Abstract
Pneumocystis pneumonia (PcP) is a disease produced by the opportunistic infection of the fungus Pneumocystis jirovecii. As delayed or unsuitable treatments increase the risk of mortality, the development of rapid and accurate diagnostic tools for PcP are of great importance. Unfortunately, current standard methods present severe limitations and are far from adequate. In this work, a time-competitive, sensitive and selective biosensor based on DNA-gated nanomaterials for the identification of P. jirovecii is presented. The biosensor consists of a nanoporous anodic alumina (NAA) scaffold which pores are filled with a dye reporter and capped with specific DNA oligonucleotides. In the presence of P. jirovecii genomic DNA, the gated biosensor is open, and the cargo is delivered to the solution where it is monitored through fluorescence spectroscopy. The use of capping oligonucleotides able to form duplex or triplex with P. jirovecii DNA is studied. The final diagnostic tool shows a limit of detection (LOD) of 1 nM of target complementary DNA and does not require previous amplification steps. The method was applied to identify DNA from P. jirovecii in unmodified bronchoalveolar lavage, nasopharyngeal aspirates, and sputum samples in 60 min. This is a promising alternative method for the routinely diagnosis of Pneumocystis pneumonia.
1. Introduction
Pneumocystis jirovecii is an opportunistic fungus that can cause asymptomatic or mild infection in healthy people and fulminating pneumonia in immunocompromised individuals [1]. Pneumocystis spp. can affect many species of mammals (rodent, horses, and primates) and presents strong specificity for host species. The fungus was identified as a human pathogen in the 1940s when it was detected in the lungs of children affected with interstitial pneumonia of plasmatic cells [2]. Usually, in humans, the pathogen is disseminated from person to person producing asymptomatic or subclinical infection; however, when it is transmitted to an immunocompromised host, a severe pneumonia can take place (Pneumocystis pneumonia, PcP) [3]. In the 1980s, the high incidence of acquired immunodeficiency syndrome (AIDS) lead to a markedly interest in PcP due to the high incidence in those patients. Nowadays, the widespread use of immunosuppressive treatments such as corticosteroid therapies, chemotherapies, or biological therapies, has increased the incidence of P. jirovecii infections. In addition, recent studies have provided evidence of transplacental transmission in humans and stablished a relation between P. jirovecii colonization in preterm infants and neonatal Respiratory Distress Syndrome risk [4,5].
In the last decade, several published works have allowed a high progress in the knowledge of the Pneumocystis pathobiology; however, morbidity and mortality due to PcP still remains high, especially in developing countries. One of the main challenges in the diagnosis of PcP is the absence of a specific clinical manifestation, which difficult diagnosis and makes it stand on non-specific symptoms [6]. Therefore, a proper diagnosis of Pneumocystis pneumonia involves correct recognition of the fungus. Unfortunately, the current standard methods for P. jirovecii detection are far from adequate, and further aggravated by the impossibility to keep continuous fungus ex vivo axenic culture [7].
Classical methodologies for the identification of P. jirovecii comprise microscopic observation of stained biological samples that reveal the characteristic morphology of Pneumocystis cystic and/or trophic forms. These techniques provide sensitivity and specificity values between 60–75% and 80–90%, respectively; however, they remain time-consuming and require high experienced staff and specific equipment [8,9,10]. Some alternatives for PcP diagnosis involve the detection of blood biomarkers that indicate the host-pathogen interaction, including (bio)molecules such as (1,3)-β-d-glucan (BG), lactate dehydrogenase (LDH), Krebs von den Lungen-6 antigen (KL-6), and adenosylmethionine (SAM) [10]. Nonetheless, despite their good sensitivity (70–95%), the detected metabolites are not strictly specific to P. jirovecii infection, which significantly reduces the specificity values (50–70%) [10]. On the other hand, serological methods detect the antibodies produced by the patient as consequence of the presence of the fungus, but despite their greater selectivity, they are expensive, require long-time procedures, and there is still a need to better characterize the immune response to obtain better standardized protocols [9,11,12].
Among molecular techniques, PCR assays have revealed to be highly efficient, allowing even the early detection of the fungus in samples from patients that were confirmed negative by microscopic examination [8]. During the last 15 years, numerous PCR techniques for the detection of genomic DNA from P. jirovecii have been reported, obtaining sensitivity and specificity values close to 90–100%. The most remarkable studies employ both conventional PCR and its variants nested-PCR, and real time PCR, and they are applied to recognize target gene such as the mitochondrial large subunit RNA (mtLSUrRNA), the internally transcribed spacer (ITS) or 18S or 5S ribosomal RNA (18S rRNA or 5S rRNA) [8]. However, despite its great potential, PCR is not exempt from limitations such us high susceptibility to polymerase inhibitors or cross contamination [13].
In the last decades, new diagnostic platforms based upon nanotechnology have become a reality thanks to their ability to provide sensitive, accurate and rapid results. For instance, nanotechnology has burst into Pneumocystis diagnosis by developing serological biosensors that uses gold nanoparticles (AuNPs) to detect anti-P. jirovecii antibodies by a colorimetric assay in a lateral flow immunoassay [14,15]. Nevertheless, the development of nanosensors for PcP detection is still an incipient area. However, this is a stimulating field of research and recent advances have demonstrated that the design of new hybrid organic–inorganic systems in a single entity can benefit from both, the potential of nanomaterials and the recognition, selectivity, and sensitivity properties of biomolecules. In that regard, several hybrid systems have been extensively applied in sensing and drug delivery applications [16,17,18,19,20,21,22,23].
Among the great variety of nanomaterials available, nanoporous anodic alumina (NAA) has been widely used because of its biocompatibility, high surface area, high loading capacity, and easy modification of the surface. Moreover, NAA supports can be easily prepared by cost-competitive and well-known production techniques [24]. NAA have additionally been used to develop capped stimuli-responsive systems, in which the mesoporous support is loaded with a cargo and capped with a biomolecule, so that only target molecules are able to induce cargo release. In NAA, oligonucleotides as capping systems have recently been demonstrated to be excellent candidates to detect and quantify a range of molecules and biomolecules [24,25]. Some of these tools rely on the use of DNA as caps and base their recognition mechanism on duplex hybridization interactions (DNA-DNA or DNA-RNA) between the probe and the target. Nevertheless, innovative triple-helix hybridization formats based on Hoogsteen and reverse-Hoogsteen base pairs to the Watson-Crick duplex are also interesting approaches that have demonstrated increased recognition efficiency.
Triplex formation is observed at certain polypurine-polypyrimidine sequences that are widely found in the human genome, particularly at promoter regions [26,27,28,29]. Several published studies have described the triplex-stabilizing features of 8-aminopurines and the use of parallel and antiparallel tail-clamps to improve the efficacy of triplex recognition with complementary RNA and DNA targets [30,31]. On that regard, this triplex hybridization has been effectively applied to develop biosensors. For example, Carrascosa et al. and Aviñó et al. developed a sensitive and label-free method for the detection of an mRNA from Listeria and miRNA-145 by the formation of triplex hybridization structures in SPR biosensors [32]. More recently, Wei et al. have reported a method that combines the formation of triplex DNA structures with further amplification reactions for the sensitive detection of microRNAs [33] and some of us have developed an oligonucleotide-gated mesoporous support for the identification of miRNA-145 based on the formation of triple-helix structure, allowing a sensitivity as low as 2.5 pM and accurate qualitative determination in human serum samples [34].
Based on the above, we report herein two probes for the specific identification of P. jirovecii using gated nanomaterials. The diagnostic tool is based in NAA, whose pores are loaded with the fluorophore rhodamine B and blocked with different oligonucleotide probes (single or hairpin strands, see Scheme S1) able to form duplex or triplex structures with the target gene of mtLSUrRNA of P. jirovecii DNA. Best analytical performances are obtained for the triplex hybridization with a LOD of 1 nM in less than one hour without previous DNA amplification. Likewise, this method detects P. jirovecii DNA in unmodified sputum, nasopharyngeal aspirates (NPA), and bronchoalveolar lavage (BAL) samples, showing its great potential in point-of-care applications.
2. Materials and Methods
2.1. General Techniques
A ZEISS Ultra 55 microscope was employed to perform field emission scanning electron microscopy (FSEM) and energy dispersive X-ray spectroscopy (EDX) analyses. Fluorescence spectroscopy measurements were carried out on a Synergy H1 microplate reader (BioTek, Winooski, VT, USA).
2.2. Chemicals
Tris(hydroxymethyl)aminomethane (TRIS), hydrochloric acid, (3-aminopropyl)triethoxysilane (APTES), and rhodamine B, were obtained from Sigma-Aldrich Quimica (Madrid, Spain). NAA scaffolds were purchased from InRedox (Longmont, CO, USA).
2.3. Synthesis of Oligonucleotides
In this study it was used the gene encoding the mitochondrial large-subunit of P. jirovecii (mt LSU rRNA). Design of oligonucleotides and in silico analysis is described in the Electronic Supplementary Information (Scheme S1). Table 1 shows the sequences of the oligonucleotides employed in this study. The synthesis took place by the well-defined phosphoramidite solid phase protocol [32]. For this, oligonucleotides were assembled on controlled pore glass (CPG) scaffolds by consecutive incorporation of the suitable phosphoramidites employing an automated Applied Biosystems 394 DNA synthesizer (Foster City, CA, USA). Then, the scaffolds were treated overnight with concentrated ammonia at 55 °C. Finally, synthetized oligonucleotides were purified by Glen-PackTM DNA cartridges (Glen Research, Sterling, VA, USA) and mass spectrometry was used to analyze them (Table S1, ESI).

Table 1.
Sequences of the used oligonucleotides.
2.4. Synthesis of Nanomaterials S0, S1 (Duplex), S2 (Clamp), and S3 (Control)
For the synthesis of S0, 10 individual NAA scaffolds (2 mm of diameter each one) were submerged in a solution of rhodamine B in CH3CN (1 mM, 8 mL). The mixture was stirred at room temperature for 24 h. Then the functionalization of the surface with aminopropyl moieties was carried out by adding 40 µL per individual NAA scaffold of (3-aminopropyl)triethoxysilane (1.25 µM) and stirring the mixture at room temperature for 6 h. For the synthesis of the sensing probes S1, S2, and S3, different pieces of solid S0 were capped by adding to each support 10 µL of the corresponding oligonucleotide (O1, O2, or O3, respectively) (100 µM) in a final volume of 250 µL of hybridization buffer (20 mM Tris-HCl, 37.5 mM MgCl2, pH 7.5). The mixtures were shaken for 60 min at 37 °C. The capped materials were rinsed with hybridization buffer to remove the unbounded oligonucleotide.
2.5. Release Kinetics
The response of the gated materials was determined by measuring the fluorescence of the dye released from the pores to the solution in the presence of the target complementary DNA. In a common experiment, two independent supports of each material S1, S2, and S3 were immersed in 900 µL of hybridization buffer. Then, 100 µL of the complementary DNA (10 µM) was added to one of the supports while 100 µL of hybridization buffer was added to the other. Both solutions were stirred at 37 °C and aliquots were taken at scheduled times. Finally, dye delivery was determined by registering the fluorescence of the rhodamine B in the solution at 575 nm (λexc = 555 nm).
2.6. Real Media Experiments
The response of S2 was assessed in a more realistic context. For that, 500 µL of sputum, bronchoalveolar lavage (BAL) and nasopharyngeal aspirate (NPA) samples were artificially inoculated with 100 µL of DNA from P. jirovecii (10 µM) and added to two independent S2 supports in a final volume of 1 mL in hybridization buffer. Solutions were maintained at 37 ˚C and rhodamine B released from the pores was measured at 575 nm (λexc = 555 nm) after 60 min.
2.7. Response to Different Target Concentrations
The response of the material S2 to different concentrations of the target DNA was assessed. For this, six independent supports were submerged separately in 900 µL of hybridization buffer and 100 µL of 10-fold diluted target solutions were added to each one, reaching final concentrations from 10−5 to 1 µM. After 60 min at 37 °C, the released rhodamine B from the porous was measured at 575 nm (λexc 555 nm).
2.8. Selectivity to Possible Interferents
The selectivity of the method was determined for S2 by carrying out cargo delivery experiments in the presence of 100 µL of DNA (10 ng/µL) from other microorganisms (Aspergillus spp., Candida albicans, Candida tropicalis, Candida auris, Schizosaccharomyces pompe, and Taphrina deformans) in a final reaction volume of 1 mL of hybridization buffer. In the same experiment, 100 µL of complementary DNA from P. jirovecii at 10 ng/µL was used as a positive control and 100 µL of hybridization buffer as a negative control. Mixtures were maintained in agitation for 60 min at 37 °C and the delivered rhodamine B was determined by measuring the fluorescence in the solution (λexc = 555 nm, λem = 585 nm).
2.9. Validation of the Method in Clinical Samples
In order to determine the clinical applicability of the gated nanosensor, the performance of S2 was evaluated in a more realistic media. First, the system was used to analyze 8 sputa and 4 bronchoalveolar lavages (BAL) from infected and non-infected patients from the Hospital Universitari i Politècnic La Fe. In another experiment, 21 nasopharyngeal aspirates (NPA) from colonized and non-colonized newborn infants from the Hospital Universitario Virgen del Rocío were analyzed using S2 nanomaterials. For both assays, 250 µL of each sample were added to individual S2 solids in a final volume of 400 µL of hybridization buffer. After 60 min at 37 °C, dye released from the pores to the solution was measured by fluorescence spectroscopy at 575 nm (λexc 555 nm).
3. Results and Discussion
3.1. Synthesis and Characterization of the Biosensors
Figure 1 depicts the synthetic process followed to obtain the biosensors and their performance in the presence of the target DNA. NAA scaffolds were selected as inorganic supports to develop the capped nanomaterials. First, pores were filled with the dye rhodamine B which has demonstrated excellent properties for the preparation of optical gated nanosensors applied to the detection of different analytes [18,19,20,21,22,23,24,25,26]. In a second step, the outer surface of the NAA support was functionalized with the linker (3-aminopropyl)triethoxysilane, yielding S0. At a neutral pH, aminopropyl moieties are partially protonated (and therefore positively charged) being able to electrostatically interact with negatively charged oligonucleotides. Based on this, the interaction of oligonucleotides O1, O2, or O3 with the amino-functionalized S0, yielded the gated sensing solids S1, S2, and S3. O1 consists of a single strand oligonucleotide that hybridizes with the DNA sequence forming a duplex structure, O2 is a clamp that forms a triplex hybridization structure, whereas O3 is also a clamp but results in a duplex conformation.
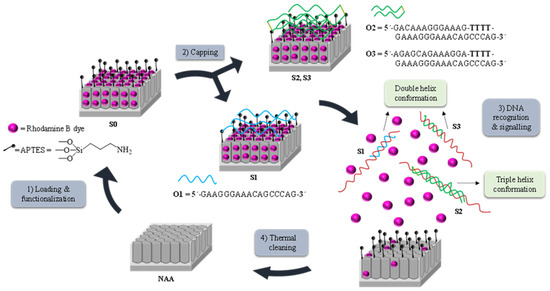
Figure 1.
Representation of the preparation and sensing process: (1) Loading and functionalization of the nanoporous anodic alumina (NAA) support; (2) Capping with the oligonucleotides O1, O2, or O3, to obtain solids S1, S2, and S3, respectively; (3) Release of the cargo in the presence of P. jirovecii genomic DNA by forming (A) duplex structures when using solids S1 or S3 or (B) triplex hybridization structures when using S2 and (4) Calcination prior to the reutilization of the scaffold.
The recognition and detection mechanism of the loaded and capped S1, S2, and S3 materials takes place as follows. In the absence of target analyte, the oligonucleotides O1, O2, or O3, electrostatically attached to the external surface of the inorganic scaffold, were expected to be bulky enough to block pores and to inhibit dye delivery. On the contrary, due to the higher affinity of the oligonucleotide probes for the target than for the aminopropyl moieties, it was expected that in the presence of the analyte, the capping oligonucleotides were selectively displaced from the surface resulting in DNA target-probe hybridization, pore opening and dye diffusion from the pores to the aqueous solution. Finally, the used solids were calcined to remove organic matter, allowing supports to be used again (Figure 1).
The starting NAA supports were commercially obtained from InRedox® (Longmont, CO, USA). The supports are anodic aluminum oxide (AAO) films grown on a 0.1 mm thick aluminum layer with a pore density of 9·1011 cm−2. Pore entrance has a funnel-like form which gradually decreases from a larger size (20–30 nm) at the top of the funnel to a 5 nm size at the end. Pores have a profundity of ca. 10 µm. The starting NAA scaffold and S1, S2, and S3 were characterized following standard techniques such as FESEM and EDX analysis. FESEM images of the starting NAA scaffolds confirmed the expected non-ordered porous structure, whereas typical imagens of S1, S2, and S3 showed the presence of an organic layer attributed to the capping oligonucleotides (Figure 2). Furthermore, in S1, S2, and S3, the porous structure of the NAA scaffolds can still be seen in some places where the surface was not completely covered, demonstrating that the loading, functionalization, and capping steps did not modify the NAA structure. This behavior agrees with that observed in previous studies carried out with the same or similar materials [35,36,37].
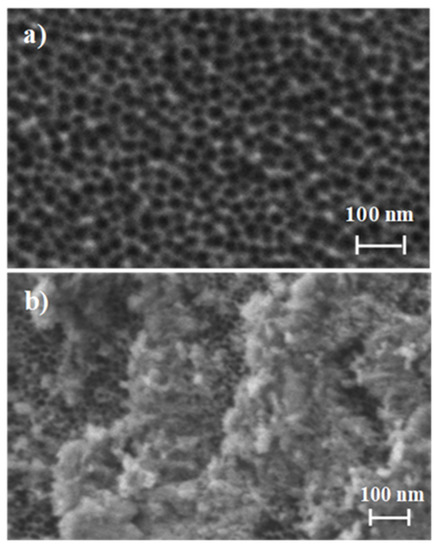
Figure 2.
Representation of the preparation and sensing process: (a) Loading and functionalization of the NAA support; (b) Capping with the oligonucleotides.
The organic content of the different prepared materials (S0, S1, S2, and S3) was studied by energy dispersive X-ray spectroscopy (EDX) (Table 2). After dye loading and surface functionalization with aminopropyl moieties, solid S0 shows a high C/Al content, which was reduced in S1, S2, and S3 due to the partial rhodamine B release that takes place during the gating process. Moreover, the existence of nitrogen in all final solids is indicative of the presence of aminopropyl groups and oligonucleotides. The occurrence of the latter is also confirmed by the presence of phosphorous atoms.

Table 2.
Atomic elements ratio in the different prepared solids by energy dispersive X-ray spectroscopy (EDX) studies.
3.2. Delivery Kinetics
In a first step, the response of the nanoprobes S1, S2, and S3 to the presence of the target sequence O4 was studied. In a typical experiment, two individual solids of each material S1–S3 were submerged in hybridization buffer. Then, 100 μL of the complementary oligonucleotide sequence (O4, 1 ng μL−1) were transferred to one of the supports while 100 µL of buffer were added to the other. Mixtures were stirred at 37 °C and at predetermined times (i.e., 15, 30, 45, and 60 min) the fluorescence of the solution was measured to quantify the amount of the dye delivered from the pores to the aqueous phase. As an example, Figure 3 shows the delivery profile for solid S2. When the target sequence O4 was present, a remarkable dye delivery from the pores to the solution was observed as a result of the oligonucleotide-DNA hybridization to form a DNA triplex structure (curve b), pore opening and dye release. The maximum dye diffusion from the pores to the solution was reached at 60 min and this value was considered the 100% of delivered dye. On the contrary, in the lack of the O4 sequence, a very low dye is released, what indicates tight pore blockage by the probes electrostatically attached to the scaffold surface (curve a).
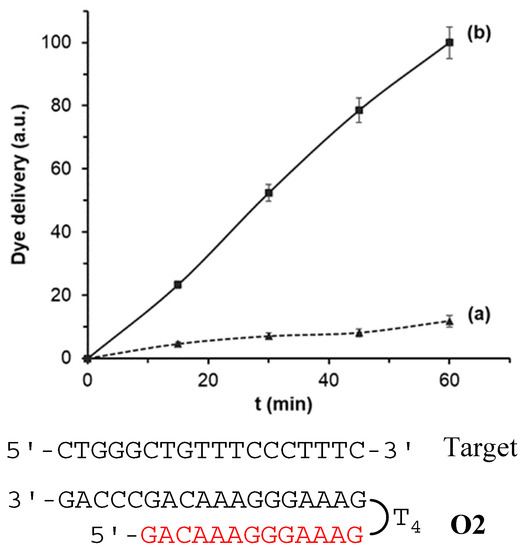
Figure 3.
Rhodamine B delivery from S2 (a) in the absence and (b) in the presence of 1 nM of complementary O4 target sequence in a solution of hybridization buffer (20 mM Tris-HCl, 37.5 mM MgCl2, pH 7.5).
Similar delivery profiles were observed for solids S1 and S3, also indicating adequate performance for these solids in the stages of DNA recognition and dye release (Figure S1, ESI). In terms of blockage ability, all three developed systems were able to keep the pore entrances closed regardless the capping oligonucleotide was a clamp (S2 and S3) or a single strand, obtaining in all cases a very low residual dye delivery (lower than 20%) in the absence of O4 target sequence.
All three nanodevices were also able to deliver the cargo in the presence of target sequence O4. However, differences in terms of the amount of dye delivered were observed when comparing the formation of triplex (S2) and duplex hybridization (S1 and S3) structures. The obtained results showed that nanomaterial S2 provided a faster response and the highest cargo release, reaching a first order constant rate of the release kinetics of 1.70 and an amount of ca. 2.8 ng of delivered Rhodamine B, calculated by interpolating the fluorescence of the amount of dye diffused from the pores to the solution after 60 min into a calibration curve of Rhodamine B. On the other hand, for S1 and S3 materials, the uncapping mechanism and subsequent dye release was slower (constant rates of 1.16 and 1.14, respectively), and the amount of dye delivered at 60 min was lower (by ca. 20%). This in tentatively attributed to a more stable, favorable and rapid formation of the triplex structure than those obtained by the duplex conformations (S1 and S3). These results agree with others found in the literature in which the efficiency and stability of triplex hybridization structures were higher than those obtained by duplex conformations [33,34] Therefore, further studies were carried out using S2 as sensing material.
3.3. Analytical Performance: Sensitivity and Specificity Studies
New diagnostic tools should be evaluated in terms of limit of detection (LOD) and specificity. The LOD in S2 was determined via the study of the response of the gated material to the presence of different amounts of target sequence O4. Based on the proposed sensing mechanism, the amount of released dye depends on the number of opened pores, which in turn depends on the number of target molecules that will induce the displacement of the capping molecules. Results depicted in Figure 4A show that the amount of delivered dye was directly proportional to the target DNA concentration, which agrees with the uncapping protocol detailed above. A LOD of 1 ± 0.1 nM was calculated based on the intersection point of the two slopes of the represented curve (Figure 4A). This LOD value is in the range of other reported protocols for the detection of P. jirovecii genomic DNA. Thus, for instance, several works based on the diagnosis of PcP by real time PCR reported LODs of 50–200 copies/µL, which corresponds to a DNA concentration range from ca. 250 pM to 1 nM. However, our system is faster, simpler, and requires no especial equipment or trained personnel [38,39,40,41,42,43].
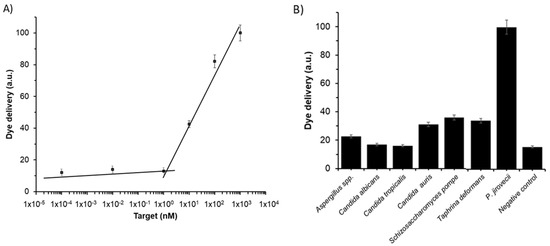
Figure 4.
Release of rhodamine B from solid S2 (A) in the presence of different concentrations of O4 and (B) in the presence of 1 ng/µL of genomic DNA from Aspergillus spp., C. albicans, C. tropicalis, C. auris, S. pompe, T. deformans, P. jirovecii, and 100 µL of hybridization buffer as a negative control. Assays were performed at 60 min in hybridization buffer at pH 7.5.
The specificity of the system was evaluated by assessing the response of S2 to the presence of genomic DNA from other fungi considered as possible interferents, such as Aspergillus spp., C. albicans, C. tropicalis, C. auris, S. pompe, and T. deformans. For that, the amount of dye delivered from the pores of seven independent S2 supports was measured in the presence of 100 μL of DNA at 10 ng/µL of each possible interferent. Moreover, 100 µL of DNA from target O4 at 10 ng/µL and 100 µL of hybridization buffer were added to S2 as positive and negative control, respectively. As it is shown in Figure 4B, target sequence O4, present in P. jirovecii genomic DNA, is the only capable of induce a notable pore opening and rhodamine B release, whereas all the other fungi induced less response and lower dye release. That results confirmed the high selectivity of the system, even in the presence of DNA from S. pompe and T. deformans, which belongs to the same subphylum as Pneumocystis genus [44].
In a step forward, the robustness of the system was determined by evaluating the behavior of S2 to the presence of the target sequence O4 (0.1 ng/µL) in different respiratory fluids such as sputum, bronchoalveolar lavage (BAL) and nasopharyngeal aspirate (NPA) (Figure S2, ESI). In all media, the presence of O4 produced a selective displacement of the oligonucleotide in S2, pore uncapping and dye delivery, whereas in the absence of target, a poor dye release was observed. A lower signal intensity was found in the sputum than in the BAL, NPA, and buffer media (60% vs. 100%), probably due to the higher complexity of the sputum samples. Nevertheless, the amount of dye delivered in all cases and the differences with respect to control samples (not having target O4 sequence) is significant, allowing an accurate fungi DNA detection in all the samples (Figure S2, ESI). These results show that the proposed method may be a promising alternative to classical procedures for the detection of DNA from P. jirovecii in realistic environments.
In addition, it is worth mentioning that previous studies have demonstrated the possible re-use of these type of gated nanomaterials [25,36]. In this respect, used NAA supports were calcined to remove organic matter and reused up to 3 times for the detection of O4 obtaining similar results.
3.4. P. jirovecii Detection in Clinical Samples
Once the method was analytically studied, a clinical validation is required to determine the real utility of the sensing material S2 as a tool for the diagnosis of PcP produced by the infection of P. jirovecii as a possible alternative to existing conventional methods. The gold standard procedure used in most hospitals to diagnose PcP uses extraction of the genomic DNA from the fungus and its subsequent amplification by PCR techniques.
In the present study, 12 respiratory samples consisting of sputum and BAL samples from infected and non-infected patients from Hospital Universitari i Politècnic La Fe, were analyzed using S2 material and the hospital reference method (i.e., PCR). For that, samples were isolated from patients with respiratory symptoms as part of routine diagnosis treatment. All samples were analyzed by PCR for the detection of P. jirovecii genomic DNA. Extraction of DNA from the samples was carried out by the QIAamp DNA Blood and Tissue kit and PCR reactions were performed using the VIASURE P. jirovecii Real Time PCR Detection Kit. Following this procedure, ten samples were confirmed as positive and two resulted negative. In parallel, the same samples (i.e., unmodified sputum or BAL samples) were tested using S2. For that, 12 individual S2 supports were immersed in 150 μL of hybridization buffer, then 250 μL of each raw sample were directly added without any previous treatment and the fluorescence in the solution, due to rhodamine delivery, was measured after 60 min. Results were considered positives when the fluorescence intensity was higher than the average fluorescence of the negative controls plus three times their standard deviation. As shown in Table 2 (samples 1 to 12), the results show a total coincidence between both methods, obtaining a sensitivity and a specificity of the 100%, and therefore, positive and negative predictive values of 100%, demonstrating an excellent analytical and clinical results when using S2 as diagnostic tool.
On the other hand, S2 was employed to evaluate 21 NPA samples from newborn infants with and without primary Pneumocystis infection from Hospital Universitario Virgen del Rocío. First, all samples were analyzed by Nested-PCR of the Pneumocystis mtLSU rRNA gene as described elsewhere [6]. Briefly, DNA from P. jirovecii was extracted using a QIAmp DNA Mini Kit with proteinase K digestion at 56 °C. In the first round of amplification, the external primers pAZ102-E and pAZ102-H were used. This yielded a 346-base pair (bp) fragment. The second round utilized the internal primers pAZ102-X and pAZ102-Y and yielded a 260-bp product. Both rounds of PCR comprised 35 amplification cycles. Specific amplification was corroborated by melting curve analysis (Tm: 76–77 °C) and amplicons were analyzed by electrophoresis on a 1.5% agarose gel containing ethidium bromide. Following this procedure, 11 samples were confirmed as positive and 10 resulted negative.
Nested-PCR positive samples were quantified by qPCR. Real-time quantitative PCR was performed in a CFX96 real time system (BIO-RAD) using AceQ qPCR SYBR® Green Master Mix (Vazyme) in a final volume of 10 µL containing 0.2 mM of primers pAZ102-X and pAZ102-Y and 2 µL of DNA sample. Serial dilutions of the target sequence cloned in the pGEM-T Easy Vector (Promega) were used to generate standard curves. After quantification, all the samples showed less than 102 copies/µL, which is equivalent to less than 25,000 copies per mL of NPA. That concentration confirms the P. jirovecii colonization condition and agrees with normal concentrations found in NPA samples from newborns with primary Pneumocystis infection [45].
In parallel, the same samples (i.e., unmodified NPA samples) were analyzed using S2. For that, 21 individual S2 supports were submerged in 150 μL of hybridization buffer and 250 μL of of each raw sample were directly added without any previous treatment. Finally, the fluorescence of the solution was measured after 60 min and results were considered positives when the fluorescence intensity was higher than the average fluorescence of the negative controls plus three times their standard deviation. As it is depicted in Table 3 (samples 13 to 33), results showed the coincidence between two methods in 16 samples and a total of 5 incongruent values that corresponded to two false positives and three false negatives, gave a sensitivity of 70%, a specificity of 78%, and positive and negative predictive values of 78% and 70%, respectively. The incongruences observed between the results obtained by PCR and S2 materials might be due several causes such as sample handling, DNA degradation or the very low concentration of pathogen present in NPA samples (calculated as less than 25,000 copies per mL of NPA). Nevertheless, it is noteworthy the high competitiveness of the gated biosensor as rapid diagnostic tool compared to the standard reference procedures, additionally providing easiness to use and lower cost, and avoiding sample treatments such as DNA extraction steps or amplification reactions [46].

Table 3.
Detection of P. jirovecii in clinical samples using the reference method (PCR) and S2 material. Each sample was analyzed by triplicate.
4. Conclusions
Early diagnosis of PcP is decisive for a suitable treatment and for a better prognosis. In that regard, rapid and accurate diagnostic tools are important for an early identification of the pathogen and to reduce the risk of mortality associated. However, standard diagnosis of PcP in remote or less developed countries or in resource-limited settings is difficult as usually it is necessary the use of costly well-equipped laboratories and trained personnel. In this scenario, the design low-cost easy-to-use diagnostic systems and point-of-care devices from readily accessible bio-fluids constitute a potential solution. In this study, a new method for the detection of P. jirovecii genomic DNA by using gated nanomaterials is proposed. In the developed tool, the pores of a NAA scaffold are loaded with a dye and gated with oligonucleotides that selectively recognizes a sequence of the genomic DNA from the fungi. The sensing mechanism relies in the recognition between the gating oligonucleotides and the P. jirovecii genomic DNA that induces a displacement of the capping oligonucleotide, pore uncapping and cargo delivery. In addition, the use of different gating oligonucleotides in the form of single strands or clamps to obtain duplex or triple-helix hybridization structures is also studied. From the three evaluated nanomaterials (S1, S2, and S3), the solid S2 provides the highest rhodamine B release and the faster response, probably due to more efficient formation of a triplex with respect to duplex conformations (S1 and S3). S2 showed a LOD 1 nM in hybridization buffer, which is equivalent to other state-of-the-art reported detection systems. Moreover, the probe presented a high selectivity to P. jirovecii, since no response was observed in the presence of DNA from other microorganisms. Finally, the gated material S2 is tested in unmodified sputum and bronchoalveolar lavages samples from infected and non-infected patients to diagnose PcP, obtaining a sensitivity and a selectivity of 100%. In the same way, S2 is used to analyze nasopharyngeal aspirate samples from infants to detect primary infected individuals with a sensitivity and a selectivity of 70% and 78%, respectively. In both cases, S2 detects P. jirovecii in 60 min without any sample treatment or the use of amplification reactions. This diagnostic tool has great potential for the further development of point-of-care devices for the simple and accurate detection of P. jirovecii from different biofluids avoiding PCR and using a small amount of sample, which could represent a high gain in the fight against PcP.
Supplementary Materials
The following are available online at https://www.mdpi.com/2309-608X/6/4/292/s1, Scheme S1: Scheme of the duplexes formed by oligonucleotides O1 and O3 with the target, and the triplex formed by oligonucleotide O2 and the target sequence, Table S1: Mass spectrometry analysis of the oligonucleotides, Figure S1: Amount of rhodamine B released from the pores of solids S1 (A) and S3 (B) when 1 nM of DNA from P. jirovecii was (a) absent and (b) present in a solution of hybridization buffer (20 mM Tris-HCl, 37.5 mM MgCl2, pH 7.5), Figure S2: Delivery of rhodamine B from material S2 in buffer, sputum, BAL, and NPA fluids in the presence of complementary DNA from P. jirovecii (1 nM).
Author Contributions
L.P.: investigation, methodology, writing—original draft; A.A.: investigation, resources, writing—review and editing; R.E.: investigation, resources, writing—review and editing, A.R.-G.: resources, writing—review and editing; J.P.: resources, writing—review and editing; V.F.: resources, writing—review and editing; E.J.C.: resources, writing—review and editing; E.A.: investigation, supervision, funding acquisition, writing—review and editing; R.M.-M.: conceptualization, supervision, funding acquisition, writing—review and editing; and S.S.-F.: investigation, supervision, methodology; writing—original draft, writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Spanish Government, (projects RTI2018-100910-B-C41 (MCUI/AEI/FEDER, UE) and CTQ2017-84415-R), the Generalitat Valenciana (project PROMETEO/2018/024) and CIBER-BBN (projects NANOPATH and CANDI-EYE).
Acknowledgments
L.P. thanks UPV for her predoctoral fellowship. S.S. thanks the Instituto de Salud Carlos III and the European Social Fund for the financial support “Sara Borrell” (CD16/000237). The authors also thank the Electron Microscopy Service at the UPV and ICTS NANBIOSIS oligonucleotide synthesis platform (U29) for support. The authors would like to thank Jordi Luque Font, from the IRTA Institute of Agrifood Research and Technology-Cabrils, and Josep Armengol from the Instituto Agroforestal Mediterráneo-Universitat Politècnica de València for the supply of T. deformans samples. The fungal culture of S. pompe was supplied by Piet W.J. de Groot from the Regional Center for Biomedical Research, Castilla-La Mancha Science & Technology Park, University of Castilla–La Mancha.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Fauchier, T.; Hasseine, L.; Gari-Toussaint, M.; Casanova, V.; Marty, P.M.; Pomares, C. Detection of Pneumocystis jirovecii by Quantitative PCR To Differentiate Colonization and Pneumonia in Immunocompromised HIV-Positive and HIV-Negative Patients. J. Clin. Microbiol. 2016, 54, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Van de Meer, G.; Brug, S.L. Infection Àpneumocystis Chez L’homme et Chez Les Animax. Annales de la Société Belge de Médecine Tropicale 1942, 22, 301–308. [Google Scholar]
- Skalski, J.H.; Kottom, T.J.; Limper, A.H. Pathobiology of Pneumocystis pneumonia: Life cycle, cell wall and cell signal transduction. FEMS Yeast Res. 2015, 15, fov046. [Google Scholar] [CrossRef] [PubMed]
- Dellière, S.; Gits-Muselli, M.; Bretagne, S.; Alanio, A. Outbreak-Causing Fungi: Pneumocystis jirovecii. Mycopathologia 2020, 185, 783–800. [Google Scholar] [CrossRef] [PubMed]
- Montes-Cano, M.A.; Chabe, M.; Fontillon-Alberdi, M.; De La Horra, C.; Respaldiza, N.; Medrano, F.J.; Varela, J.M.; Dei-Cas, E.; Calderón, E.J. Vertical transmission of Pneumocystis jirovecii in humans. Emergy Infect. Dis. 2009, 15, 125–127. [Google Scholar] [CrossRef] [PubMed]
- Jarboui, M.A.; Sellami, A.; Cheikhrouhou, F.; Makni, F.; Ben Arab, N.; Ben Jemaa, M.; Ayadi, A.; Sellami, H. Molecular diagnosis of Pneumocystis jirovecipneumonia in immunocompromised patients. Mycoses 2009, 53, 329–333. [Google Scholar]
- Durand-Joly, I.; Chabã, M.; Soula, F.; Delhaes, L.; Camus, D.; Dei-Cas, E.; Chabe, M. Molecular diagnosis ofPneumocystispneumonia. FEMS Immunol. Med. Microbiol. 2005, 45, 405–410. [Google Scholar] [CrossRef]
- Song, Y.; Ren, Y.; Wang, X.; Li, R. The biosynthesis pathway of Swainsonine, a new anticancer drug from three endophytic fungi. Med. Mycol. 2016, 57, 111–116. [Google Scholar] [CrossRef]
- Tomás, A.L.; Matos, O. Pneumocystis jirovecii Pneumonia: Current Advances in Laboratory Diagnosis. OBM Genet. 2018, 2, 1. [Google Scholar] [CrossRef]
- Urabe, N.; Sakamoto, S.; Sano, G.; Ito, A.; Sekiguchi, R.; Homma, S. Serial change in serum biomarkers during treatment of Non-HIV Pneumocystis pneumonia. J. Infect. Chemother. 2019, 25, 936–942. [Google Scholar] [CrossRef]
- Esteves, F.; Calé, S.; Badura, R.; De Boer, M.; Maltez, F.; Calderón, E.; Van Der Reijden, T.; Márquez-Martín, E.; Antunes, F.; Matos, O. Diagnosis of Pneumocystis pneumonia: Evaluation of four serologic biomarkers. Clin. Microbiol. Infect. 2015, 21, 379.e1–379.e10. [Google Scholar] [CrossRef] [PubMed]
- Tomás, A.L.; Cardoso, F.; Esteves, F.; Matos, O. Serological diagnosis of pneumocystosis: Production of a synthetic recombinant antigen for immunodetection of Pneumocystis jirovecii. Sci. Rep. 2016, 6, 36287–36295. [Google Scholar] [CrossRef] [PubMed]
- Arvanitis, M.; Anagnostou, T.; Fuchs, B.B.; Caliendo, A.M.; Mylonakis, E. Molecular and Nonmolecular Diagnostic Methods for Invasive Fungal Infections. Clin. Microbiol. Rev. 2014, 27, 490–526. [Google Scholar] [CrossRef] [PubMed]
- Tomás, A.L.; Cardoso, F.; Pinto, M.; de Almeida, M.P.; de Sousa, B.; Pereira, E. In Proceedings of the 28th European Congress of Clinical Microbiology and Infectious Diseases, Madrid, Spain, 21–24 April 2018. Available online: https://www.eurekalert.org/pub_releases/2018-04/esoc-2ec041018.php (accessed on 29 October 2020).
- Tomás, A.L.; De Almeida, M.P.; Cardoso, F.; Pinto, M.; Pereira, E.; Franco, R.; Matos, O. Development of a Gold Nanoparticle-Based Lateral-Flow Immunoassay for Pneumocystis Pneumonia Serological Diagnosis at Point-of-Care. Front. Microbiol. 2019, 10, 2917. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, A.; Aznar, E.; Martínez-Máñez, R.; Sancenón, F. New Advances in In Vivo Applications of Gated Mesoporous Silica as Drug Delivery Nanocarriers. Small 2020, 16, e1902242. [Google Scholar] [CrossRef] [PubMed]
- Aznar, E.; Coll, C.; Marcos, M.D.; Martínez-Máñez, R.; Sancenón, F.; Soto, J.; Amorós, P.; Cano, J.; Ruiz, E. Controlled delivery systems using antibody-capped mesoporous nanocontainers. Chem. Eur. J. 2009, 15, 6877–6888. [Google Scholar] [CrossRef]
- Sancenón, F.; Pascual, L.; Oroval, M.; Aznar, E.; Martínez-Máñez, R. Gated Silica Mesoporous Materials in Sensing Applications. ChemistryOpen 2015, 4, 418–437. [Google Scholar] [CrossRef] [PubMed]
- Mondragón, L.; Mas, N.; Ferragud, V.; De La Torre, C.; Agostini, A.; Martínez-Máñez, R.; Sancenón, F.; Amorós, P.; Pérez-Payá, E.; Orzáez, M. Enzyme-Responsive Intracellular-Controlled Release Using Silica Mesoporous Nanoparticles Capped with ε-Poly-l-lysine. Chem. A Eur. J. 2014, 20, 5271–5281. [Google Scholar] [CrossRef]
- Giménez, C.; De La Torre, C.; Gorbe, M.; Aznar, E.; Sancenón, F.; Murguía, J.R.; Martínez-Máñez, R.; Marcos, M.D.; Amorós, P. Gated Mesoporous Silica Nanoparticles for the Controlled Delivery of Drugs in Cancer Cells. Langmuir 2015, 31, 3753–3762. [Google Scholar] [CrossRef]
- Aznar, E.; Villalonga, R.; Giménez, C.; Sancenón, F.; Marcos, M.D.; Martínez-Máñez, R.; Díez, P.; Pingarrón, J.M.; Amorós, P. Glucose-triggered release using enzyme-gated mesoporous silica nanoparticles. Chem. Commun. 2013, 49, 6391–6393. [Google Scholar] [CrossRef]
- Ouyang, C.; Zhang, S.; Xue, C.; Yu, X.; Xu, H.; Wang, Z.; Lu, Y.; Wu, Z.-S. Precision-Guided Missile-Like DNA Nanostructure Containing Warhead and Guidance Control for Aptamer-Based Targeted Drug Delivery into Cancer Cells in Vitro and in Vivo. J. Am. Chem. Soc. 2020, 142, 1265–1277. [Google Scholar] [CrossRef] [PubMed]
- Argoubi, W.; Sánchez, A.; Parrado, C.; Raouafi, N.; Villalonga, R. Label-free electrochemical aptasensing platform based on mesoporous silica thin film for the detection of prostate specific antigen. Sens. Actuators B Chem. 2018, 255, 309–315. [Google Scholar] [CrossRef]
- Llopis-Lorente, A.; Lozano-Torres, B.; Bernardos, A.; Martínez-Máñez, R.; Sancenón, F. Mesoporous silica materials for controlled delivery based on enzymes. J. Mater. Chem. B 2017, 5, 3069–3083. [Google Scholar] [CrossRef] [PubMed]
- Pla, L.; Xifre-Perez, E.; Ribes, À.; Aznar, E.; Marcos, M.D.; Marsal, L.F.; Martínez-Máñez, R.; Sancenón, F. A Mycoplasma Genomic DNA Probe using Gated Nanoporous Anodic Alumina. ChemPlusChem 2017, 82, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Ribes, A.; Aznar, E.; Santiago-Felipe, S.; Xifre-Perez, E.; Tormo-Mas, M.Á.; Pemán, J.; Marsal, L.F.; Martínez-Máñez, R. Selective and Sensitive Probe Based in Oligonucleotide-Capped Nanoporous Alumina for the Rapid Screening of Infection Produced by Candida albicans. ACS Sens. 2019, 4, 1291–1298. [Google Scholar] [CrossRef] [PubMed]
- Goñi, J.R.; Vaquerizas, R.E.-G.A.S.J.M.; Dopazo, J.; Orozco, M. Exploring the reasons for the large density of triplex-forming oligonucleotide target sequences in the human regulatory regions. BMC Genomic 2006, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Frank-Kamenetskii, M.D.; Mirkin, S.M. Triplex DNA structures. Annu. Rev. Biochem. 1995, 64, 65–95. [Google Scholar] [CrossRef] [PubMed]
- Goñi, J.R.; De La Cruz, X.; Orozco, M. Triplex-forming oligonucleotide target sequences in the human genome. Nucleic Acids Res. 2004, 32, 354–360. [Google Scholar] [CrossRef]
- Aviñó, A.; Frieden, M.; Morales, J.C.; García de la Torre, B.; Güimil García, R.; Azorín, F.; Gelpí, J.L.; Orozco, M.; González, C.; Eritja, R. Properties of triple helices formed by parallel-stranded hairpins containing 8-aminopurines. Nucleic Acids Res. 2002, 30, 2609–2619. [Google Scholar] [CrossRef]
- Nadal, A.; Eritja, R.; Esteve, T.; Pla, M. “Parallel” and “Antiparallel Tail-Clamps” Increase the Efficiency of Triplex Formation with Structured DNA and RNA Targets. ChemBioChem 2005, 6, 1034–1042. [Google Scholar] [CrossRef]
- Carrascosa, L.G.; Gómez-Montes, S.; Aviñó, A.; Nadal, A.; Pla, M.; Eritja, R.; Lechuga, L.M. Sensitive and label-free biosensing of RNA with predicted secondary structures by a triplex affinity capture method. Nucleic Acids Res. 2012, 40, e56. [Google Scholar] [CrossRef] [PubMed]
- Aviñó, A.; Huertas, C.S.; Lechuga, L.M.; Eritja, R. Sensitive and label-free detection of miRNA-145 by triplex formation. Anal. Bioanal. Chem. 2016, 408, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Chen, G.; Jia, X.; Mao, X.; Chen, T.; Mao, D.; Zhang, W.; Xiong, W. Exponential amplification reaction and triplex DNA mediated aggregation of gold nanoparticles for sensitive colorimetric detection of microRNA. Anal. Chim. Acta 2020, 1095, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Ribes, À.; Santiago-Felipe, S.; Aviñó, A.; Candela-Noguera, V.; Eritja, R.; Sancenón, F.; Martínez-Máñez, R.; Aznar, E. Design of oligonucleotide-capped mesoporous silica nanoparticles for the detection of miRNA-145 by duplex and triplex formation. Sens. Actuators B Chem. 2018, 277, 598–603. [Google Scholar] [CrossRef]
- Pascual, L.; Baroja, I.; Aznar, E.; Sancenón, F.; Marcos, M.D.; Murguia, J.R.; Amoros, P.; Rurack, K.; Martínez-Máñez, R. Oligonucleotide-capped mesoporous silica nanoparticles as DNA-responsive dye delivery systems for genomic DNA detection. Chem. Commun. 2015, 51, 1414–1416. [Google Scholar] [CrossRef]
- Oroval, M.; Coll, C.; Bernardos, A.; Marcos, M.D.; Martínez-Máñez, R.; Shchukin, D.G.; Sancenón, F. Selective Fluorogenic Sensing of As(III) Using Aptamer-Capped Nanomaterials. ACS Appl. Mater. Interfaces 2017, 9, 11332–11336. [Google Scholar] [CrossRef]
- Vojkuvka, L.; Marsal, L.F.; Ferré-Borrull, J.; Formentin, P.; Pallares, J. Self-ordered porous alumina membranes with large lattice constant fabricated by hard anodization. Superlattices Microstruct. 2008, 44, 577–582. [Google Scholar] [CrossRef]
- Matsumura, Y.; Tsuchido, Y.; Yamamoto, M.; Nakano, S.; Nagao, M. Development of a fully automated PCR assay for the detection of Pneumocystis jirovecii using the GENECUBE system. Med. Mycol. J. 2019, 57, 841–847. [Google Scholar] [CrossRef]
- Yang, S.-L.; Wen, Y.-H.; Wu, Y.-S.; Wang, M.-C.; Chang, P.-Y.; Yang, S.; Lu, J.-J. Diagnosis of Pneumocystis pneumonia by real-time PCR in patients with various underlying diseases. J. Microbiol. Immunol. Infect. 2019, 5, 785–790. [Google Scholar] [CrossRef]
- Moodley, B.; Tempia, S.; Frean, J.A. Comparison of quantitative real-time PCR and direct immunofluorescence for the detection of Pneumocystis jirovecii. PLoS ONE 2017, 12, e0180589. [Google Scholar] [CrossRef]
- Alanio, A.; Desoubeaux, G.; Sarfati, C.; Hamane, S.; Bergeron, A.; Azoulay, É.; Molina, J.; DeRouin, F.; Menotti, J. Real-time PCR assay-based strategy for differentiation between active Pneumocystis jirovecii pneumonia and colonization in immunocompromised patients. Clin. Microbiol. Infect. 2011, 17, 1531–1537. [Google Scholar] [CrossRef] [PubMed]
- Marimuthu, S. Development of a Real-time PCR assay for Pneumocystis jirovecii on the Luminex ARIES® Platform. Univ. Louisev. J. Respir. Infect. 2019, 3, 5. [Google Scholar] [CrossRef]
- Cissé, O.H.; Almeida, J.M.G.C.F.; Fonseca, Á.; Kumar, A.A.; Salojärvi, J.; Overmyer, K.; Hauser, P.M.; Pagni, M. Genome Sequencing of the Plant Pathogen Taphrina deformans, the Causal Agent of Peach Leaf Curl. MBio 2013, 4, e00055-13. [Google Scholar] [CrossRef]
- Rojas, P.; Friaza, V.; García, E.; De La Horra, C.; Vargas, S.L.; Calderón, E.J.; Pavón, A. Early Acquisition of Pneumocystis jirovecii Colonization and Potential Association with Respiratory Distress Syndrome in Preterm Newborn Infants. Clin. Infect. Dis. 2017, 65, 976–981. [Google Scholar] [CrossRef] [PubMed]
- Kidd, S.E.; Chen, S.C.-A.; Meyer, W.; Halliday, C.L. A New Age in Molecular Diagnostics for Invasive Fungal Disease: Are We Ready? Front. Microbiol. 2020, 10, 2903. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).