Google Scholar] [CrossRef]
- Peres Da Silva, R.; Puccia, R.; Rodrigues, M.L.; Oliveira, D.L.; Joffe, L.S.; César, G.V.; Nimrichter, L.; Goldenberg, S.; Alves, L.R. Extracellular vesicle-mediated export of fungal RNA. Sci.Rep. 2015, 5, 7763. [Google Scholar] [CrossRef]
- Wolf, J.M.; Espadas-Moreno, J.; Luque-Garcia, J.L.; Casadevall, A. Interaction of Cryptococcus neoformans Extracellular Vesicles with the Cell Wall. Eukaryot. Cell 2014, 13, 10. [Google Scholar] [CrossRef]
- Oliveira, D.L.; Nakayasu, E.; Joffe, L.S.; Guimaraes, A.; Sobreira, T.; Nosanchuk, J.D.; Cordero, R.J.B.; Frases, S.; Casadevall, A.; Almeida, I.C.; et al. Characterization of Yeast Extracellular Vesicles: Evidence for the Participation of Different Pathways of Cellular Traffic in Vesicle Biogenesis. PLoS ONE 2010, 5, e11113. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nimrichter, L.; Oliveira, D.L.; Nosanchuk, J.; Casadevall, A. Vesicular Trans-Cell Wall Transport in Fungi: A Mechanism for the Delivery of Virulence-Associated Macromolecules? Lipid Insights 2008, 2, LPI.S1000-40. [Google Scholar] [CrossRef]
- Vallejo, M.C.; Nakayasu, E.; Matsuo, A.L.; Sobreira, T.; Longo, L.V.G.; Ganiko, L.; Almeida, I.; Puccia, R. Vesicle and Vesicle-Free Extracellular Proteome of Paracoccidioides brasiliensis: Comparative Analysis with Other Pathogenic Fungi. J. Proteome Res. 2012, 11, 1676–1685. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Franzen, A.J.; Nimrichter, L.; Miranda, K. Vesicular mechanisms of traffic of fungal molecules to the extracellular space. Curr. Opin. Microbiol. 2013, 16, 414–420. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Travassos, L.R.; Miranda, K.R.; Franzen, A.J.; Rozental, S.; De Souza, W.; Alviano, C.S.; Barreto-Bergter, E. Human Antibodies against a Purified Glucosylceramide from Cryptococcus neoformans Inhibit Cell Budding and Fungal Growth. Infect. Immun. 2000, 68, 7049–7060. [Google Scholar] [CrossRef]
- Wolf, J.M.; Espadas, J.; Luque-Garcia, J.; Reynolds, T.; Casadevall, A. Lipid Biosynthetic Genes Affect Candida albicans Extracellular Vesicle Morphology, Cargo, and Immunostimulatory Properties. Eukaryot. Cell 2015, 14, 745–754. [Google Scholar] [CrossRef]
- Feng, D.; Zhao, W.-L.; Ye, Y.-Y.; Bai, X.-C.; Liu, R.-Q.; Chang, L.-F.; Zhou, Q.; Sui, S.-F. Cellular Internalization of Exosomes Occurs Through Phagocytosis. Traffic 2010, 11, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Fitzner, D.; Schnaars, M.; Van Rossum, D.; Krishnamoorthy, G.; Dibaj, P.; Bakhti, M.; Regen, T.; Hanisch, U.-K.; Simons, M. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J. Cell Sci. 2011, 124, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat.Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Huang-Doran, I.; Zhang, C.-Y.; Vidal-Puig, A. Extracellular Vesicles: Novel Mediators of Cell Communication in Metabolic Disease. Trends Endocrinol. Metab. 2017, 28, 3–18. [Google Scholar] [CrossRef]
- Laulagnier, K.; Grand, D.; Dujardin, A.; Hamdi, S.; Vincent-Schneider, H.; Lankar, D.; Salles, J.-P.; Bonnerot, C.; Perret, B.; Record, M. PLD2 is enriched on exosomes and its activity is correlated to the release of exosomes. FEBS Lett. 2004, 572, 11–14. [Google Scholar] [CrossRef]
- Laulagnier, K.; Motta, C.; Hamdi, S.; Roy, S.; Fauvelle, F.; Pageaux, J.-F.; Kobayashi, T.; Salles, J.-P.; Perret, B.; Bonnerot, C.; et al. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem. J. 2004, 380, 161–171. [Google Scholar] [CrossRef]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH Is a Key Factor for Exosome Traffic in Tumor Cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef]
- Jacomin, A.-C.; Fauvarque, M.-O.; Taillebourg, E. A functional endosomal pathway is necessary for lysosome biogenesis in Drosophila. BMC Cell Biol. 2016, 17, 36. [Google Scholar] [CrossRef]
- Goode, B.L.; Eskin, J.A.; Wendland, B. Actin and Endocytosis in Budding Yeast. Genetics 2015, 199, 315–358. [Google Scholar] [CrossRef]
- Carroll, S.Y.; Stimpson, H.E.M.; Weinberg, J.; Toret, C.P.; Sun, Y.; Drubin, D.G. Analysis of yeast endocytic site formation and maturation through a regulatory transition point. MBoC 2012, 23, 657–668. [Google Scholar] [CrossRef]
- Stimpson, H.E.M.; Toret, C.P.; Cheng, A.T.; Pauly, B.S.; Drubin, D.G. Early-Arriving Syp1p and Ede1p Function in Endocytic Site Placement and Formation in Budding Yeast. MBoC 2009, 20, 4640–4651. [Google Scholar] [CrossRef] [PubMed]
- Wendland, B. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 1999, 18, 4383–4393. [Google Scholar] [CrossRef] [PubMed]
- Wendland, B.; Emr, S.D. Pan1p, Yeast eps15, Functions as a Multivalent Adaptor That Coordinates Protein–Protein Interactions Essential for Endocytosis. J. Cell Biol. 1998, 141, 71–84. [Google Scholar] [CrossRef]
- Sun, Y.; Leong, N.T.; Wong, T.; Drubin, D.G. A Pan1/End3/Sla1 complex links Arp2/3-mediated actin assembly to sites of clathrin-mediated endocytosis. MBoC 2015, 26, 3841–3856. [Google Scholar] [CrossRef] [PubMed]
- Feliciano, D.; Di Pietro, S.M. SLAC, a complex between Sla1 and Las17, regulates actin polymerization during clathrin-mediated endocytosis. MBoC 2012, 23, 4256–4272. [Google Scholar] [CrossRef] [PubMed]
- Raths, S.; Riezman, H. The END3 Gene Encodes a Protein that Is Required for the Internalization Step of Endocytosis and for Actin Cytoskeleton Organization in Yeast. Mol. Biol. Cell 1994, 5, 15. [Google Scholar]
- Whitworth, K.; Bradford, M.K.; Camara, N.; Wendland, B. Targeted Disruption of an EH-domain Protein Endocytic Complex, Pan1-End3: Pan1 Central Region Binds C-terminal End3 Repeats. Traffic 2014, 15, 43–59. [Google Scholar] [CrossRef]
- Tang, H.Y.; Cai, M. The EH-domain-containing protein Pan1 is required for normal organization of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996, 16, 4897–4914. [Google Scholar] [CrossRef]
- Sun, Y.; Leong, N.T.; Jiang, T.; Tangara, A.; Darzacq, X.; Drubin, D.G. Switch-like Arp2/3 activation upon WASP and WIP recruitment to an apparent threshold level by multivalent linker proteins in vivo. eLife 2017, 6, e29140. [Google Scholar] [CrossRef]
- Moreno-Ruiz, E.; Galán-Díez, M.; Zhu, W.; Fernández-Ruiz, E.; d’Enfert, C.; Filler, S.G.; Cossart, P.; Veiga, E. Candida albicans internalization by host cells is mediated by a clathrin-dependent mechanism. Cell. Microbiol. 2009, 11, 1179–1189. [Google Scholar] [CrossRef]
- Martin, R.; Hellwig, D.; Schaub, Y.; Bauer, J.; Walther, A.; Wendland, J. Functional analysis ofCandida albicans genes whoseSaccharomyces cerevisiae homologues are involved in endocytosis. Yeast 2007, 24, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Asleson, C.M.; Bensen, E.S.; Gale, C.A.; Melms, A.-S.; Kurischko, C.; Berman, J. Candida albicans INT1-Induced Filamentation in Saccharomyces cerevisiae Depends on Sla2p. Mol. Cell. Biol. 2001, 21, 1272–1284. [Google Scholar] [CrossRef] [PubMed]
- Oberholzer, U.; Nantel, A.; Berman, J.; Whiteway, M. Transcript Profiles of Candida albicans Cortical Actin Patch Mutants Reflect Their Cellular Defects: Contribution of the Hog1p and Mkc1p Signaling Pathways. Eukaryot. Cell 2006, 5, 1252–1265. [Google Scholar] [CrossRef] [PubMed]
- Weissman, Z.; Shemer, R.; Conibear, E.; Kornitzer, D. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol. Microbiol. 2008, 69, 201–217. [Google Scholar] [CrossRef]
- Gale, C.A.; Leonard, M.D.; Finley, K.R.; Christensen, L.; McClellan, M.; Abbey, D.; Kurischko, C.; Bensen, E.; Tzafrir, I.; Kauffman, S.; et al. SLA2 mutations cause SWE1-mediated cell cycle phenotypes in Candida albicans and Saccharomyces cerevisiae. Microbiology 2009, 155, 3847–3859. [Google Scholar] [CrossRef][Green Version]
- Reijnst, P.; Jorde, S.; Wendland, J. Candida albicans SH3-domain proteins involved in hyphal growth, cytokinesis, and vacuolar morphology. Curr. Genet. 2010, 56, 309–319. [Google Scholar] [CrossRef]
- Zeng, G.; Wang, Y.-M.; Wang, Y. Cdc28–Cln3 phosphorylation of Sla1 regulates actin patch dynamics in different modes of fungal growth. MBoC 2012, 23, 3485–3497. [Google Scholar] [CrossRef]
- Walther, A.; Wendland, J. Polarized Hyphal Growth in Candida albicans Requires the Wiskott-Aldrich Syndrome Protein Homolog Wal1p. Eukaryot. Cell 2004, 3, 471–482. [Google Scholar] [CrossRef][Green Version]
- Prosser, D.C.; Drivas, T.G.; Maldonado-Báez, L.; Wendland, B. Existence of a novel clathrin-independent endocytic pathway in yeast that depends on Rho1 and formin. J. Cell Biol. 2011, 195, 657–671. [Google Scholar] [CrossRef]
- Prosser, D.C.; Pannunzio, A.E.; Brodsky, J.L.; Thorner, J.; Wendland, B.; O’Donnell, A.F. α-Arrestins participate in cargo selection for both clathrin-independent and clathrin-mediated endocytosis. J. Cell Sci. 2015, 128, 4220–4234. [Google Scholar] [CrossRef]
- Epp, E.; Nazarova, E.; Regan, H.; Douglas, L.M.; Konopka, J.B.; Vogel, J.; Whiteway, M. Clathrin- and Arp2/3-Independent Endocytosis in the Fungal Pathogen Candida albicans. mBio 2013, 4, e00476-13. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.A.; Robbins, N.; Xie, J.L.; Ketela, T.; Cowen, L.E. Functional Genomic Analysis of Candida albicans Adherence Reveals a Key Role for the Arp2/3 Complex in Cell Wall Remodelling and Biofilm Formation. PLoS Genet. 2016, 12, e1006452. [Google Scholar] [CrossRef] [PubMed]
- Epp, E.; Walther, A.; Lépine, G.; Leon, Z.; Mullick, A.; Raymond, M.; Wendland, J.; Whiteway, M. Forward genetics in Candida albicans that reveals the Arp2/3 complex is required for hyphal formation, but not endocytosis. Mol. Microbiol. 2010, 75, 1182–1198. [Google Scholar] [CrossRef] [PubMed]
- Langemeyer, L.; Fröhlich, F.; Ungermann, C. Rab GTPase Function in Endosome and Lysosome Biogenesis. Trends Cell Biol. 2018, 28, 957–970. [Google Scholar] [CrossRef] [PubMed]
- Toshima, J.Y.; Toshima, J.; Kaksonen, M.; Martin, A.C.; King, D.S.; Drubin, D.G. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent -factor derivatives. Proc. Natl. Acad. Sci. USA 2006, 103, 5793–5798. [Google Scholar] [CrossRef] [PubMed]
- Bowers, K.; Stevens, T.H. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2005, 1744, 438–454. [Google Scholar] [CrossRef] [PubMed]
- Schellmann, S.; Pimpl, P. Coats of endosomal protein sorting: Retromer and ESCRT. Curr. Opin. PlantBiol. 2009, 12, 670–676. [Google Scholar] [CrossRef]
- Mellman, I. The importance of being acid: The role of acidification in intracellular membrane traffic. J. Exp. Biol. 1992, 172, 39. [Google Scholar]
- Nishi, T.; Forgac, M. The vacuolar (H+)-ATPases—Nature’s most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 2002, 3, 94–103. [Google Scholar] [CrossRef]
- Nass, R.; Rao, R. Novel Localization of a Na+/H+ Exchanger in a Late Endosomal Compartment of Yeast: IMPLICATIONS FOR VACUOLE BIOGENESIS. J. Biol. Chem. 1998, 273, 21054–21060. [Google Scholar] [CrossRef]
- Bowers, K.; Levi, B.P.; Patel, F.I.; Stevens, T.H. The Sodium/Proton Exchanger Nhx1p Is Required for Endosomal Protein Trafficking in the Yeast Saccharomyces cerevisiae. MBoC 2000, 11, 4277–4294. [Google Scholar] [CrossRef] [PubMed]
- Brett, C.L.; Tukaye, D.N.; Mukherjee, S.; Rao, R. The Yeast Endosomal Na+(K)+/H+ Exchanger Nhx1 Regulates Cellular pH to Control Vesicle Trafficking. MBoC 2005, 16, 1396–1405. [Google Scholar] [CrossRef] [PubMed]
- Riezman, H. Endocytosis in yeast: Several of the yeast secretory mutants are defective in endocytosis. Cell 1985, 40, 1001–1009. [Google Scholar] [CrossRef]
- Voříšek, J. Functional morphology of the secretory pathway organelles in yeast. Microsc. Res. Tech. 2000, 51, 530–546. [Google Scholar] [CrossRef]
- Burston, H.E.; Maldonado-Báez, L.; Davey, M.; Montpetit, B.; Schluter, C.; Wendland, B.; Conibear, E. Regulators of yeast endocytosis identified by systematic quantitative analysis. J. Cell Biol. 2009, 185, 1097–1110. [Google Scholar] [CrossRef]
- Rooij, I.I.S.-D.; Allwood, E.G.; Aghamohammadzadeh, S.; Hettema, E.H.; Goldberg, M.W.; Ayscough, K.R. A role for the dynamin-like protein Vps1 during endocytosis in yeast. J. Cell Sci. 2010, 123, 3496–3506. [Google Scholar] [CrossRef]
- Palanisamy, S.K.A.; Ramirez, M.A.; Lorenz, M.; Lee, S.A. Candida albicans PEP12 Is Required for Biofilm Integrity and In Vivo Virulence. Eukaryot. Cell 2010, 9, 266–277. [Google Scholar] [CrossRef][Green Version]
- Bernardo, S.M.; Lee, S.A. Candida albicans SUR7 contributes to secretion, biofilm formation, and macrophage killing. BMC Microbiol. 2010, 10, 133. [Google Scholar] [CrossRef]
- Lettner, T.; Zeidler, U.; Gimona, M.; Hauser, M.; Breitenbach, M.; Bito, A. Candida albicans AGE3, the Ortholog of the S. cerevisiae ARF-GAP-Encoding Gene GCS1, Is Required for Hyphal Growth and Drug Resistance. PLoS ONE 2010, 5, e11993. [Google Scholar] [CrossRef]
- Bonhomme, J.; Chauvel, M.; Goyard, S.; Roux, P.; Rossignol, T.; d’Enfert, C. Contribution of the glycolytic flux and hypoxia adaptation to efficient biofilm formation by Candida albicans: Biofilm formation and adaptation to hypoxia in C. albicans. Mol. Microbiol. 2011, 80, 995–1013. [Google Scholar] [CrossRef]
- Heilmann, C.J.; Sorgo, A.G.; Siliakus, A.R.; Dekker, H.L.; Brul, S.; De Koster, C.G.; De Koning, L.J.; Klis, F.M. Hyphal induction in the human fungal pathogen Candida albicans reveals a characteristic wall protein profile. Microbiology 2011, 157, 2297–2307. [Google Scholar] [CrossRef] [PubMed]
- Sorgo, A.G.; Heilmann, C.J.; Dekker, H.L.; Bekker, M.; Brul, S.; De Koster, C.G.; De Koning, L.J.; Klis, F.M. Effects of Fluconazole on the Secretome, the Wall Proteome, and Wall Integrity of the Clinical Fungus Candida albicans. Eukaryot. Cell 2011, 10, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Fox, E.P.; Nett, J.E.; Sorrells, T.R.; Mitrovich, Q.M.; Hernday, A.D.; Tuch, B.B.; Andes, D.R.; Johnson, A.D. A Recently Evolved Transcriptional Network Controls Biofilm Development in Candida albicans. Cell 2012, 148, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Heider, M.R.; Munson, M. Exorcising the Exocyst Complex: Exorcising the Exocyst Complex. Traffic 2012, 13, 898–907. [Google Scholar] [CrossRef]
- TerBush, D.R.; Maurice, T.; Roth, D.; Novick, P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996, 15, 6483–6494. [Google Scholar] [CrossRef]
- Soll, D.R.; Herman, M.A.; Staebell, M.A. The Involvement of Cell Wall Expansion in the Two Modes of Mycelium Formation of Candida albicans. Microbiology 1985, 131, 2367–2375. [Google Scholar] [CrossRef]
- Weiner, A.; Orange, F.; Lacas-Gervais, S.; Rechav, K.; Ghugtyal, V.; Bassilana, M.; Arkowitz, R.A. On-site secretory vesicle delivery drives filamentous growth in the fungal pathogen Candida albicans. Cell. Microbiol. 2018, 21, e12963. [Google Scholar] [CrossRef]
- Crampin, H. Candida albicans hyphae have a Spitzenkorper that is distinct from the polarisome found in yeast and pseudohyphae. J. Cell Sci. 2005, 118, 2935–2947. [Google Scholar] [CrossRef]
- Harris, S.D.; Read, N.D.; Roberson, R.W.; Shaw, B.; Seiler, S.; Plamann, M.; Momany, M. Polarisome Meets Spitzenko¨rper: Microscopy, Genetics, and Genomics Converge. Eukaryot. Cell 2005, 4, 5. [Google Scholar] [CrossRef]
- Sudbery, P.E. Growth of Candida albicans hyphae. Nat.Rev. Microbiol 2011, 9, 737–748. [Google Scholar] [CrossRef]
- Douglas, L.M.; Konopka, J.B. Plasma membrane organization promotes virulence of the human fungal pathogen Candida albicans. J. Microbiol. 2016, 54, 178–191. [Google Scholar] [CrossRef]
- Warenda, A.J.; Konopka, J.B. Septin Function in Candida albicans Morphogenesis□D. Mol. Biol. Cell 2002, 13, 15. [Google Scholar] [CrossRef] [PubMed]
- Bishop, A.; Lane, R.; Beniston, R.; Chapa-y-Lazo, B.; Smythe, C.; Sudbery, P. Hyphal growth in Candida albicans requires the phosphorylation of Sec2 by the Cdc28-Ccn1/Hgc1 kinase. EMBO J. 2010, 29, 2930–2942. [Google Scholar] [CrossRef] [PubMed]
- Caballero-Lima, D.; Sudbery, P.E. In Candida albicans, phosphorylation of Exo84 by Cdk1-Hgc1 is necessary for efficient hyphal extension. MBoC 2014, 25, 1097–1110. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Wang, Y.; Wang, Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 2004, 23, 1845–1856. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.-D.; Lee, R.T.H.; Wang, Y.-M.; Lin, Q.-S.; Wang, Y. Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 2007, 26, 3760–3769. [Google Scholar] [CrossRef] [PubMed]
- Sinha, I.; Wang, Y.-M.; Philp, R.; Li, C.-R.; Yap, W.H.; Wang, Y. Cyclin-Dependent Kinases Control Septin Phosphorylation in Candida albicans Hyphal Development. Dev. Cell 2007, 13, 421–432. [Google Scholar] [CrossRef]
- Guo, P.P.; Yong, J.Y.A.; Wang, Y.M.; Li, C.R. Sec15 links bud site selection to polarised cell growth and exocytosis in Candida albicans. Sci.Rep. 2016, 6, 26464. [Google Scholar] [CrossRef]
- Walch-Solimena, C.; Collins, R.N.; Novick, P.J. Sec2p Mediates Nucleotide Exchange on Sec4p and Is Involved in Polarized Delivery of Post-Golgi Vesicles. J. Cell Biol. 1997, 137, 1495–1509. [Google Scholar] [CrossRef]
- Medkova, M.; France, Y.E.; Coleman, J.; Novick, P. The rab Exchange Factor Sec2p Reversibly Associates with the Exocyst. Mol. Biol. Cell 2006, 17, 2757–2769. [Google Scholar]
- Dong, G.; Medkova, M.; Novick, P.; Reinisch, K.M. A Catalytic Coiled Coil: Structural Insights into the Activation of the Rab GTPase Sec4p by Sec2p. Mol. Cell 2007, 25, 455–462. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bassilana, M.; Blyth, J.; Arkowitz, R.A. Cdc24, the GDP-GTP Exchange Factor for Cdc42, Is Required for Invasive Hyphal Growth of Candida albicans. Eukaryot. Cell 2003, 2, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Ushinsky, S.C.; Harcus, D.; Ash, J.; Dignard, D.; Marcil, A.; Morchhauser, J.; Thomas, D.Y.; Whiteway, M.; Leberer, E. CDC42 Is Required for Polarized Growth in Human Pathogen Candida albicans. Eukaryot. Cell 2002, 1, 95–104. [Google Scholar] [CrossRef]
- VandenBerg, A.L.; Ibrahim, A.S.; Edwards, J.E.; Toenjes, K.A.; Johnson, D.I. Cdc42p GTPase Regulates the Budded-to-Hyphal-Form Transition and Expression of Hypha-Specific Transcripts in Candida albicans. Eukaryot. Cell 2004, 3, 724–734. [Google Scholar] [CrossRef]
- Brand, A.C.; Morrison, E.; Milne, S.; Gonia, S.; Gale, C.A.; Gow, N.A.R. Cdc42 GTPase dynamics control directional growth responses. Proc. Natl. Acad. Sci. USA 2014, 111, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Court, H.; Sudbery, P. Regulation of Cdc42 GTPase Activity in the Formation of Hyphae in Candida albicans□D. Mol. Biol. Cell 2007, 18, 17. [Google Scholar] [CrossRef]
- Silva, P.M.; Puerner, C.; Seminara, A.; Bassilana, M.; Arkowitz, R.A. Secretory Vesicle Clustering in Fungal Filamentous Cells Does Not Require Directional Growth. Cell Rep. 2019, 28, 2231–2245.e5. [Google Scholar] [CrossRef]
- Nishikawa, A.; Poster, J.B.; Jigami, Y.; Dean, N. Molecular and Phenotypic Analysis of CaVRG4, Encoding an Essential Golgi Apparatus GDP-Mannose Transporter. J. Bacteriol. 2002, 184, 29–42. [Google Scholar] [CrossRef]
- Rida, P.C.G.; Nishikawa, A.; Won, G.Y.; Dean, N. Yeast-to-Hyphal Transition Triggers Formin-dependent Golgi Localization to the Growing Tip in Candida albicans. MBoC 2006, 17, 4364–4378. [Google Scholar] [CrossRef]
- Caballero-Lima, D.; Kaneva, I.N.; Watton, S.P.; Sudbery, P.E.; Craven, C.J. The Spatial Distribution of the Exocyst and Actin Cortical Patches Is Sufficient To Organize Hyphal Tip Growth. Eukaryot. Cell 2013, 12, 998–1008. [Google Scholar] [CrossRef]
- Taheri-Talesh, N.; Horio, T.; Araujo-Bazán, L.; Dou, X.; Espeso, E.A.; Peñalva, M.A.; Osmani, S.A.; Oakley, B.R. The Tip Growth Apparatus of Aspergillus nidulans. MBoC 2008, 19, 1439–1449. [Google Scholar] [CrossRef] [PubMed]
- Valkonen, M.; Kalkman, E.R.; Saloheimo, M.; Penttilä, M.; Read, N.D.; Duncan, R.R. Spatially Segregated SNARE Protein Interactions in Living Fungal Cells. J. Biol. Chem. 2007, 282, 22775–22785. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.D.; Free, S.J.; Levina, N.N.; Keränen, S.; Heath, I.B. Two divergent plasma membrane syntaxin-like SNAREs, nsyn1 and nsyn2, contribute to hyphal tip growth and other developmental processes in Neurospora crassa. Fungal Genet. Biol. 2003, 40, 271–286. [Google Scholar] [CrossRef]
- Rasheed, M.; Kumar, N.; Kaur, R. Global Secretome Characterization of the Pathogenic Yeast Candida glabrata. J. Proteome Res. 2020, 19, 49–63. [Google Scholar] [CrossRef]
- Pitarch, A.; Sánchez, M.; Nombela, C.; Gil, C. Sequential Fractionation and Two-dimensional Gel Analysis Unravels the Complexity of the Dimorphic Fungus Candida albicans Cell Wall Proteome. Mol. Cell Proteom. 2002, 1, 967–982. [Google Scholar] [CrossRef]
- Urban, C.; Sohn, K.; Lottspeich, F.; Brunner, H.; Rupp, S. Identification of cell surface determinants in Candida albicans reveals Tsa1p, a protein differentially localized in the cell. FEBS Lett. 2003, 544, 228–235. [Google Scholar] [CrossRef]
- Alloush, H.M.; Lopez-Ribot, J.L.; Masten, B.J.; Chaffin, W.L. 3-Phosphoglycerate kinase: A glycolytic enzyme protein present in the cell wall of Candida albicans. Microbiology 1997, 143, 321–330. [Google Scholar] [CrossRef]
- Nombela, C.; Gil, C.; Chaffin, W.L. Non-conventional protein secretionin yeast. Trends Microbiol. 2006, 14, 15–21. [Google Scholar] [CrossRef]
- Miura, N.; Ueda, M. Evaluation of Unconventional Protein Secretion by Saccharomyces cerevisiae and other Fungi. Cells 2018, 7, 128. [Google Scholar] [CrossRef]
- Wolf, J.M.; Casadevall, A. Challenges posed by extracellular vesicles from eukaryotic microbes. Curr. Opin. Microbiol. 2014, 22, 73–78. [Google Scholar] [CrossRef]
- Moller, I.; Jung, M.; Beatrix, B.; Levy, R.; Kreibich, G.; Zimmermann, R.; Wiedmann, M.; Lauring, B. A general mechanism for regulation of access to the translocon: Competition for a membrane attachment site on ribosomes. Proc. Natl. Acad. Sci. USA 1998, 95, 13425–13430. [Google Scholar] [CrossRef] [PubMed]
- Wiedmann, B.; Prehn, S. The nascent polypeptide-associated complex (NAC) of yeast functions in the targeting process of ribosomes to the ER membrane. FEBS Lett. 1999, 458, 51–54. [Google Scholar] [CrossRef]
- Reimann, B.; Bradsher, J.; Franke, J.; Hartmann, E.; Wiedmann, M.; Prehn, S.; Wiedmann, B. Initial characterization of the nascent polypeptide-associated complex in yeast. Yeast 1999, 15, 397–407. [Google Scholar] [CrossRef]
- Kellis, M.; Birren, B.W.; Lander, E.S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 2004, 428, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Franke, J.; Reimann, B.; Hartmann, E.; Köhler, M.; Wiedmann, B. Evidence for a nuclear passage of nascent polypeptide-associated complex subunits in yeast. J. Cell Sci. 2001, 114, 2641. [Google Scholar]
- Sorgo, A.G.; Heilmann, C.J.; Brul, S.; De Koster, C.G.; Klis, F.M. Beyond the wall: Candida albicans secret(e)s to survive. FEMS Microbiol. Lett. 2013, 338, 10–17. [Google Scholar] [CrossRef]
- Butler, G.; Rasmussen, M.D.; Lin, M.F.; Santos, M.A.S.; Sakthikumar, S.; Munro, C.A.; Rheinbay, E.; Grabherr, M.; Forche, A.; Reedy, J.L.; et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 2009, 459, 657–662. [Google Scholar] [CrossRef]
- Whiteway, M.; Tebung, W.A.; Choudhury, B.I.; Rodríguez-Ortiz, R. Metabolic regulation in model ascomycetes—Adjusting similar genomes to different lifestyles. Trends Genet. 2015, 31, 445–453. [Google Scholar] [CrossRef]
- Ihmels, J. Rewiring of the Yeast Transcriptional Network through the Evolution of Motif Usage. Science 2005, 309, 938–940. [Google Scholar] [CrossRef]
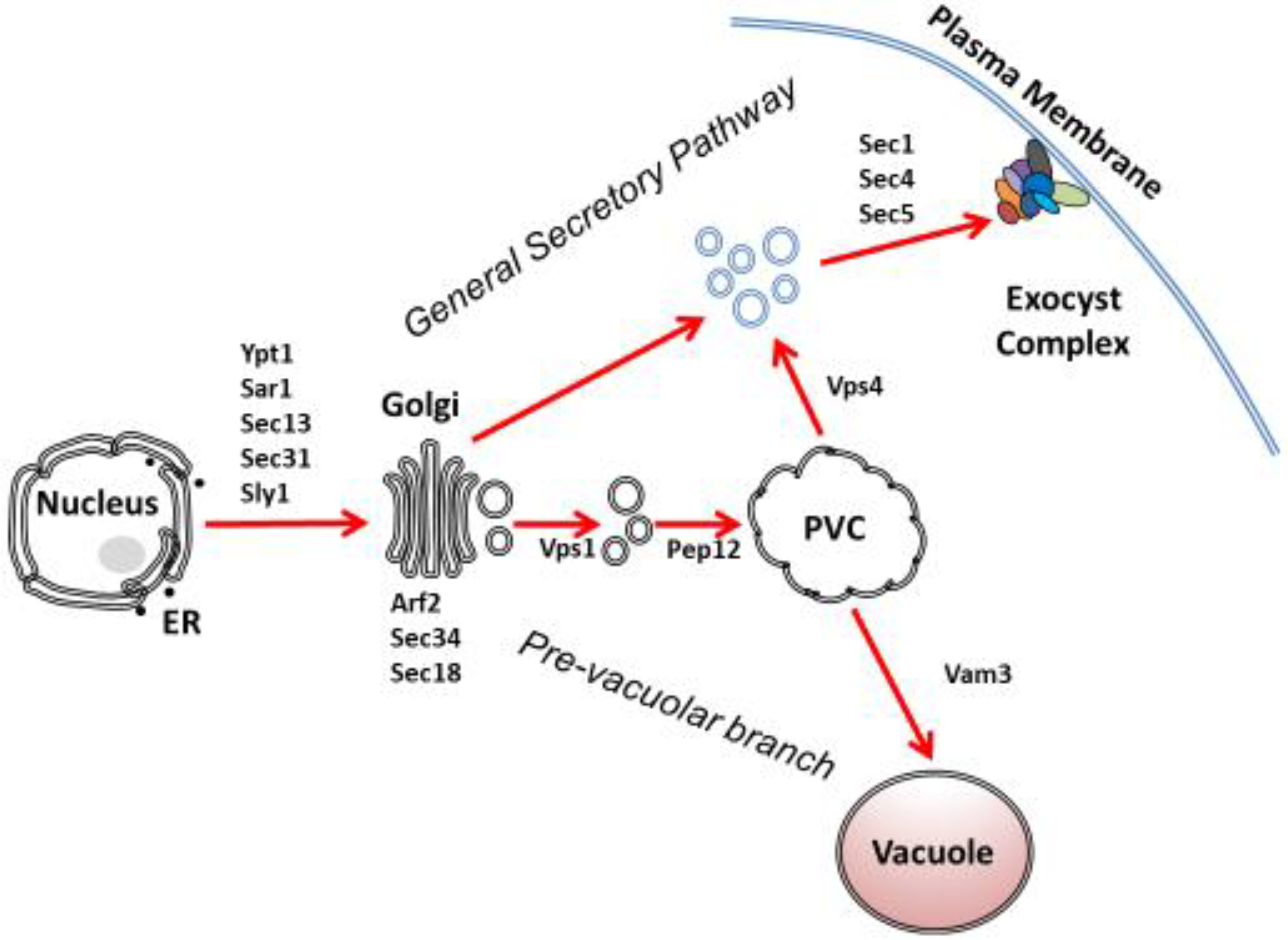


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rollenhagen, C.; Mamtani, S.; Ma, D.; Dixit, R.; Eszterhas, S.; Lee, S.A. The Role of Secretory Pathways in Candida albicans Pathogenesis. J. Fungi 2020, 6, 26. https://doi.org/10.3390/jof6010026
Rollenhagen C, Mamtani S, Ma D, Dixit R, Eszterhas S, Lee SA. The Role of Secretory Pathways in Candida albicans Pathogenesis. Journal of Fungi. 2020; 6(1):26. https://doi.org/10.3390/jof6010026
Chicago/Turabian StyleRollenhagen, Christiane, Sahil Mamtani, Dakota Ma, Reva Dixit, Susan Eszterhas, and Samuel A. Lee. 2020. "The Role of Secretory Pathways in Candida albicans Pathogenesis" Journal of Fungi 6, no. 1: 26. https://doi.org/10.3390/jof6010026
APA StyleRollenhagen, C., Mamtani, S., Ma, D., Dixit, R., Eszterhas, S., & Lee, S. A. (2020). The Role of Secretory Pathways in Candida albicans Pathogenesis. Journal of Fungi, 6(1), 26. https://doi.org/10.3390/jof6010026




