Transcriptional Remodeling Patterns in Murine Dendritic Cells Infected with Paracoccidioides brasiliensis: More Is Not Necessarily Better
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Cells and Growth Conditions
2.2. Mouse Strains and Bone Marrow-Derived Cells Differentiation
2.3. Ex Vivo Infection of Dendritic Cells from PCM-Resistant and -Susceptible Mouse Strains
2.4. Cytokine and Chemokine Measurements
2.5. Ex Vivo Infection of Resistant and Susceptible BMMs and BMDCs with P. brasiliensis for LC3 Immunofluorescence
2.6. Immunolocalization of LC3 in Infected BMMs and BMDCs
2.7. RNA Isolation
2.8. Sequencing Parameters
2.9. RNA-Seq Data Analysis
2.10. Data Access
2.11. RNA-Seq Validation by Quantitative PCR (RT-qPCR)
2.12. Statistical Analysis
3. Results
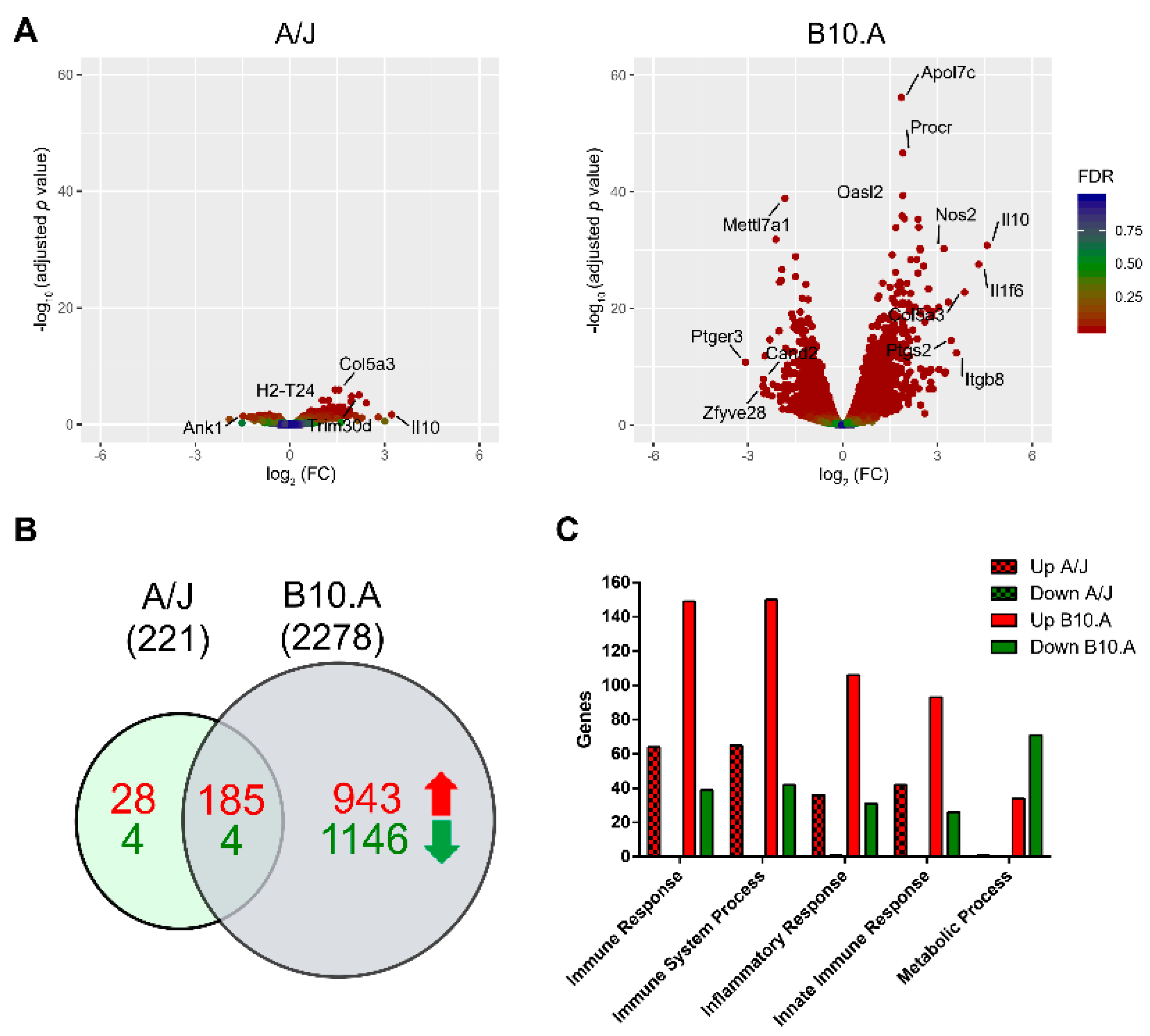
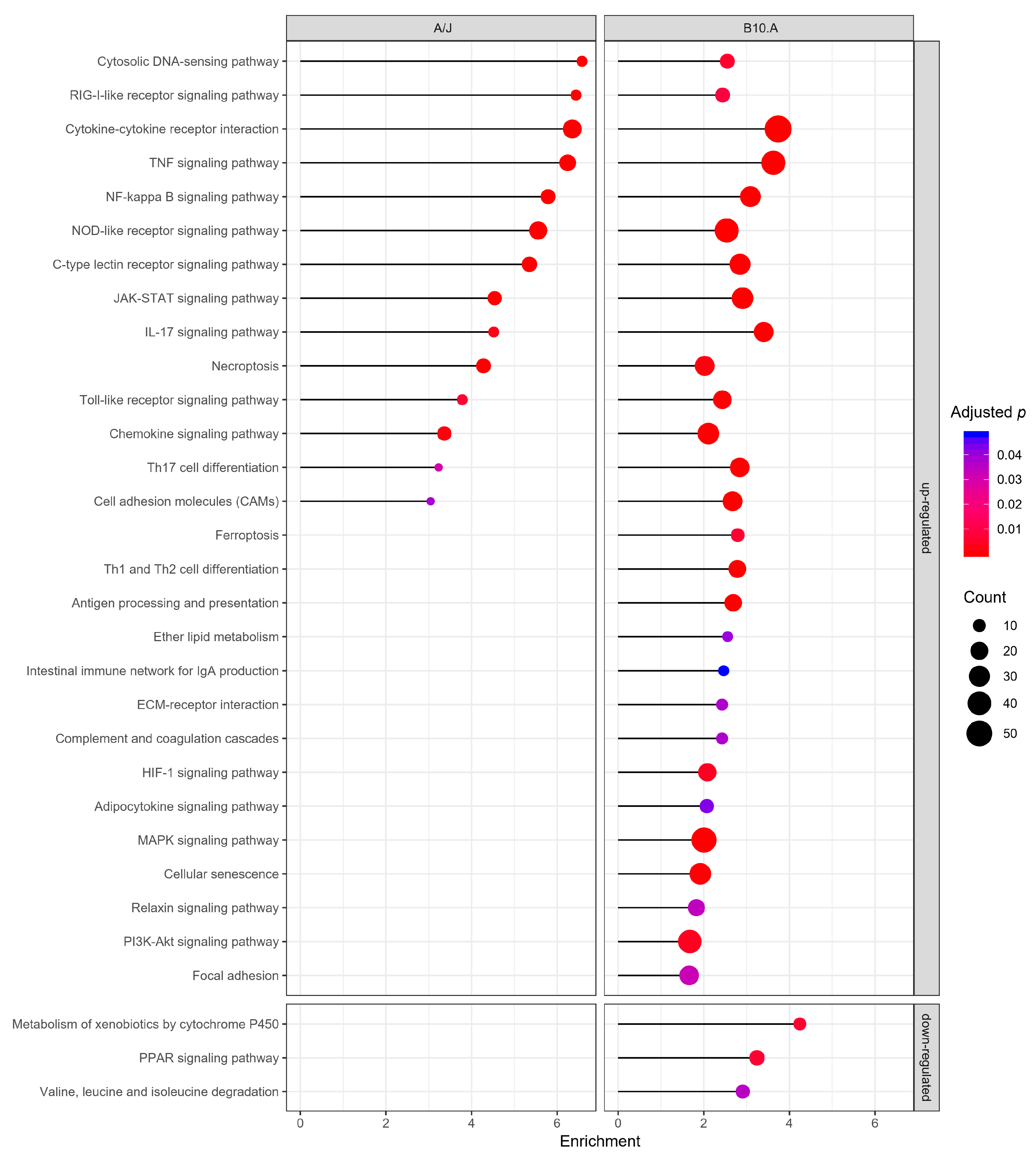
3.1. P. brasiliensis Infection Triggers Widespread Transcriptional Remodeling in a PCM-Susceptible Mouse Strain
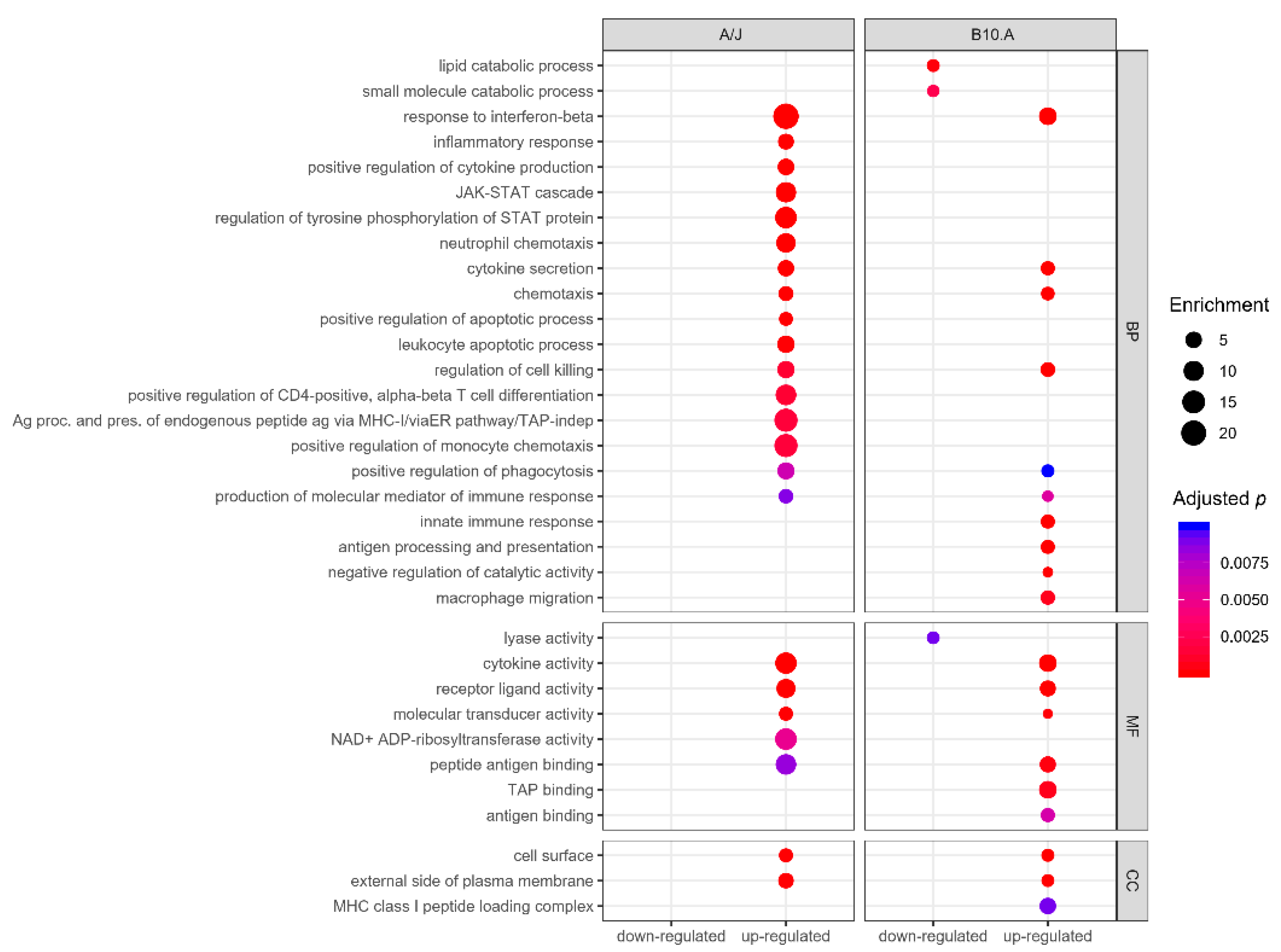
3.2. PCM-Resistant Mouse Strain Reveals a Precise and Coordinated Immune Response upon P. brasiliensis Infection
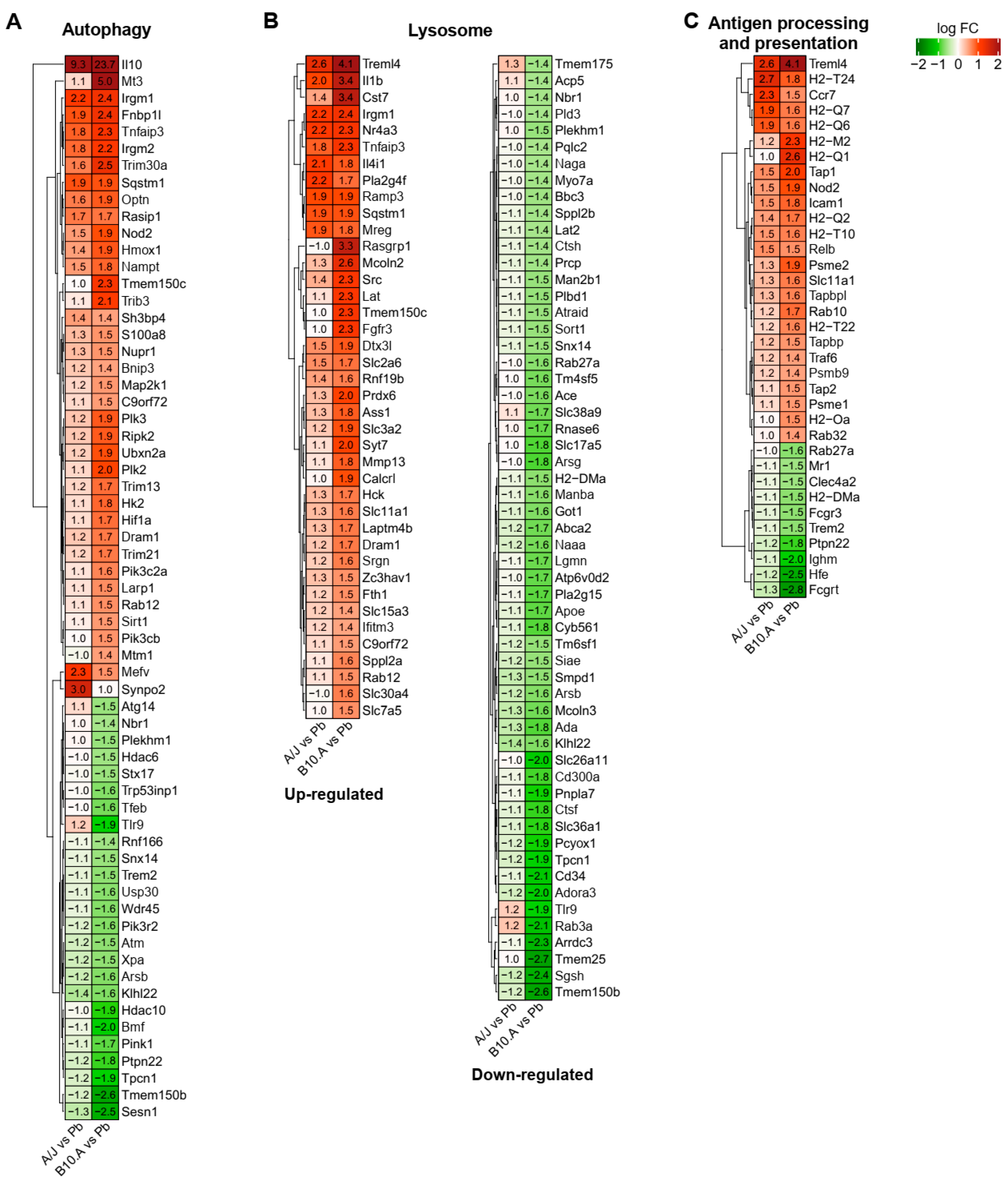
3.3. Resistant and Susceptible Strains had Significant Differences in the Modulation of Genes Related to Antigen Presentation, Autophagy, and Lysosome Function
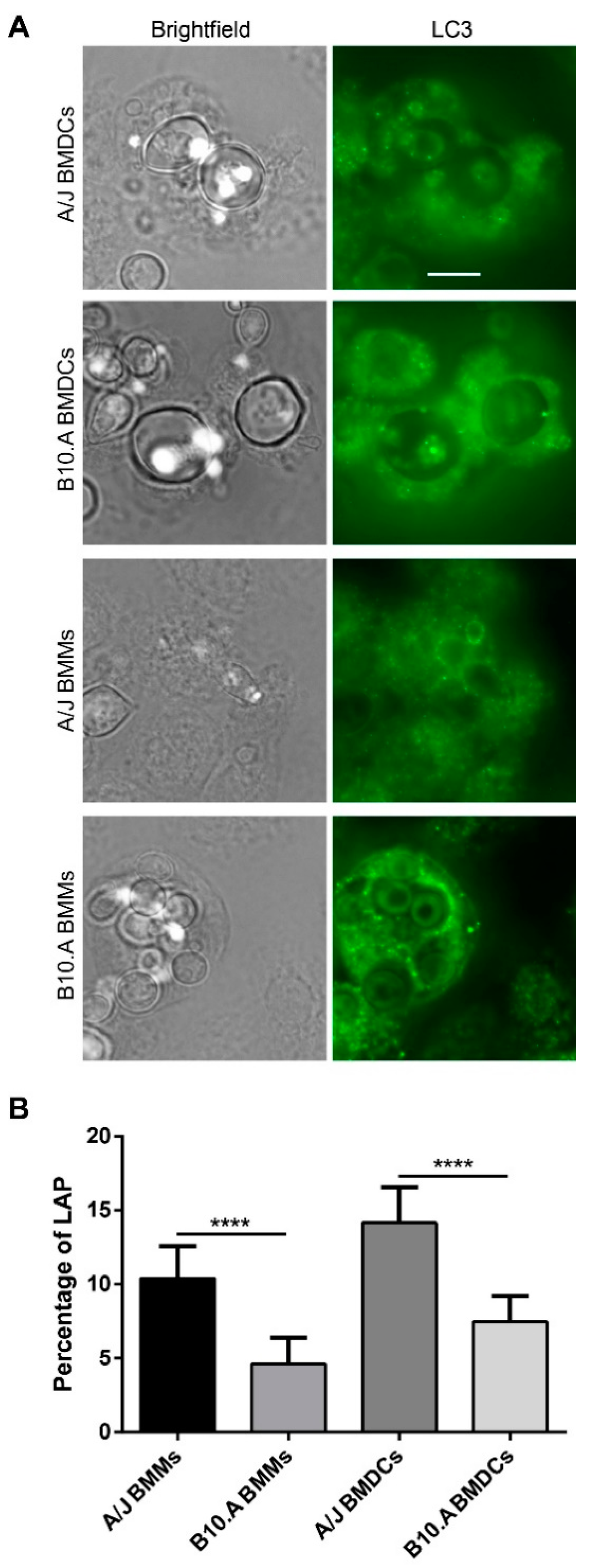
3.4. PCM-Susceptible Mouse Strain Shows a Deficiency in Performing LC3-Associated Phagocytosis of P. brasiliensis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Martinez, R. New trends in paracoccidioidomycosis epidemiology. J. Fungi 2017, 3, 1. [Google Scholar] [CrossRef]
- De Macedo, P.M.; De Melo Teixeira, M.; Barker, B.M.; Zancopé-Oliveira, R.M.; Almeida-Paes, R.; Do Valle, A.C.F. Clinical features and genetic background of the sympatric species Paracoccidioides brasiliensis and Paracoccidioides americana. PLoS Negl. Trop. Dis. 2019, 13. [Google Scholar] [CrossRef]
- Negroni, R. Paracoccidioidomycosis. In Hunter’s Tropical Medicine and Emerging Infectious Diseases; Elsevier: Cham, Switzerland, 2020; pp. 674–677. ISBN 978-0-323-55512-8. [Google Scholar]
- Griffiths, J.; Colombo, A.L.; Denning, D.W. The case for paracoccidioidomycosis to be accepted as a neglected tropical (Fungal) disease. PLoS Negl. Trop. Dis. 2019, 13. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, J.N.; Peçanha, P.M.; Colombo, A.L. Paracoccidioidomycosis in immunocompromised patients: A literature review. J. Fungi 2019, 5, 2. [Google Scholar] [CrossRef]
- Prado, M.; da Silva, M.B.; Laurenti, R.; Travassos, L.R.; Taborda, C.P. Mortality due to systemic mycoses as a primary cause of death or in association with AIDS in Brazil: A review from 1996 to 2006. Mem. Inst. Oswaldo Cruz 2009, 104, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, K.S.; Lopes, J.D.; Almeida, S.R. Regulation of T helper cell differentiation in vivo by GP43 from Paracoccidioides brasiliensis provided by different antigen-presenting cells. Scand. J. Immunol. 2003, 58, 290–297. [Google Scholar] [CrossRef]
- Ferreira, K.S.; Bastos, K.R.; Russo, M.; Almeida, S.R. Interaction between Paracoccidioides brasiliensis and Pulmonary Dendritic Cells Induces Interleukin-10 Production and Toll-Like Receptor–2 Expression: Possible Mechanisms of Susceptibility. J. Infect. Dis. 2007, 196, 1108–1115. [Google Scholar] [CrossRef][Green Version]
- de Araújo, E.F.; Medeiros, D.H.; Galdino, N.A.d.L.; Condino-Neto, A.; Calich, V.L.G.; Loures, F.V. Tolerogenic Plasmacytoid Dendritic Cells Control Paracoccidioides brasiliensis Infection by Inducting Regulatory T Cells in an IDO-Dependent Manner. PLoS Pathog. 2016, 12. [Google Scholar] [CrossRef]
- Moreira, A.L.E.; Oliveira, M.A.P.; Silva, L.O.S.; Inácio, M.M.; Bailão, A.M.; Parente-Rocha, J.A.; Cruz-Leite, V.R.M.; Paccez, J.D.; de Almeida Soares, C.M.; Weber, S.S.; et al. Immunoproteomic Approach of Extracellular Antigens From Paracoccidioides Species Reveals Exclusive B-Cell Epitopes. Front. Microbiol. 2020, 10. [Google Scholar] [CrossRef]
- Camacho, E.; Niño-Vega, G.A. Paracoccidioides Spp.: Virulence Factors and Immune-Evasion Strategies. Mediat. Inflamm. 2017, 2017. [Google Scholar] [CrossRef]
- de Castro, L.F.; Ferreira, M.C.; da Silva, R.M.; Blotta, M.H.d.S.L.; Longhi, L.N.A.; Mamoni, R.L. Characterization of the immune response in human paracoccidioidomycosis. J. Infect. 2013, 67, 470–485. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, H.C.; Assato, P.A.; Marcos, C.M.; Scorzoni, L.; De Paula E Silva, A.C.A.; Da Silva, J.d.F.; Singulani, J.d.L.; Alarcon, K.M.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Paracoccidioides-host interaction: An overview on recent advances in the paracoccidioidomycosis. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Calich, V.L.G.; Mamoni, R.L.; Loures, F.V. Regulatory T cells in paracoccidioidomycosis. Virulence 2019, 10, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Calich, V.L.G.; Da Costa, T.A.; Felonato, M.; Arruda, C.; Bernardino, S.; Loures, F.V.; Ribeiro, L.R.R.; De Cássia Valente-Ferreira, R.; Pina, A. Innate immunity to Paracoccidioides brasiliensis infection. Mycopathologia 2008, 165, 223–236. [Google Scholar] [CrossRef]
- Iovannitti, C.A.; Finquelievich, J.L.; Negroni, R.; Elías Costa, M.R. Histopathological evolution of experimental paracoccidioidomycosis in Wistar rats. Zent. Bakteriol. 1999, 289, 211–216. [Google Scholar] [CrossRef]
- Scorzoni, L.; de Lucas, M.P.; de Lacorte Singulani, J.; de Oliveira, H.C.; Assato, P.A.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Evaluation of Caenorhabditis elegans as a host model for Paracoccidioides brasiliensis and Paracoccidioides lutzii. Pathog. Dis. 2018, 76. [Google Scholar] [CrossRef]
- Albuquerque, P.; Nicola, A.M.; Magnabosco, D.A.G.; da Silveira Derengowski, L.; Crisóstomo, L.S.; Xavier, L.C.G.; de Oliveira Frazão, S.; Guilhelmelli, F.; De Oliveira, M.A.; do Nascimento Dias, J.; et al. A hidden battle in the dirt: Soil amoebae interactions with Paracoccidioides spp. PLoS Negl. Trop. Dis. 2019, 13, e0007742. [Google Scholar] [CrossRef]
- Garcia Calich, V.L.; Singer-Vermes, L.M.; Siqueira, A.M.; Burger, E. Susceptibility and resistance of inbred mice to Paracoccidioides brasiliensis. Br. J. Exp. Pathol. 1985, 66, 585–594. [Google Scholar]
- Singer-Vermes, L.M.; Caldeira, C.B.; Burger, E.; Calich, V.L.G. Experimental murine paracoccidioidomycosis: Relationship among the dissemination of the infection, humoral and cellular immune responses. Clin. Exp. Immunol. 1993, 94, 75–79. [Google Scholar] [CrossRef]
- Calich, V.L.G.; Singer-Vermes, L.M.; Russo, M.; Vaz, C.A.; Burger, E. Immunogenetics in paracoccidioidomycosis. In Paracoccidioidomycosis; Franco, M., Lacaz, C.S., Restrepo-Moreno, A., Del Negro, G.E., Eds.; CRC Press: Boca Raton, FL, USA, 1994; pp. 151–173. ISBN 0-8493-4868-4. [Google Scholar]
- Cano, L.E.; Singer-Vermes, L.M.; Vaz, C.A.C.; Russo, M.; Calich, V.L.G. Pulmonary paracoccidioidomycosis in resistant and susceptible mice: Relationship among progression of infection, bronchoalveolar cell activation, cellular immune response, and specific isotype patterns. Infect. Immun. 1995, 63, 1777–1783. [Google Scholar] [CrossRef]
- Calich, V.L.G.; Kashino, S.S. Cytokines produced by susceptible and resistant mice in the course of Paracoccidioides brasiliensis infection. Braz. J. Med. Biol. Res. 1998, 31, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Nishikaku, A.S.; Molina, R.F.S.; Albe, B.P.; Cunha, C.d.S.; Scavone, R.; Pizzo, C.R.P.; de Camargo, Z.P.; Burger, E. Immunolocalization of IFN-gamma in the lesions of resistant and susceptible mice to Paracoccidioides brasiliensis infection. FEMS Immunol. Med. Microbiol. 2011, 63, 281–288. [Google Scholar] [CrossRef] [PubMed]
- De Souza Silva, C.; Tavares, A.H.; Sousa Jeronimo, M.; Soares De Lima, Y.; Da Silveira Derengowski, L.; Lorenzetti Bocca, A.; Silva-Pereira, I. The Effects of Paracoccidioides brasiliensis Infection on GM-CSF- and M-CSF-Induced Mouse Bone Marrow-Derived Macrophage from Resistant and Susceptible Mice Strains. Mediat. Inflamm. 2015, 2015. [Google Scholar] [CrossRef]
- Pina, A.; Bernardino, S.; Calich, V.L.G. Alveolar macrophages from susceptible mice are more competent than those of resistant mice to control initial Paracoccidioides brasiliensis infection. J. Leukoc. Biol. 2008, 83, 1088–1099. [Google Scholar] [CrossRef]
- Almeida, S.R.; Lopes, J.D. The low efficiency of dendritic cells and macrophages from mice susceptible to Paracoccidioides brasiliensis in inducing a Th1 response. Braz. J. Med. Biol. Res. 2001, 34, 529–537. [Google Scholar] [CrossRef]
- Ferreira, K.S.; Almeida, S.R. Immunization of susceptible mice with gp43-pulsed dendritic cells induce an increase of pulmonary Paracoccidioidomycosis. Immunol. Lett. 2006, 103, 121–126. [Google Scholar] [CrossRef]
- Alves da Costa, T.; Di Gangi, R.; Martins, P.; Longhini, A.L.F.; Zanucoli, F.; de Oliveira, A.L.R.; Stach-Machado, D.R.; Burger, E.; Verinaud, L.; Thomé, R. Protection against Paracoccidioides brasiliensis infection in mice treated with modulated dendritic cells relies on inhibition of interleukin-10 production by CD8+ T cells. Immunology 2015, 146, 486–495. [Google Scholar] [CrossRef]
- Fernandes, R.; Rodrigues, D.; Romagnoli, G.; Vieira, I.; Dias-Melicio, L.; Angela, R.; Soares, M. In vitro challenge of human dendritic cells with Paracoccidioides brasiliensis induces preferential generation of Treg cells. Med. Res. Arch. 2017, 5. [Google Scholar] [CrossRef]
- Silva, L.B.R.; Taira, C.L.; Dias, L.S.; Souza, A.C.O.; Nosanchuk, J.D.; Travassos, L.R.; Taborda, C.P. Experimental therapy of Paracoccidioidomycosis using p10-primed monocyte-derived dendritic cells isolated from infected mice. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- de Araújo, E.F.; Feriotti, C.; de Lima Galdino, N.A.; Preite, N.W.; Calich, V.L.G.; Loures, F.V. The IDO-AhR axis controls Th17/Treg immunity in a pulmonary model of fungal infection. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef]
- Goihman-Yahr, M.; Pine, L.; Albornoz, M.C.; Yarzabal, L.; De Gomez, M.H.; Martin, B.S.; Ocanto, A.; Molina, T.; Convit, J. Studies on plating efficiency and estimation of viability of suspensions of Paracoccidioides brasiliensis yeast cells. Mycopathologia 1980, 71, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Singer-Vermes, L.M.; Sakamoto, T.N.; Vaz, C.A.C. Influence of the genetic pattern and sex of mice in experimental paracoccidioidomycosis. Clin. Exp. Immunol. 1995, 101, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lutz, M.B.; Kukutsch, N.; Ogilvie, A.L.; Rößner, S.; Koch, F.; Romani, N.; Schuler, G. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J. Immunol. Methods 1999, 223, 77–92. [Google Scholar] [CrossRef]
- Tavares, A.H.; Derengowski, L.S.; Ferreira, K.S.; Silva, S.S.; Macedo, C.; Bocca, A.L.; Passos, G.A.; Almeida, S.R.; Silva-Pereira, I. Murine dendritic cells transcriptional modulation upon Paracoccidioides brasiliensis infection. PLoS Negl. Trop. Dis. 2012, 6. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, I.M.; Fraga, C.L.F.; Amaral, A.C.; Souza, A.C.O.; Jerônimo, M.S.; Correa, J.R.; Magalhães, K.G.; Inácio, C.A.; Ribeiro, A.M.; Burguel, P.H.; et al. Distinct patterns of yeast cell morphology and host responses induced by representative strains of Paracoccidioides brasiliensis (Pb18) and Paracoccidioides lutzii (Pb01). Med. Mycol. 2016, 54, 177–188. [Google Scholar] [CrossRef]
- Tavares, A.H.F.P.; Silva, S.S.; Dantas, A.; Campos, É.G.; Andrade, R.V.; Maranhão, A.Q.; Brígido, M.M.; Passos-Silva, D.G.; Fachin, A.L.; Teixeira, S.M.R.; et al. Early transcriptional response of Paracoccidioides brasiliensis upon internalization by murine macrophages. Microbes Infect. 2007, 9, 583–590. [Google Scholar] [CrossRef]
- Silva, S.S.; Tavares, A.H.F.P.; Passos-Silva, D.G.; Fachin, A.L.; Teixeira, S.M.R.; Soares, C.M.A.; Carvalho, M.J.A.; Bocca, A.L.; Silva-Pereira, I.; Passos, G.A.S.; et al. Transcriptional response of murine macrophages upon infection with opsonized Paracoccidioides brasiliensis yeast cells. Microbes Infect. 2008, 10, 12–20. [Google Scholar] [CrossRef]
- Nicola, A.M.; Albuquerque, P.; Martinez, L.R.; Dal-Rosso, R.A.; Saylor, C.; De Jesus, M.; Nosanchuk, J.D.; Casadevall, A. Macrophage autophagy in immunity to Cryptococcus neoformans and Candida albicans. Infect. Immun. 2012, 80, 3065–3076. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef]
- Andrews, S.; Krueger, F.; Seconds-Pichon, A.; Biggins, F.; Wingett, S. FastQC. A Quality Control Tool for High Throughput Sequence Data 2015. Available online: https://qubeshub.org/resources/fastqc (accessed on 12 June 2019).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Blake, J.A.; Christie, K.R.; Dolan, M.E.; Drabkin, H.J.; Hill, D.P.; Ni, L.; Sitnikov, D.; Burgess, S.; Buza, T.; Gresham, C.; et al. Gene ontology consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Spandidos, A.; Wang, X.; Wang, H.; Seed, B. PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2009, 38. [Google Scholar] [CrossRef]
- Panganiban, R.P.; Vonakis, B.M.; Ishmael, F.T.; Stellato, C. Coordinated post-transcriptional regulation of the chemokine system: Messages from CCL2. J. Interf. Cytokine Res. 2014, 34, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Das, A.S.; Basu, A.; Kumar, R.; Borah, P.K.; Bakshi, S.; Sharma, M.; Duary, R.K.; Ray, P.S.; Mukhopadhyay, R. Post-transcriptional regulation of C-C motif chemokine ligand 2 expression by ribosomal protein L22 during LPS-mediated inflammation. FEBS J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Wang, Q.; Morgan, S.; Si, Y.; Ravichander, A.; Dou, C.; Kent, K.C.; Liu, B. Protein kinase C-δ (PKCδ) regulates proinflammatory chemokine expression through cytosolic interaction with the NF-κB subunit p65 in vascular smooth muscle cells. J. Biol. Chem. 2014, 289, 9013–9026. [Google Scholar] [CrossRef] [PubMed]
- Nakatsumi, H.; Matsumoto, M.; Nakayama, K.I. Noncanonical Pathway for Regulation of CCL2 Expression by an mTORC1-FOXK1 Axis Promotes Recruitment of Tumor-Associated Macrophages. Cell Rep. 2017, 21, 2471–2486. [Google Scholar] [CrossRef] [PubMed]
- Bansal, K.; Sinha, A.Y.; Ghorpade, D.S.; Togarsimalemath, S.K.; Patil, S.A.; Kaveri, S.V.; Balaji, K.N.; Bayry, J. Src homology 3-interacting domain of Rv1917c of Mycobacterium tuberculosis induces selective maturation of human dendritic cells by regulating PI3K-MAPK-NF-κB signaling and drives Th2 immune responses. J. Biol. Chem. 2010, 285, 36511–36522. [Google Scholar] [CrossRef]
- Ji, W.-T.; Liu, H. PI3K-Akt Signaling and Viral Infection. Recent Pat. Biotechnol. 2008, 2, 218–226. [Google Scholar] [CrossRef]
- Roca, H.; Varsos, Z.S.; Mizutani, K.; Pienta, K.J. CCL2, survivin and autophagy: New links with implications in human cancer. Autophagy 2008, 4, 969–971. [Google Scholar] [CrossRef]
- Knight, M.; Stanley, S. HIF-1α as a central mediator of cellular resistance to intracellular pathogens. Curr. Opin. Immunol. 2019, 60, 111–116. [Google Scholar] [CrossRef]
- Kotsias, F.; Cebrian, I.; Alloatti, A. Antigen processing and presentation. In International Review of Cell and Molecular Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2019; Volume 348, pp. 69–121. ISBN 9780128183519. [Google Scholar]
- Ma, J.; Becker, C.; Lowell, C.A.; Underhill, D.M. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J. Biol. Chem. 2012, 287, 34149–34156. [Google Scholar] [CrossRef]
- Martinez, J.; Malireddi, R.K.S.; Lu, Q.; Cunha, L.D.; Pelletier, S.; Gingras, S.; Orchard, R.; Guan, J.L.; Tan, H.; Peng, J.; et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat. Cell Biol. 2015, 17, 893–906. [Google Scholar] [CrossRef]
- Huang, J.H.; Liu, C.Y.; Wu, S.Y.; Chen, W.Y.; Chang, T.H.; Kan, H.W.; Hsieh, S.T.; Ting, J.P.Y.; Wu-Hsieh, B.A. NLRX1 Facilitates Histoplasma capsulatum-Induced LC3-Associated Phagocytosis for Cytokine Production in Macrophages. Front. Immunol. 2018, 9, 2761. [Google Scholar] [CrossRef]
- Pereira de Oliveira Júnior, G.; Renney de Sousa, H.; César de Melo Gorgo-, K.; Karla dos Santos Borges, T.; Teixeira Rangel, K.; Fabricant, S.; Koser Gustavo, C.; Fraga Friaça, L.; Rossi Neto, A.; Andrés Hurtado, F.; et al. Molecular mechanisms of LC3-associated phagocytosis in the macrophage response to Paracoccidioides spp. bioRxiv 2020. [Google Scholar] [CrossRef]
- Sanjuan, M.A.; Dillon, C.P.; Tait, S.W.G.; Moshiach, S.; Dorsey, F.; Connell, S.; Komatsu, M.; Tanaka, K.; Cleveland, J.L.; Withoff, S.; et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 2007, 450, 1253–1257. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.P.; Cavalcante, R.d.S.; Marques, S.A.; Marques, M.E.A.; Venturini, J.; Sylvestre, T.F.; Paniago, A.M.M.; Pereira, A.C.; da Silva, J.d.F.; Fabro, A.T.; et al. Paracoccidioidomycosis: Current Perspectives from Brazil. Open Microbiol. J. 2017, 11, 224–282. [Google Scholar] [CrossRef] [PubMed]
- Pina, A.; de Araujo, E.F.; Felonato, M.; Loures, F.V.; Feriotti, C.; Bernardino, S.; Barbuto, J.A.M.; Calich, V.L.G. Myeloid dendritic cells (DCs) of mice susceptible to paracoccidioidomycosis suppress T cell responses whereas myeloid and plasmacytoid DCs from resistant mice induce effector and regulatory T cells. Infect. Immun. 2013, 81, 1064–1077. [Google Scholar] [CrossRef]
- Daynes, R.A.; Jones, D.C. Emerging roles of PPARs in inflammation and immunity. Nat. Rev. Immunol. 2002, 2, 748–759. [Google Scholar] [CrossRef]
- Ricote, M.; Glass, C.K. PPARs and molecular mechanisms of transrepression. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2007, 1771, 926–935. [Google Scholar] [CrossRef]
- Pina, A.; Saldiva, P.H.N.; Cano Restrepo, L.E.; Calich, V.L.G. Neutrophil role in pulmonary paracoccidioidomycosis depends on the resistance pattern of hosts. J. Leukoc. Biol. 2006, 79, 1202–1213. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.R.; Dias-Melicio, L.A.; Calvi, S.A.; Peraçoli, M.T.S.; Soares, A.M.V.C. Paracoccidioides brasiliensis killing by IFN-γ, TNF-α and GM-CSF activated human neutrophils: Role for oxygen metabolites. Med. Mycol. 2007, 45, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Acorci-Valério, M.J.; Bordon-Graciani, A.P.; Dias-Melicio, L.A.; De Assis Golim, M.; Nakaira-Takahagi, E.; De Campos Soares, Â.M.V. Role of TLR2 and TLR4 in human neutrophil functions against Paracoccidioides brasiliensis. Scand. J. Immunol. 2010, 71, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Puerta-Arias, J.D.; Pino-Tamayo, P.A.; Arango, J.C.; González, Á. Depletion of neutrophils promotes the resolution of pulmonary inflammation and fibrosis in Mice infected with Paracoccidioides brasiliensis. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Pino-Tamayo, P.A.; Puerta-Arias, J.D.; Lopera, D.; Urán-Jiménez, M.E.; González, Á. Depletion of Neutrophils Exacerbates the Early Inflammatory Immune Response in Lungs of Mice Infected with Paracoccidioides brasiliensis. Mediat. Inflamm. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Kashino, S.S.; Fazioli, R.d.A.; Moscardi-Bacchi, M.; Franco, M.; Singer-Vermes, L.M.; Burger, E.; Calich, V.L.G. Effect of macrophage blockade on the resistance of inbred mice to Paracoccidioides brasiliensis infection. Mycopathologia 1995, 130, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Brummer, E.; Hanson, L.H.; Restrepo, A.; Stevens, D.A. Intracellular multiplication of Paracoccidioides brasiliensis in macrophages: Killing and restriction of multiplication by activated macrophages. Infect. Immun. 1989, 57, 2289–2294. [Google Scholar] [CrossRef] [PubMed]
- Delamarre, L.; Pack, M.; Chang, H.; Mellman, I.; Trombetta, E.S. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science 2005, 307, 1630–1634. [Google Scholar] [CrossRef]
- Deretic, V.; Saitoh, T.; Akira, S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013, 13, 722–737. [Google Scholar] [CrossRef]
- Tam, J.M.; Mansour, M.K.; Acharya, M.; Sokolovska, A.; Timmons, A.K.; Lacy-Hulbert, A.; Vyas, J.M. The role of autophagy-related proteins in Candida albicans infections. Pathogens 2016, 5, 34. [Google Scholar] [CrossRef]
- Oikonomou, V.; Renga, G.; De Luca, A.; Borghi, M.; Pariano, M.; Puccetti, M.; Paolicelli, G.; Stincardini, C.; Costantini, C.; Bartoli, A.; et al. Autophagy and LAP in the fight against fungal infections: Regulation and therapeutics. Mediat. Inflamm. 2018, 2018. [Google Scholar] [CrossRef]
- Heckmann, B.L.; Green, D.R. LC3-associated phagocytosis at a glance. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef]
- Matsuzawa-Ishimoto, Y.; Hwang, S.; Cadwell, K. Autophagy and Inflammation. Annu. Rev. Immunol. 2018, 36, 73–101. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, J.; Zeng, Z.; Que, W.; Zhou, L. Knockdown of DEPTOR induces apoptosis, increases chemosensitivity to doxorubicin and suppresses autophagy in RPMI-8226 human multiple myeloma cells in vitro. Int. J. Mol. Med. 2013, 31, 1127–1134. [Google Scholar] [CrossRef]
- Braverman, J.; Sogi, K.M.; Benjamin, D.; Nomura, D.K.; Stanley, S.A. HIF-1α Is an Essential Mediator of IFN-γ–Dependent Immunity to Mycobacterium tuberculosis. J. Immunol. 2016, 197, 1287–1297. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, D.; Zapf, D.; Lohse, B.; Fecher, R.A.; Deepe, G.S.; Rupp, J. The HIF-1/LC3-II axis impacts fungal immunity in human macrophages. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed]
- Quäschling, T.; Friedrich, D.; Deepe, G.S.; Rupp, J. Crosstalk Between Autophagy and Hypoxia-Inducible Factor-1α in Antifungal Immunity. Cells 2020, 9, 2150. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Pirofski, L.-A. What is a host? Attributes of individual susceptibility. Infect. Immun. 2018, 86. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de-Souza-Silva, C.M.; Hurtado, F.A.; Tavares, A.H.; de Oliveira, G.P., Jr.; Raiol, T.; Nishibe, C.; Agustinho, D.P.; Almeida, N.F.; Walter, M.E.M.T.; Nicola, A.M.; et al. Transcriptional Remodeling Patterns in Murine Dendritic Cells Infected with Paracoccidioides brasiliensis: More Is Not Necessarily Better. J. Fungi 2020, 6, 311. https://doi.org/10.3390/jof6040311
de-Souza-Silva CM, Hurtado FA, Tavares AH, de Oliveira GP Jr., Raiol T, Nishibe C, Agustinho DP, Almeida NF, Walter MEMT, Nicola AM, et al. Transcriptional Remodeling Patterns in Murine Dendritic Cells Infected with Paracoccidioides brasiliensis: More Is Not Necessarily Better. Journal of Fungi. 2020; 6(4):311. https://doi.org/10.3390/jof6040311
Chicago/Turabian Stylede-Souza-Silva, Calliandra M., Fabián Andrés Hurtado, Aldo Henrique Tavares, Getúlio P. de Oliveira, Jr., Taina Raiol, Christiane Nishibe, Daniel Paiva Agustinho, Nalvo Franco Almeida, Maria Emília Machado Telles Walter, André Moraes Nicola, and et al. 2020. "Transcriptional Remodeling Patterns in Murine Dendritic Cells Infected with Paracoccidioides brasiliensis: More Is Not Necessarily Better" Journal of Fungi 6, no. 4: 311. https://doi.org/10.3390/jof6040311
APA Stylede-Souza-Silva, C. M., Hurtado, F. A., Tavares, A. H., de Oliveira, G. P., Jr., Raiol, T., Nishibe, C., Agustinho, D. P., Almeida, N. F., Walter, M. E. M. T., Nicola, A. M., Bocca, A. L., Albuquerque, P., & Silva-Pereira, I. (2020). Transcriptional Remodeling Patterns in Murine Dendritic Cells Infected with Paracoccidioides brasiliensis: More Is Not Necessarily Better. Journal of Fungi, 6(4), 311. https://doi.org/10.3390/jof6040311







