Lipid Species in the GI Tract are Increased by the Commensal Fungus Candida albicans and Decrease the Virulence of Clostridioides difficile
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. GI Colonization in Mice
2.3. C. difficile Toxin Titer Assay
2.4. Histology
2.5. Metabolomic Analysis of Murine Cecum Contents
2.6. Microbiota Analysis
2.7. Cytokine Gene Transcription
2.8. C. difficile Gene Transcription
2.9. Statistical Analysis
3. Results
3.1. Altered Levels of Unsaturated Fatty Acids and Other Lipid Species in the Cecum Contents of C. albicans-Colonized Mice
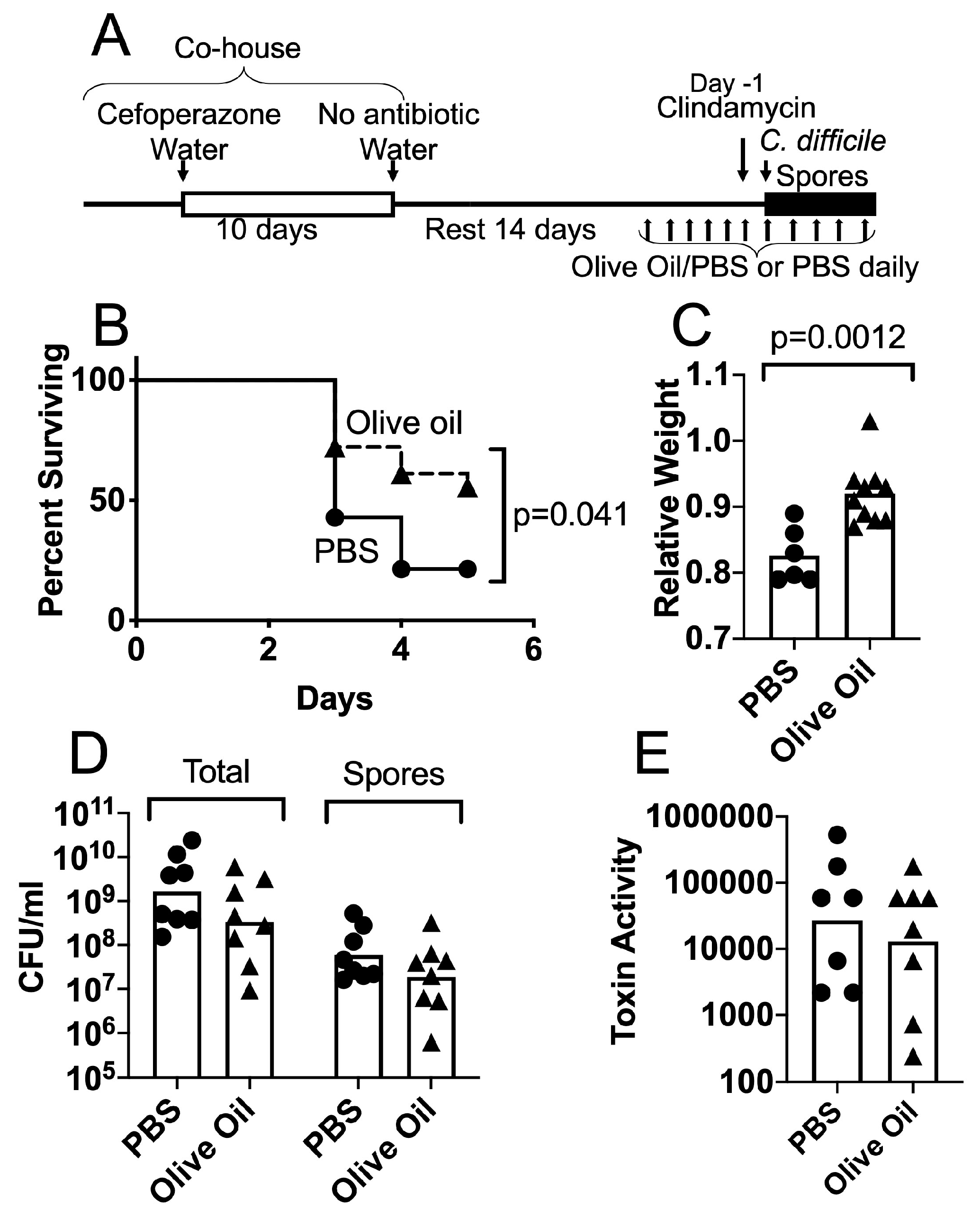
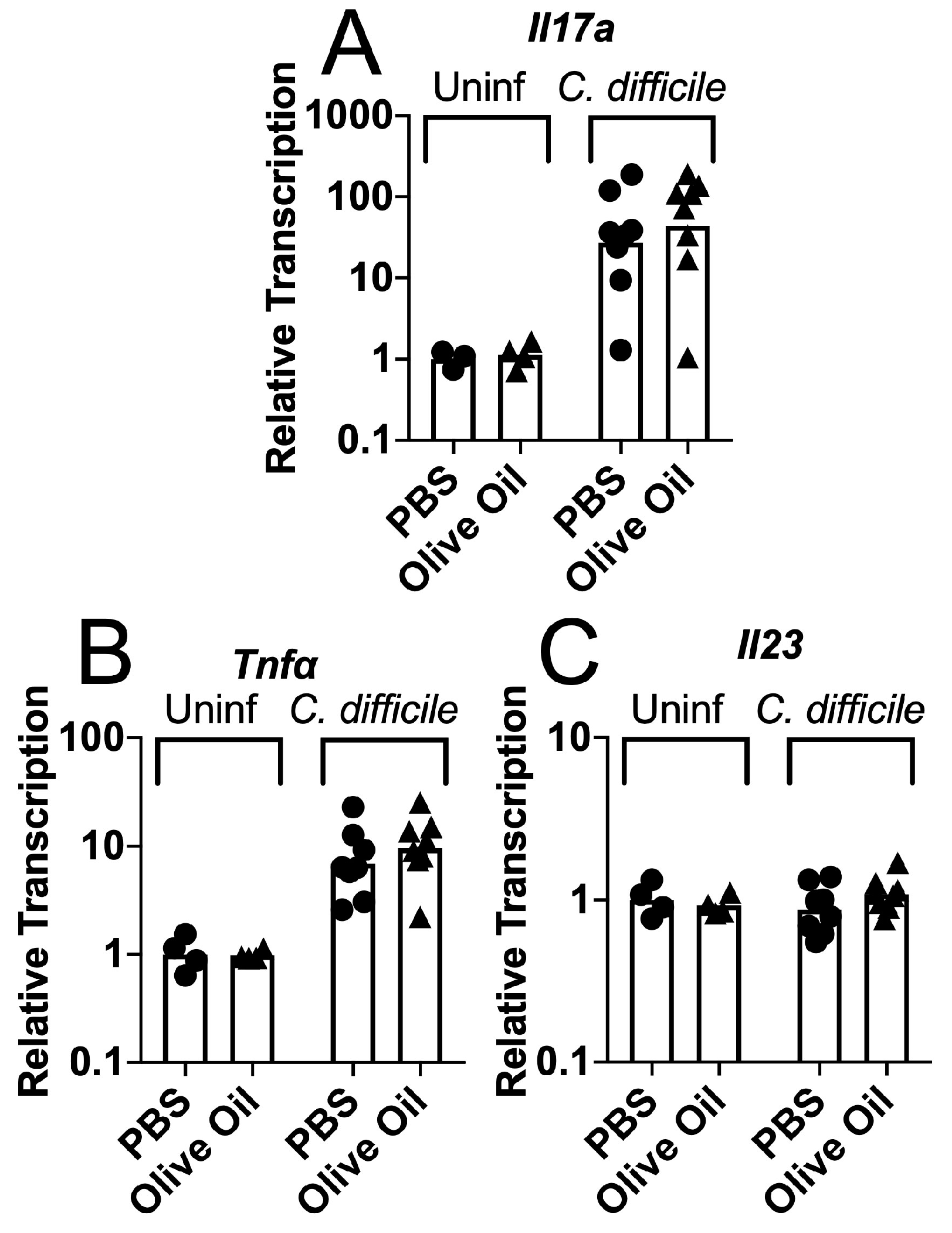
3.2. Protective Effects of Olive Oil Feeding on Antibiotic-Treated Mice Challenged with C. difficile Spores
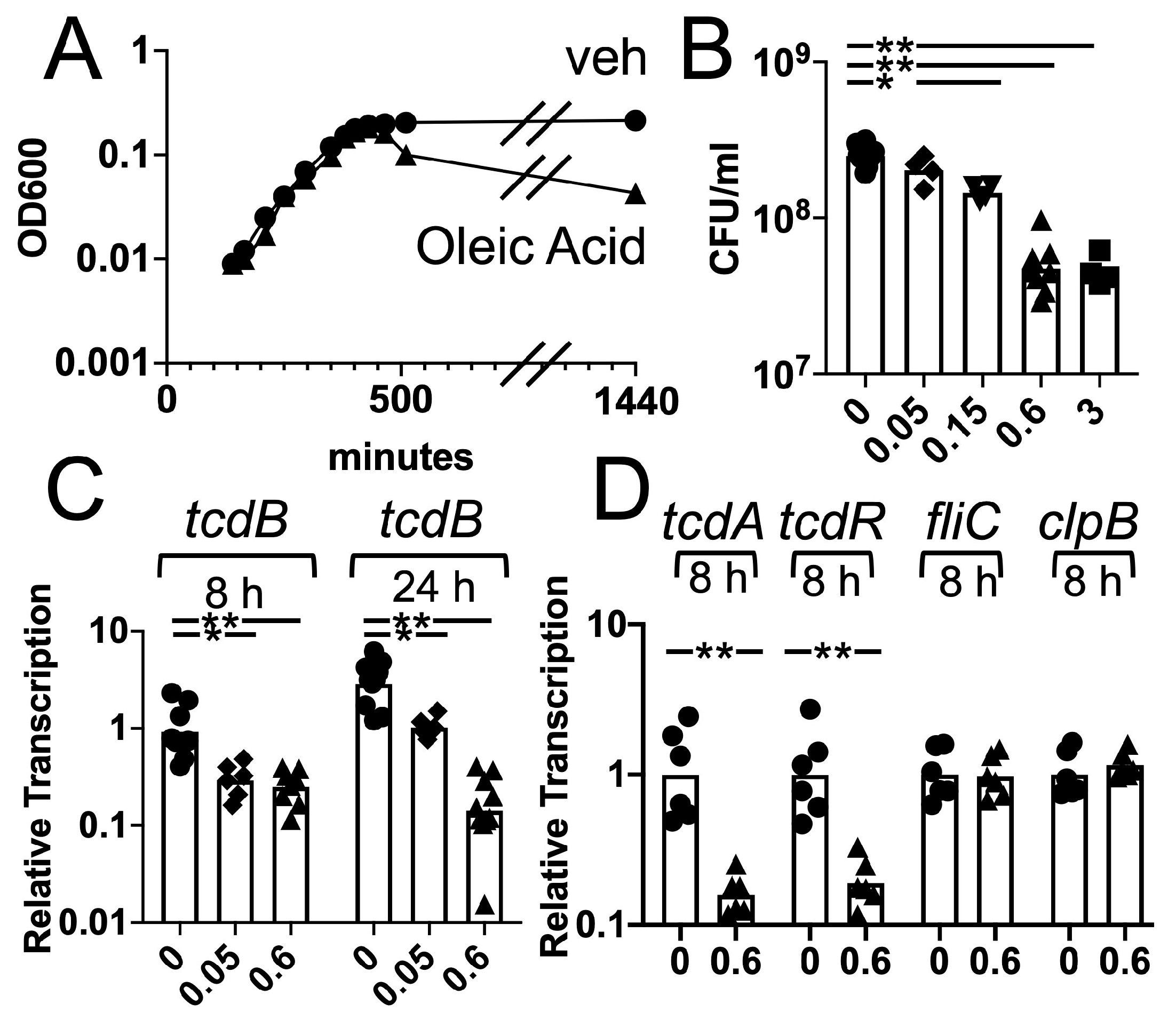
3.3. Effects of Oleic Acid on C. difficile Growth and Toxin Gene Transcription
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cole, G.T.; Lynn, K.T.; Seshan, K.R.; Pope, L.M. Gastrointestinal and systemic candidosis in immunocompromised mice. J. Med. Vet. Mycol. 1989, 27, 363–380. [Google Scholar] [CrossRef] [PubMed]
- Ekenna, O.; Sherertz, R.J. Factors affecting colonization and dissemination of Candida albicans from the gastrointestinal tract of mice. Infect. Immun. 1987, 55, 1558–1563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, M.J. Regulation of Candida albicans populations in the gastrointestinal tract: Mechanisms and significance in GI and systemic candidiasis. Curr Top. Med. Mycol. 1989, 3, 315–402. [Google Scholar] [PubMed]
- Kennedy, M.J.; Volz, P.A. Ecology of Candida albicans gut colonization: Inhibition of Candida adhesion, colonization, and dissemination from the gastrointestinal tract by bacterial antagonism. Infect. Immun. 1985, 49, 654–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hopke, A.; Brown, A.J.P.; Hall, R.A.; Wheeler, R.T. Dynamic Fungal Cell Wall Architecture in Stress Adaptation and Immune Evasion. Trends Microbiol. 2018, 26, 284–295. [Google Scholar] [CrossRef]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netea, M.G.; Joosten, L.A.; van der Meer, J.W.; Kullberg, B.J.; van de Veerdonk, F.L. Immune defence against Candida fungal infections. Nat. Rev. Immunol. 2015, 15, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Plato, A.; Hardison, S.E.; Brown, G.D. Pattern recognition receptors in antifungal immunity. Semin. Immunopathol. 2015, 37, 97–106. [Google Scholar] [CrossRef] [Green Version]
- Romani, L.; Zelante, T.; Palmieri, M.; Napolioni, V.; Picciolini, M.; Velardi, A.; Aversa, F.; Puccetti, P. The cross-talk between opportunistic fungi and the mammalian host via microbiota’s metabolism. Semin. Immunopathol. 2015, 37, 163–171. [Google Scholar] [CrossRef]
- Allison, D.L.; Willems, H.M.; Jayatilake, J.A.; Bruno, V.M.; Peters, B.M.; Shirtliff, M.E. Candida-Bacteria Interactions: Their Impact on Human Disease. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- De Sordi, L.; Mühlschlegel, F.A. Quorum sensing and fungal-bacterial interactions in Candida albicans: A communicative network regulating microbial coexistence and virulence. FEMS. Yeast Res. 2009, 9, 990–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabra-Rizk, M.A.; Meiller, T.F.; James, C.E.; Shirtliff, M.E. Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob. Agents Chemother. 2006, 50, 1463–1469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogan, D.A.; Kolter, R. Pseudomonas-Candida interactions: An ecological role for virulence factors. Science 2002, 296, 2229–2232. [Google Scholar] [CrossRef]
- Hogan, D.A.; Vik, A.; Kolter, R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 2004, 54, 1212–1223. [Google Scholar] [CrossRef] [PubMed]
- Morales, D.K.; Hogan, D.A. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 2010, 6, e1000886. [Google Scholar] [CrossRef]
- Erb Downward, J.R.; Falkowski, N.R.; Mason, K.L.; Muraglia, R.; Huffnagle, G.B. Modulation of Post-Antibiotic Bacterial Community Reassembly and Host Response by Candida albicans. Sci. Rep. 2013, 3, 2191. [Google Scholar] [CrossRef] [Green Version]
- Bartlett, J.G. Management of Clostridium difficile infection and other antibiotic-associated diarrhoeas. Eur J. Gastroenterol. Hepatol. 1996, 8, 1054–1061. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.P.; LaMont, J.T. Clostridium difficile infection. Annu Rev. Med. 1998, 49, 375–390. [Google Scholar] [CrossRef]
- McCollum, D.L.; Rodriguez, J.M. Detection, treatment, and prevention of Clostridium difficile infection. Clin. Gastroenterol. Hepatol. 2012, 10, 581–592. [Google Scholar] [CrossRef]
- Bartlett, J.G. Antibiotic-associated colitis. Dis Mon. 1984, 30, 1–54. [Google Scholar]
- Bartlett, J.G. Treatment of antibiotic-associated pseudomembranous colitis. Rev. Infect. Dis 1984, 6, S235–S241. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, J.G. Antibiotic-associated pseudomembranous colitis. Rev. Infect. Dis. 1979, 16, 530–539. [Google Scholar] [CrossRef]
- Theriot, C.M.; Bowman, A.A.; Young, V.B. Antibiotic-Induced Alterations of the Gut Microbiota Alter Secondary Bile Acid Production and Allow for Clostridium difficile Spore Germination and Outgrowth in the Large Intestine. mSphere 2016, 1, e00045-15. [Google Scholar] [CrossRef] [Green Version]
- Dicks, L.M.T.; Mikkelsen, L.S.; Brandsborg, E.; Marcotte, H. Clostridium difficile, the Difficult “Kloster” Fuelled by Antibiotics. Curr. Microbiol. 2018, 76, 774–782. [Google Scholar] [CrossRef]
- Lessa, F.C.; Mu, Y.; Bamberg, W.M.; Beldavs, Z.G.; Dumyati, G.K.; Dunn, J.R.; Farley, M.M.; Holzbauer, S.M.; Meek, J.I.; Phipps, E.C.; et al. Burden of Clostridium difficile Infection in the United States. N. Engl. J. Med. 2015, 372, 825–834. [Google Scholar] [CrossRef] [Green Version]
- Gerding, D.N.; Lessa, F.C. The epidemiology of Clostridium difficile infection inside and outside health care institutions. Infect. Dis. Clin. North. Am. 2015, 29, 37–50. [Google Scholar] [CrossRef]
- Dubberke, E.R.; Olsen, M.A. Burden of Clostridium difficile on the healthcare system. Clin. Infect. Dis. 2012, 55, S88–S92. [Google Scholar] [CrossRef]
- Sebaihia, M.; Wren, B.W.; Mullany, P.; Fairweather, N.F.; Minton, N.; Stabler, R.; Thomson, N.R.; Roberts, A.P.; Cerdeno-Tarraga, A.M.; Wang, H.; et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 2006, 38, 779–786. [Google Scholar] [CrossRef]
- Tenover, F.C.; Tickler, I.A.; Persing, D.H. Antimicrobial-resistant strains of Clostridium difficile from North America. Antimicrob. Agents Chemother. 2012, 56, 2929–2932. [Google Scholar] [CrossRef] [Green Version]
- He, M.; Miyajima, F.; Roberts, P.; Ellison, L.; Pickard, D.J.; Martin, M.J.; Connor, T.R.; Harris, S.R.; Fairley, D.; Bamford, K.B.; et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat. Genet. 2013, 45, 109–113. [Google Scholar] [CrossRef]
- Abou Chakra, C.N.; Pepin, J.; Sirard, S.; Valiquette, L. Risk factors for recurrence, complications and mortality in Clostridium difficile infection: A systematic review. PLoS ONE 2014, 9, e98400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alfa, M.; Harding, G.; Ronald, A.; Light, R.; Macfarlane, N.; Olson, N.; Degagne, P.; Kasdorf, K.; Simor, A.; Macdonald, K.; et al. Diarrhea recurrence in patients with Clostridium difficile-associated diarrhea: Role of concurrent antibiotics. Can. J. Infect. Dis. 1999, 10, 287–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.K.; Kim, K.H.; Lee, S.H.; Lee, S.J. Risk factors for recurrence of Clostridium difficile infection: Effect of vancomycin-resistant enterococci colonization. J. Korean Med. Sci. 2011, 26, 859–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lupse, M.; Flonta, M.; Cioara, A.; Filipescu, I.; Todor, N. Predictors of first recurrence in Clostridium difficile-associated disease. A study of 306 patients hospitalized in a Romanian tertiary referral center. J. Gastrointestin Liver Dis. 2013, 22, 397–403. [Google Scholar] [PubMed]
- Petrella, L.A.; Sambol, S.P.; Cheknis, A.; Nagaro, K.; Kean, Y.; Sears, P.S.; Babakhani, F.; Johnson, S.; Gerding, D.N. Decreased Cure and Increased Recurrence Rates for Clostridium difficile Infection Caused by the Epidemic C. difficile BI Strain. Clin. Infect. Dis. 2012, 55, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, M.H.; Fawley, W.N.; Settle, C.D.; Davidson, A. Recurrence of symptoms in Clostridium difficile infection--relapse or reinfection? J. Hosp. Infect. 1998, 38, 93–100. [Google Scholar] [CrossRef]
- Kuehne, S.A.; Cartman, S.T.; Heap, J.T.; Kelly, M.L.; Cockayne, A.; Minton, N.P. The role of toxin A and toxin B in Clostridium difficile infection. Nature 2010, 467, 711–713. [Google Scholar] [CrossRef] [Green Version]
- Voth, D.E.; Ballard, J.D. Clostridium difficile toxins: Mechanism of action and role in disease. Clin. Microbiol. Rev. 2005, 18, 247–263. [Google Scholar] [CrossRef] [Green Version]
- Sorg, J.A.; Sonenshein, A.L. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 2008, 190, 2505–2512. [Google Scholar] [CrossRef] [Green Version]
- Wilson, K.H.; Kennedy, M.J.; Fekety, F.R. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 1982, 15, 443–446. [Google Scholar] [CrossRef] [Green Version]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theriot, C.M.; Koenigsknecht, M.J.; Carlson, P.E., Jr.; Hatton, G.E.; Nelson, A.M.; Li, B.; Huffnagle, G.B.; Li, J.Z.; Young, V.B. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014, 5, 3114. [Google Scholar] [CrossRef] [PubMed]
- Buffie, C.G.; Bucci, V.; Stein, R.R.; McKenney, P.T.; Ling, L.; Gobourne, A.; No, D.; Liu, H.; Kinnebrew, M.; Viale, A.; et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015, 517, 205–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorg, J.A.; Sonenshein, A.L. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 2010, 192, 4983–4990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hryckowian, A.J.; Van Treuren, W.; Smits, S.A.; Davis, N.M.; Gardner, J.O.; Bouley, D.M.; Sonnenburg, J.L. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat. Microbiol. 2018, 3, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Manian, F.A.; Bryant, A. Does Candida Species Overgrowth Protect Against Clostridium difficile Infection? Clin. Infect. Dis. 2013, 56, 464–465. [Google Scholar] [CrossRef] [Green Version]
- Shiuan Hsu, M.; Tay Wang, J.; Kuei Huang, W.; Ching Liu, Y.; Chwen Chang, S. Prevalence and clinical features of Clostridium difficile-associated diarrhea in a tertiary hospital in northern Taiwan. J. Microbiol. Immunol. Infect. 2006, 39, 242–248. [Google Scholar]
- Raponi, G.; Visconti, V.; Brunetti, G.; Ghezzi, M.C. Clostridium difficile infection and Candida colonization of the gut: Is there a correlation? Clin. Infect. Dis. 2014, 59, 1648–1649. [Google Scholar] [CrossRef] [Green Version]
- Nerandzic, M.M.; Mullane, K.; Miller, M.A.; Babakhani, F.; Donskey, C.J. Reduced Acquisition and Overgrowth of Vancomycin-Resistant Enterococci and Candida Species in Patients Treated With Fidaxomicin Versus Vancomycin for Clostridium difficile Infection. Clin. Infect. Dis. 2012, 55, S121–S126. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Wong, S.H.; Cheung, C.P.; Lam, K.; Lui, R.; Cheung, K.; Zhang, F.; Tang, W.; Ching, J.Y.L.; Wu, J.C.Y.; et al. Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection. Nat. Commun. 2018, 9, 3663. [Google Scholar] [CrossRef]
- Markey, L.; Shaban, L.; Green, E.R.; Lemon, K.P.; Mecsas, J.; Kumamoto, C.A. Pre-colonization with the commensal fungus Candida albicans reduces murine susceptibility to Clostridium difficile infection. Gut Microbes 2018, 9, 497–509. [Google Scholar] [CrossRef] [Green Version]
- Brown, D.H., Jr.; Giusani, A.D.; Chen, X.; Kumamoto, C.A. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 1999, 34, 651–662. [Google Scholar] [CrossRef]
- Sherman, F. Getting started with yeast. Methods Enzym. 1991, 194, 3–21. [Google Scholar] [CrossRef]
- Sorg, J.A.; Dineen, S.S. Laboratory maintenance of Clostridium difficile. Curr. Protoc. Microbiol. 2009, Chapter 9, Unit9A 1. [Google Scholar] [CrossRef]
- Maniar, A.C.; Williams, T.W.; Hammond, G.W. Detection of Clostridium difficile toxin in various tissue culture monolayers. J. Clin. Microbiol. 1987, 25, 1999–2000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Env. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Bouillaut, L.; McBride, S.M.; Sorg, J.A. Genetic manipulation of Clostridium difficile. Curr. Protoc. Microbiol. 2011, Chapter 9, Unit 9A 2. [Google Scholar] [CrossRef] [Green Version]
- Husson, M.O.; Ley, D.; Portal, C.; Gottrand, M.; Hueso, T.; Desseyn, J.L.; Gottrand, F. Modulation of host defence against bacterial and viral infections by omega-3 polyunsaturated fatty acids. J. Infect. 2016, 73, 523–535. [Google Scholar] [CrossRef]
- Panpetch, W.; Somboonna, N.; Palasuk, M.; Hiengrach, P.; Finkelman, M.; Tumwasorn, S.; Leelahavanichkul, A. Oral Candida administration in a Clostridium difficile mouse model worsens disease severity but is attenuated by Bifidobacterium. PLoS ONE 2019, 14, e0210798. [Google Scholar] [CrossRef] [PubMed]
- Samonis, G.; Gikas, A.; Anaissie, E.J.; Vrenzos, G.; Maraki, S.; Tselentis, Y.; Bodey, G.P. Prospective evaluation of effects of broad-spectrum antibiotics on gastrointestinal yeast colonization of humans. Antimicrob. Agents Chemother. 1993, 37, 51–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, J.D. Influence of antibiotics or certain intestinal bacteria on orally administered Candida albicans in germ-free and conventional mice. Infect. Immun. 1971, 4, 731–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, D.; Romo, J.A.; Lamendella, R.; Kumamoto, C.A. The role of fungi in C. difficile infection: An underappreciated transkingdom interaction. Fungal Genet. Biol. 2019, 129, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Agoramoorthy, G.; Chandrasekaran, M.; Venkatesalu, V.; Hsu, M.J. Antibacterial and antifungal activities of fatty acid methyl esters of the blind-your-eye mangrove from India. Braz. J. Microbiol. 2007, 38, 739–742. [Google Scholar] [CrossRef] [Green Version]
- Desbois, A.P.; Lawlor, K.C. Antibacterial activity of long-chain polyunsaturated fatty acids against Propionibacterium acnes and Staphylococcus aureus. Mar. Drugs 2013, 11, 4544–4557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muthamil, S.; Prasath, K.G.; Priya, A.; Precilla, P.; Pandian, S.K. Global proteomic analysis deciphers the mechanism of action of plant derived oleic acid against Candida albicans virulence and biofilm formation. Sci. Rep. 2020, 10, 5113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carabajal, M.A.; Viarengo, G.; Yim, L.; Martínez-Sanguiné, A.; Mariscotti, J.F.; Chabalgoity, J.A.; Rasia, R.M.; Véscovi, E.G. PhoQ is an unsaturated fatty acid receptor that fine-tunes Salmonella pathogenic traits. Sci. Signal. 2020, 13, eaaz3334. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.R.; Erwin, S.; Lanzas, C.; Theriot, C.M. Shifts in the Gut Metabolome and Clostridium difficile Transcriptome throughout Colonization and Infection in a Mouse Model. mSphere 2018, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mefferd, C.C.; Bhute, S.S.; Phan, J.R.; Villarama, J.V.; Do, D.M.; Alarcia, S.; Abel-Santos, E.; Hedlund, B.P. A High-Fat/High-Protein, Atkins-Type Diet Exacerbates Clostridioides (Clostridium) difficile Infection in Mice, whereas a High-Carbohydrate Diet Protects. mSystems 2020, 5. [Google Scholar] [CrossRef] [Green Version]
- Blankenship-Paris, T.L.; Walton, B.J.; Hayes, Y.O.; Chang, J. Clostridium difficile infection in hamsters fed an atherogenic diet. Vet. Pathol. 1995, 32, 269–273. [Google Scholar] [CrossRef]
- Lee, T.R.; Hsu, H.P.; Shaw, G.C. Transcriptional regulation of the Bacillus subtilis bscR-CYP102A3 operon by the BscR repressor and differential induction of cytochrome CYP102A3 expression by oleic acid and palmitate. J. Biochem. 2001, 130, 569–574. [Google Scholar] [CrossRef]
- Deakin, L.J.; Clare, S.; Fagan, R.P.; Dawson, L.F.; Pickard, D.J.; West, M.R.; Wren, B.W.; Fairweather, N.F.; Dougan, G.; Lawley, T.D. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect. Immun. 2012, 80, 2704–2711. [Google Scholar] [CrossRef] [Green Version]
- Underwood, S.; Guan, S.; Vijayasubhash, V.; Baines, S.D.; Graham, L.; Lewis, R.J.; Wilcox, M.H.; Stephenson, K. Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. J. Bacteriol. 2009, 191, 7296–7305. [Google Scholar] [CrossRef] [Green Version]
- Childress, K.O.; Edwards, A.N.; Nawrocki, K.L.; Anderson, S.E.; Woods, E.C.; McBride, S.M. The Phosphotransfer Protein CD1492 Represses Sporulation Initiation in Clostridium difficile. Infect. Immun. 2016, 84, 3434–3444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strauch, M.A.; Trach, K.A.; Day, J.; Hoch, J.A. Spo0A activates and represses its own synthesis by binding at its dual promoters. Biochimie 1992, 74, 619–626. [Google Scholar] [CrossRef]
- Lamendella, R.; Wright, J.R.; Hackman, J.; McLimans, C.; Toole, D.R.; Bernard Rubio, W.; Drucker, R.; Wong, H.T.; Sabey, K.; Hegarty, J.P.; et al. Antibiotic Treatments for Clostridium difficile Infection Are Associated with Distinct Bacterial and Fungal Community Structures. mSphere 2018, 3, e00572-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sangster, W.; Hegarty, J.P.; Schieffer, K.M.; Wright, J.R.; Hackman, J.; Toole, D.R.; Lamendella, R.; Stewart, D.B. Bacterial and Fungal Microbiota Changes Distinguish C. difficile Infection from Other Forms of Diarrhea: Results of a Prospective Inpatient Study. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romo, J.A.; Markey, L.; Kumamoto, C.A. Lipid Species in the GI Tract are Increased by the Commensal Fungus Candida albicans and Decrease the Virulence of Clostridioides difficile. J. Fungi 2020, 6, 100. https://doi.org/10.3390/jof6030100
Romo JA, Markey L, Kumamoto CA. Lipid Species in the GI Tract are Increased by the Commensal Fungus Candida albicans and Decrease the Virulence of Clostridioides difficile. Journal of Fungi. 2020; 6(3):100. https://doi.org/10.3390/jof6030100
Chicago/Turabian StyleRomo, Jesus A., Laura Markey, and Carol A. Kumamoto. 2020. "Lipid Species in the GI Tract are Increased by the Commensal Fungus Candida albicans and Decrease the Virulence of Clostridioides difficile" Journal of Fungi 6, no. 3: 100. https://doi.org/10.3390/jof6030100
APA StyleRomo, J. A., Markey, L., & Kumamoto, C. A. (2020). Lipid Species in the GI Tract are Increased by the Commensal Fungus Candida albicans and Decrease the Virulence of Clostridioides difficile. Journal of Fungi, 6(3), 100. https://doi.org/10.3390/jof6030100





