Effects of mrpigG on Development and Secondary Metabolism of Monascus ruber M7
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains, Culture Media and Growth Conditions
2.2. Cloning and Analysis of the mrpigG Gene
2.3. Deletion, Complementation and Overexpression of the mrpigG Gene
2.4. Real-Time qPCR Analysis
2.5. MP and Citrinin Analyses
3. Results
3.1. Sequence Analysis of the mrpigG Gene in M. ruber M7
3.2. Verification of the mrpigG Deletion, Complementation and Overexpression Strains
3.3. RT-qPCR Analysis of ΔmrpigG, ΔmrpigG::mrpigG, M7::PtrpC-mrpigG and M. ruber M7
3.4. Morphologies and Biomasses of ΔmrpigG, ΔmrpigG::mrpigG, M7::PtrpC-mrpigG and M. ruber M7
3.5. MP and CIT Production Analysis of ΔmrpigG, ΔmrpigG::mrpigG, M7::PtrpC-mrpigG and M. ruber M7
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Chen, W.P.; He, Y.; Zhou, Y.X.; Shao, Y.C.; Feng, Y.L.; Li, M.; Chen, F.S. Edible Filamentous Fungi from the Species Monascus: Early Traditional Fermentations, Modern Molecular Biology, and Future Genomics. Compr. Rev. Food Sci. Food Saf. 2015, 14, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.C.; Lee, G.D.; Choi, I.H. Breast meat quality of broilers fed fermented red ginseng marc powder mixed with red-koji during storage. Emir. J. Food Agric. 2016, 28, 283–287. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.L.; Shao, Y.C.; Chen, F.S. Monascus pigments. Appl. Microbiol. Biotechnol. 2012, 96, 1421–1440. [Google Scholar] [CrossRef]
- Patakova, P. Monascus secondary metabolites: Production and biological activity. J. Ind. Microbiol. Biotechnol. 2013, 40, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lei, M.; Zhou, Y.; Chen, F. A Comprehensive Analysis of the Small GTPases Ypt7 Involved in the Regulation of Fungal Development and Secondary Metabolism in Monascus ruber M7. Front. Microbiol. 2019, 10, 452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanc, P.J.; Laussac, J.P.; Lebars, J.; Lebars, P.; Loret, M.O.; Pareilleux, A.; Prome, D.; Prome, J.C.; Santerre, A.L.; Goma, G. Characterization of Monascidin-a from Monascus as Citrinin. Int. J. Food Microbiol. 1995, 27, 201–213. [Google Scholar] [CrossRef]
- Mukherjee, G.; Singh, S.K. Purification and characterization of a new red pigment from Monascus purpureus in submerged fermentation. Process Biochem. 2011, 46, 188–192. [Google Scholar] [CrossRef]
- Chen, W.; Feng, Y.; Molnar, I.; Chen, F. Nature and nurture: Confluence of pathway determinism with metabolic and chemical serendipity diversifies Monascus azaphilone pigments. Nat. Prod. Rep. 2019, 36, 561–572. [Google Scholar] [CrossRef]
- Chen, W.P.; Chen, R.; Liu, Q.P.; He, Y.; He, K.; Ding, X.L.; Kang, L.J.; Guo, X.X.; Xie, N.N.; Zhou, Y.X.; et al. Orange, red, yellow: Biosynthesis of azaphilone pigments in Monascus fungi. Chem. Sci. 2017, 8, 4917–4925. [Google Scholar] [CrossRef] [Green Version]
- Balakrishnan, B.; Chandran, R.; Park, S.H.; Kwon, H.J. A New Protein Factor in the Product Formation of Non-Reducing Fungal Polyketide Synthase with a C-Terminus Reductive Domain. J. Microbiol. Biotechnol. 2015, 25, 1648–1652. [Google Scholar] [CrossRef]
- Chen, F.S.; Hu, X.Q. Study on red fermented rice with high concentration of monacolin K and low concentration of citrinin. Int. J. Food Microbiol. 2005, 103, 331–337. [Google Scholar] [CrossRef]
- He, Y.; Liu, Q.; Shao, Y.; Chen, F. Ku70 and ku80 null mutants improve the gene targeting frequency in Monascus ruber M7. Appl. Microbiol. Biotechnol. 2013, 97, 4965–4976. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Liu, J.; Fang, Y.; Shao, Y.; Li, L.; Yu, J.H.; Chen, F. Effects of Different G-Protein alpha-Subunits on Growth, Development and Secondary Metabolism of Monascus ruber M7. Front. Microbiol. 2019, 10, 1555. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Xie, N.; He, Y.; Wang, L.; Shao, Y.; Zhao, H.; Chen, F. MpigE, a gene involved in pigment biosynthesis in Monascus ruber M7. Appl. Microbiol. Biotechnol. 2014, 98, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.C.; Ding, Y.D.; Zhao, Y.; Yang, S.; Xie, B.J.; Chen, F.S. Characteristic analysis of transformants in T-DNA mutation library of Monascus ruber. World J. Microbiol. Biotechnol. 2009, 25, 989–995. [Google Scholar] [CrossRef]
- Li, L.; He, L.; Lai, Y.; Shao, Y.; Chen, F. Cloning and functional analysis of the Gbeta gene Mgb1 and the Ggamma gene Mgg1 in Monascus ruber. J. Microbiol. 2014, 52, 35–43. [Google Scholar] [CrossRef]
- Mapari, S.A.S.; Thrane, U.; Meyer, A.S. Fungal polyketide azaphilone pigments as future natural food colorants? Trends Biotechnol. 2010, 28, 300–307. [Google Scholar] [CrossRef]
- Lee, C.L.; Wen, J.Y.; Hsu, Y.W.; Pan, T.M. The blood lipid regulation of Monascus-produced monascin and ankaflavin via the suppression of low-density lipoprotein cholesterol assembly and stimulation of apolipoprotein A1 expression in the liver. J. Microbiol. Immunol. Infect. 2018, 51, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Bachovchin, D.A.; Cravatt, B.F. The pharmacological landscape and therapeutic potential of serine hydrolases. Nat. Rev. Drug Discov. 2012, 11, 52–68. [Google Scholar] [CrossRef] [Green Version]
- Holmquist, M. Alpha/Beta-Hydrolase Fold Enzymes: Structures, Functions and Mechanisms. Curr. Protein Pept. Sci. 2000, 1, 209–235. [Google Scholar] [CrossRef]
- Bun, J.S.; Slack, M.D.; Schemenauer, D.E.; Johnson, R.J. Comparative analysis of the human serine hydrolase OVCA2 to the model serine hydrolase homolog FSH1 from S. cerevisiae. PLoS ONE 2020, 15, e0230166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, J.Z.; Cravatt, B.F. The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease. Chem. Rev. 2011, 111, 6022–6063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navia-Paldanius, D.; Savinainen, J.R.; Laitinen, J.T. Biochemical and pharmacological characterization of human alpha/beta-hydrolase domain containing 6 (ABHD6) and 12 (ABHD12). J. Lipid Res. 2012, 53, 2413–2424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.K.; Johnson, L.C.; Clodfelter, J.E.; Pemble, C.W.; Fulp, B.E.; Furdui, C.M.; Kridel, S.J.; Lowther, W.T. Crystal Structure and Substrate Specificity of Human Thioesterase 2: Insights into the Molecular Basis for the Modulation of Fatty Acid Synthase. J. Biol. Chem. 2016, 291, 3520–3530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Zhou, Y.; Yi, T.; Zhao, M.; Xie, N.; Lei, M.; Liu, Q.; Shao, Y.; Chen, F. Identification and role analysis of an intermediate produced by a polygenic mutant of Monascus pigments cluster in Monascus ruber M7. Appl. Microbiol. Biotechnol. 2016, 100, 7037–7049. [Google Scholar] [CrossRef]
- Fisch, K.M.; Skellam, E.; Ivison, D.; Cox, R.J.; Bailey, A.M.; Lazarus, C.M.; Simpson, T.J. Catalytic role of the C-terminal domains of a fungal non-reducing polyketide synthase. Chem. Commun. 2010, 46, 5331–5333. [Google Scholar] [CrossRef] [PubMed]


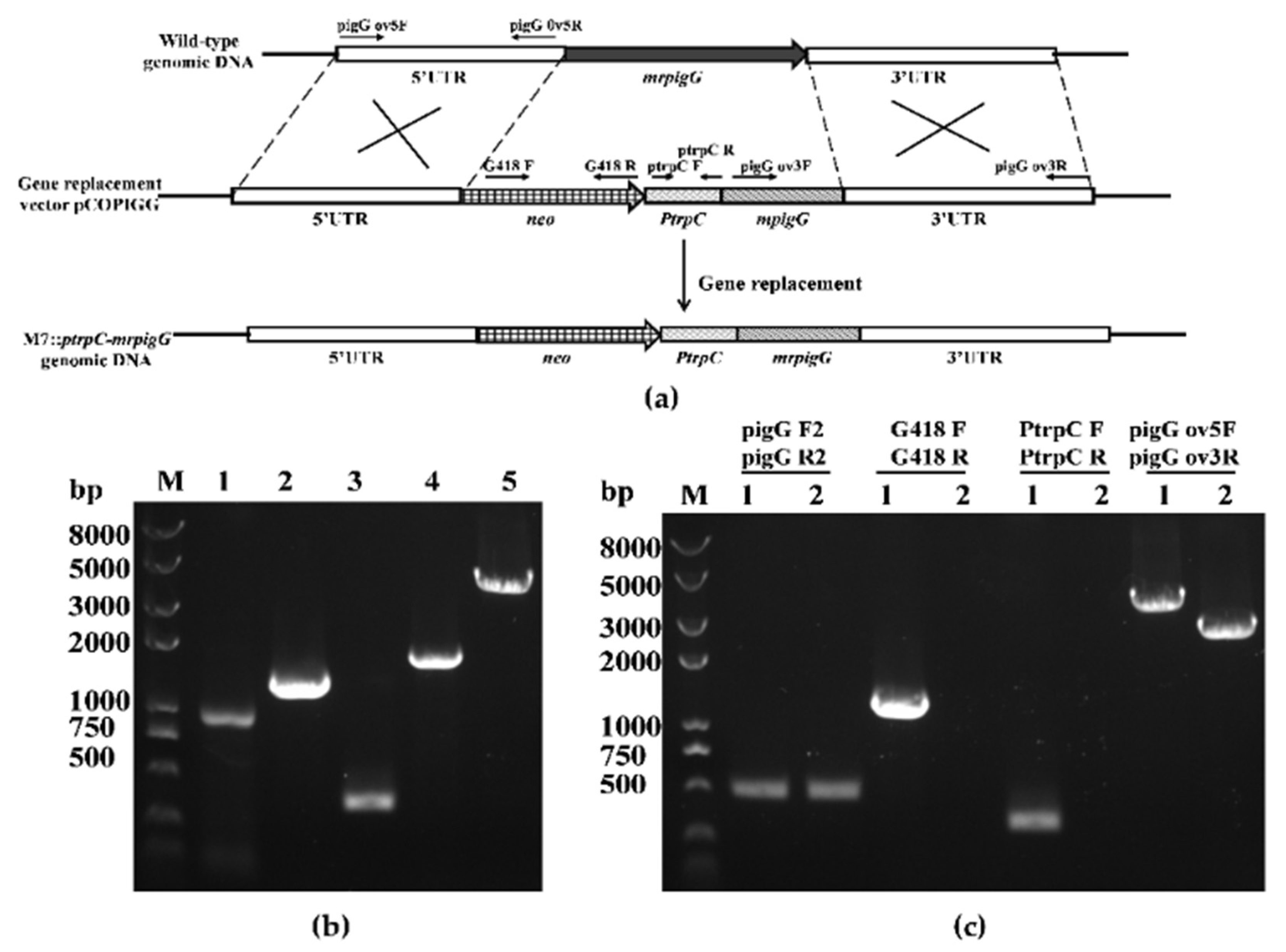
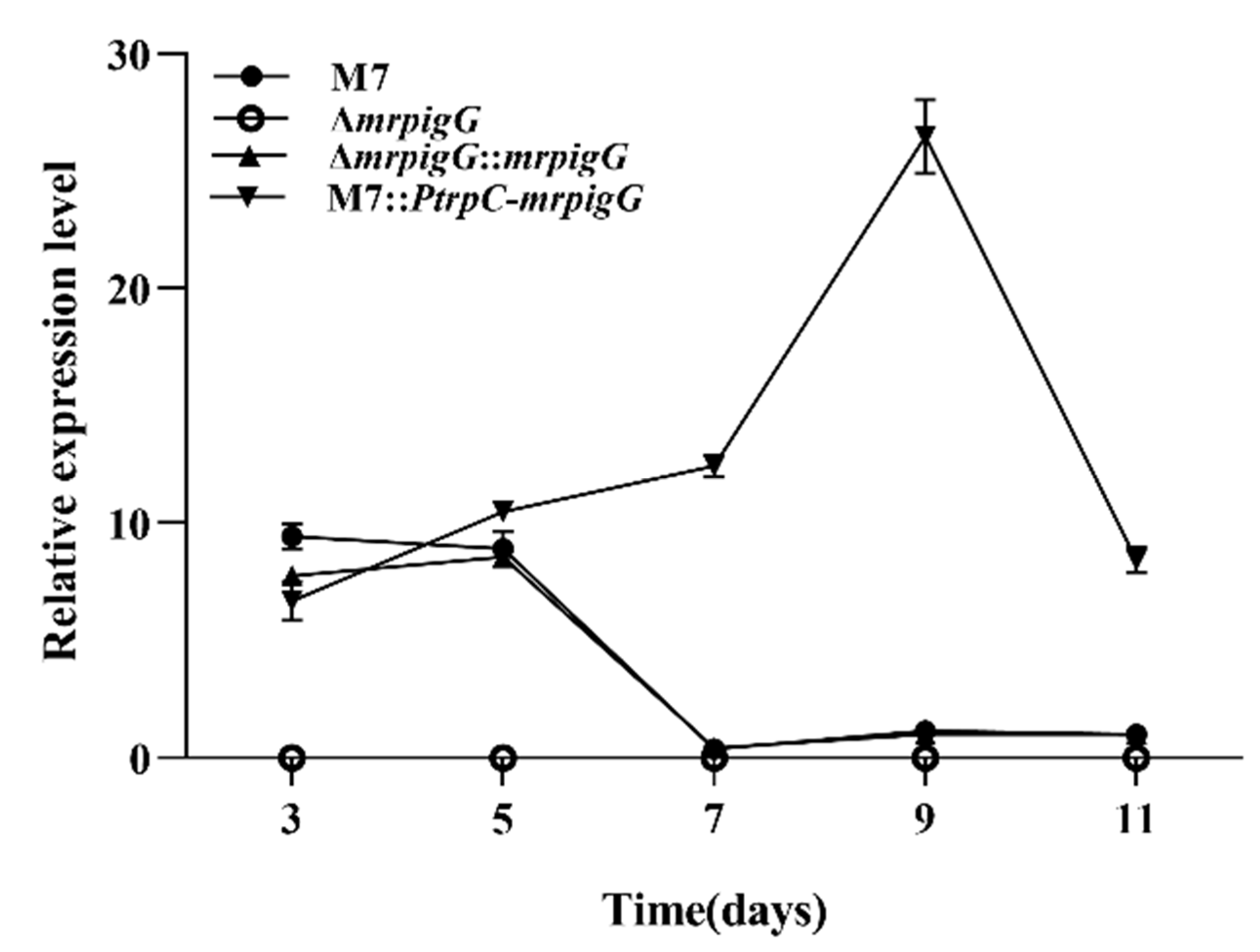
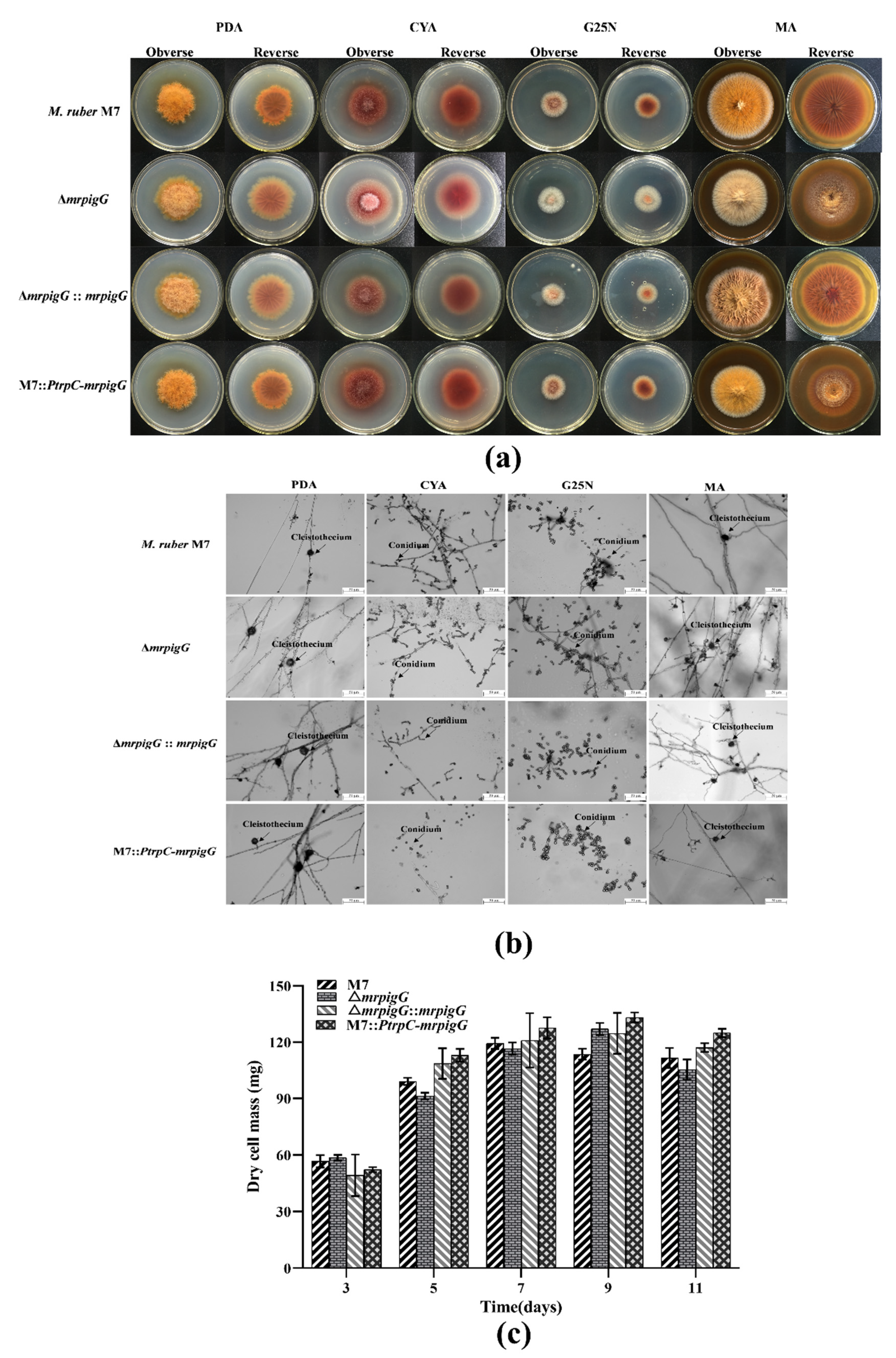



| Strain | Parent | Genotype | Source |
|---|---|---|---|
| M7 | M7 | Wild-type | Red fermented rice [11] |
| ΔmrpigG | M7 | ΔmrpigG::hph | This study |
| ΔmrpigG:: mrpigG | ΔmrpigG | ΔmrpigG::mrpigG-neo | This study |
| M7::PtrpC-mrpigG | M7 | M7::PtrpC-mrpigG-neo | This study |
| Names | Sequences (5′→3′) | Descriptions |
|---|---|---|
| pigG F1 | GATCTGCCAGAAATACTAGA | For amplification of the 1236 bp of the whole mrpigG gene |
| pigG R1 | TCGGCGAAGCAGAGGCGGAA | |
| pigG d5F | CGTCCCCCTTCTGCCCAAGA | For amplification of the 884 bp of 5′ flanking regions of the mrpigG gene |
| pigG d5R | CAATATCATCTTCTGTCGAC CCGAACTCCTTGTAGACCGA | |
| hph F | GTCGACAGAAGATGATATTG | For amplification of the 2137 bp of the hph cassette from plasmid pSKH |
| hph R | CTAGAAAGAAGGATTACCTC | |
| pigG d3F | GAGGTAATCCTTCTTTCTAGGTGCCGATCAAGACGAAGGA | For amplification of the 867 bp of 3′ flanking regions of the mrpigG gene |
| pigG d3R | CTCTTCCAGCAGGACCAACT | |
| pigG c5F | CCAGACACCGAACAGCCGCA | For amplification of the 1823 bp of 5′ flanking regions and open reading frame (ORF) of the mrpigG gene |
| pigG c5R | GGTTACGGTTCGATGGGGTTGAGTTGGGTCTCGCTGTAGGGCTGGAT | |
| G418 F | CCAACTCAACCCCATCGAACCGTAACC | For amplification of the 1221 bp of the neo cassette from plasmid pKN1 |
| G418 R | ATCATCATGCAACATGCATG | |
| pigG c3F | CATGCATGTTGCATGATGATCGATCTTCTTCGCAGACACC | For amplification of the 860 bp of 3′ flanking regions of the mrpigG gene |
| pigG c3R | CGTCACTCGCTTCCAGGTCG | |
| pigG ov5F | CAGACATACTGCTAAACTCG | For amplification of the 969 bp of 5′ flanking regions of the mrpigG gene |
| pigG ov5R | GGTTACGGTTCGATGGGGTTGAGTTGGGGTGCGGTGCTGGCGAGAGT | |
| ptrpC F | CATGCATGTTGCATGATGATGTCGACAGAAGATGATATTG | For amplification of the 373 bp of the ptrpC promoter from plasmid pKSH |
| ptrpC R | CATATCGATGCTTGGGTAGA | |
| mrpigG ov3F | TATTCTACCCAAGCATCGATATG ATGCCAGCCAACCGCTCCAG | For amplification of the 1621 bp of 3′ flanking regions and ORF of the mrpigG gene |
| pigG ov3R | TCTTCCAGCAGGACCAACTC | |
| pigG F2 | GCGCTGGCTGCGCTCAT | For amplification of the 503 bp of the partial mrpigG gene |
| pigG R2 | CCTCCCACTCCATAACCC | |
| pigG qF | GGGTGAATGGGCGGGACTA | For real-time qPCR analysis of mrpigG(144bp) |
| pigG qR | GCCAGCAATACGGCAAAGC | |
| GAPDH F | CAAGCTCACTGGCATGTCTATG | For real-time qPCR analysis of GAPDH (243bp) |
| GAPDH R | AAGTTCGAGTTGAGGGCGATA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Chen, F. Effects of mrpigG on Development and Secondary Metabolism of Monascus ruber M7. J. Fungi 2020, 6, 156. https://doi.org/10.3390/jof6030156
Li L, Chen F. Effects of mrpigG on Development and Secondary Metabolism of Monascus ruber M7. Journal of Fungi. 2020; 6(3):156. https://doi.org/10.3390/jof6030156
Chicago/Turabian StyleLi, Li, and Fusheng Chen. 2020. "Effects of mrpigG on Development and Secondary Metabolism of Monascus ruber M7" Journal of Fungi 6, no. 3: 156. https://doi.org/10.3390/jof6030156
APA StyleLi, L., & Chen, F. (2020). Effects of mrpigG on Development and Secondary Metabolism of Monascus ruber M7. Journal of Fungi, 6(3), 156. https://doi.org/10.3390/jof6030156






