Abstract
The fungal genus Fonsecaea contains etiological agents of human chromoblastomycosis, a (sub)tropical, (sub)cutaneous implantation disease caused by plant contact. The invasive potential differs significantly between species. Infections by Fonsecaea monophora are believed to originate from the environment and the species has been reported as one of the main causative agents of the disease, but also of cases of primary brain infection. The epidemiology of the disease has not been fully elucidated and questions related to its infection route and virulence are still to be clarified. The environmental species Fonsecaea erecta was isolated from organic material and living plants in endemic areas for chromoblastomycosis in Brazil. The present paper describes Agrobacterium tumefaciens-mediated transformation (AMT) of the environmental species F. erecta and the pathogenic species F. monophora. We propose the use of Agrobacterium transformation for future gene function studies related to Fonsecaea virulence and pathogenicity. We evaluated the co-cultivation ratios 1:1, 10:1 and 100:1 (Agrobacterium:conidia) at 28 °C during 72 h. pAD1625 and pCAMDsRed plasmids were inserted into both species. Confirmation of transformation was realized by hph gene amplification and Southern blot determined the amount of foreign DNA integrated into the genome. In order to evaluate a potential link between environmental and clinical strains, we obtained red fluorescent transformants after pCAMDsRed insertion. We observed by confocal fluorescence microscopy that both F. monophora and F. erecta were able to colonize the palm Bactris gasipaes, penetrating the epidermis. These results contribute to understanding the ability of Fonsecaea species to adapt to different environmental and host conditions.
1. Introduction
Chromoblastomycosis is a neglected occupational disease [1] caused by several dematiaceous fungi. The infection is chronic and involves cutaneous and subcutaneous tissues [2,3,4,5,6,7,8,9]. Late stages of the infection lead to formation of slow-growing, verrucous or tumor-like ulcerative eruptions [2,10,11,12,13,14,15]. Chromoblastomycosis may be acquired by inoculation of pathogenic agents following penetrating trauma [9]; however, the infection process and the origin of the disease are still not entirely clear [7,16,17,18].
Fonsecaea species and their relatives are asexual Ascomycetes belonging to the family Herpotrichiellaceae (order Chaetothyriales) containing numerous agents of human infection and with a significant predisposition to grow in human tissue [16,19]. Fonsecaea comprises cryptic species (Fonsecaea pedrosoi, Fonsecaea pugnacius, Fonsecaea monophora and Fonsecaea nubica) potentially causing disease [12,15], but the invasive potential differs significantly between species [20,21,22]. Fonsecaea pedrosoi and F. nubica are narrowly associated with chromoblastomycosis, while F. monophora is also involved in primary phaeohyphomycosis of the brain, and occasionally of other organs [12,23,24]. Recently, F. pugnacius was reported to develop chromoblastomycosis and subsequently disseminating to the brain in the same host [7].
An effective way to study mechanisms which these pathogens use to infect and develop the disease is to disrupt their genes in a targeted or random manner, obtaining mutants with altered virulence [25]. This represents one of the most frequently used tools to understand the molecular basis of virulence and host specificity of pathogens. Transformation mediated by Agrobacterium (AMT) is an excellent method for fungal transformation, because it has a high efficiency, usually with a single copy integrated in the DNA, does not require special equipment, and the experiment material can variably be yeast cells, mycelial fragments, or conidia [26,27]. Selective markers such as hygromycin B resistance can be used for construction of cassettes to delete one or more genes. Fluorescent reporter genes are powerful tools to signal the presence of fungi and their visualization inside host tissue, and monitor morphological alterations related to host–fungal interaction [25].
The present study aimed to transform Fonsecaea sibling species mediated by Agrobacterium tumefaciens to understand the environmental part of the life cycle, using the native palm Bactris gasipaes as a model. We describe AMT of F. erecta, an environmental species isolated from a living plant, and the pathogenic species F. monophora, transformed with pCAMDsRed and pAD1625. Our study may contribute to reveal the link between environmental and clinical strains, in order to understand how environmental species are in the process of evolution and have eliminated the ability to infect the host.
2. Materials and Methods
2.1. Strains, Plasmids, and Growth Conditions
Strains used were Fonsecaea erecta (CBS 125763) isolated from living plant and the pathogenic species F. monophora (CBS 269.37) provided by “Microbiological Collections of the Paranaense Network” (CMRP) at the University Federal of Paraná, Curitiba, Paraná, Brazil. Strains were grown on Sabouraud glucose agar (SGA), pH 5.6. We used Agrobacterium tumefaciens strain EHA 105, with (1) binary vector pCAMDsRed (DsRed-Express) which is composed of DsRed (reporter gene), hygromycin B resistance (hph) and kanamycin resistance gene for selection in bacteria [28,29]; (2) binary vector pAD1625 which has hygromycin B resistance and ampicillin gene [30,31]. Agrobacterium tumefaciens was grown at 28 °C in Luria–Bertani (LB) medium supplemented with 100 µg/mL kanamycin (pCAMDsRed) or 100 µg/mL ampicillin (pAD1625). The strains F. pedrosoi CBS 271.37 and F. pedrosoi hygromycin B resistance-carrying gGFP plasmid were used as controls [31].
2.2. Fonsecaea monophora CBS 269.37 and F. erecta CBS 125763 Susceptibility to the Dominant Selective Markers
For evaluation of the susceptibility of hygromycin B, fungi were inoculated as single points on SGA supplemented with different concentrations of hygromycin B (25, 50, 75, 100 and 150 µg/mL). Test was performed in duplicate. Plates were incubated at 28 °C and evaluated at 3, 7, 14, 21 and 30 days after inoculation. Sensitivity to hygromycin B was estimated based on mycelial growth [32]. As controls, we used F. pedrosoi CBS 271.37 and F. pedrosoi hygromycin B resistance-carrying gGFP plasmid [31].
2.3. Fonsecaea Siblings with A. tumefaciens-Mediated Transformation
Agrobacterium-mediated transformation was performed according to Florencio et al. (2018) [31] with few modifications. Conidia of F. monophora and F. erecta were inoculated in potato dextrose broth (PDB) with chloramphenicol (25 µg/mL), grown for 7 days at 28 °C at 150 rpm, centrifuged and resuspended in induction medium (IM) plus 40 mM of (2-N-morpholine)-ethane sulfonic acid (MES) and 0.2 mM of 3′,5′-dimethoxy-4′-hydroxyacetophenone (AS). A. tumefaciens was grown overnight in LB liquid medium supplemented for kanamycin (pCAM-DsRed plasmid) and ampicillin (pAD1625 plasmid), at 28 °C at 200 rpm. The bacteria were centrifuged, washed in saline solution (0.9% NaCl), resuspended in 10 mL of IM+MES+AS and grown for approximately 7 h at 28 °C/150 rpm until reaching a density 0.5 to 0.8 at OD600nm. For co-cultivation, yeast cells of F. monophora and F. erecta were mixed with A. tumefaciens cells at varying ratios (1:1, 10:1, 100:1). The cell mixtures were plated on induction medium for 3 days at 28 °C. After co-cultivation, cells were scraped off, washed with saline and inoculated on SGA plus 100 µg/mL hygromycin B and cefotaxime, and incubated at 28 °C until appearance of colonies.
2.4. Mitotic Stability of the Transformed Colonies
Mitotic stability of random colonies resistant to hygromycin B was established by single-point inoculation on SGA at 28 °C for 1 week. Subsequently, isolates were transferred another 4 times, and then grown on SGA supplemented with hygromycin B for evaluation of maintenance of the resistance marker.
2.5. PCR Assay and DNA Hybridization
Detection of the hph insertion was performed using four randomly selected transformants of each species (F. erecta and F. monophora) containing the two different plasmids (pCAMDsRed and pAD1625). Total DNA was extracted according to the method described by Vicente et al. (2008) [17]. Amplification reactions of transformants were performed using primers hph1 (5′AGCGTCTCCGACCTGATG3′) and hph2 (5′CGACGGACGCACTGACGG3′), according to Malonek and Meinhardt [33]. The wild-type strains were used as negative control and the transformed strain with gGFP [31] as positive control. To estimate the number of hph integrations on selected transformants, 38 µg of genomic DNA previously digested with BglII (NEB) was subjected separately to 1% agarose gel electrophoresis, and capillary transferred to Hybrond N+ (GE) membrane. BglII is a single cutter on T-DNA of pAD1625 and pCAMdsRED. The probe in the hybridization procedure was 0.62 kb of hph labeled with digoxigenin (PCR DIG Probe Synthesis Kit, Roche). After hybridization, the membrane was washed (Dig Wash and Block Buffer Set, Roche), and detected with alkaline phosphatase and CDP-Star conjugated anti-digoxigenin Fab antibody (CDP-Star Detection Reagent, Roche, Diagnostics GmbH, Mannheim, Germany). The signal was captured by ImageQuant LAS 4000 (GE Healthcare Bio-Sciences AB GE, Uppsala, Sweden) equipment. Genomic DNAs of F. erecta and F. monophora were used as negative controls (wild type), while pAN7.1 linearized with HindIII was the positive control.
2.6. Fluorescence Evaluation of Transformants Carrying DsRed
The fluorescence evaluation was done based on the previous described by Eckert, M. et al. [28]. Eight transformants (four strains of F. monophora and four of F. erecta) were randomly selected and cultured on SGA with 100 µg/mL hygromycin B and were incubated at 28 °C for 7 days. The red fluorescence of DsRed was detected using a laser scanning confocal microscope A1RSiMP (NIKON, Tokyo, Japan) with the following settings: 561 nm laser for excitation and 575–625 nm filter for emission. The autofluorescence was detected with the following settings: 405 nm laser for excitation/425–475 nm filter for emission of blue fluorescence, 488 nm laser for excitation/500–550 nm for emission of green fluorescence. Images were saved and processed using NIS-Elements Viewer 4.20 software (NIKON). The wild-type strains were used as negative controls.
2.7. Inoculation in Bactris Gasipaes
Bactris gasipaes was provided by the Brazilian Agricultural Research Corporation EMBRAPA. Plants were grown in vitro in MS medium [34] and inoculated with 10 µL of physiological saline solution containing transformed cells or wild-type strains (negative control) at a concentration of 102 conidia/mL for each species (F. monophora and F. erecta). The experiments were performed in duplicate per group. Inocula were applied as suspensions around plant roots [35]. Plants were incubated at room temperature and the infection was monitored by Nikon confocal microscopy.
2.8. Statistical Analysis
Analysis of variance (ANOVA) of the results was performed; when the F test was significant, subsequent comparisons between different ratios were made using Tukey’s test. SPSS Software (IBM Corp., Armonk, NY, USA) was used for statistical analysis.
3. Results
3.1. Susceptibility to the Dominant Selective Marker
Susceptibility of F. monophora (CBS 269.37) and F. erecta (CBS 125763) to hygromycin B was tested prior to transformation assays. Residual growth of F. monophora and F. erecta at concentrations of 25 µg/mL was observed and the strains were completely inhibited at concentrations of 50, 100 and 150 µg/mL. The MIC concentration was determined as 25 µg/mL of hygromycin, but 100 µg/mL was the concentration which was used for the selection of transformants mediated by A. tumefaciens.
3.2. Fonsecaea Siblings A. tumefaciens-Mediated Transformation
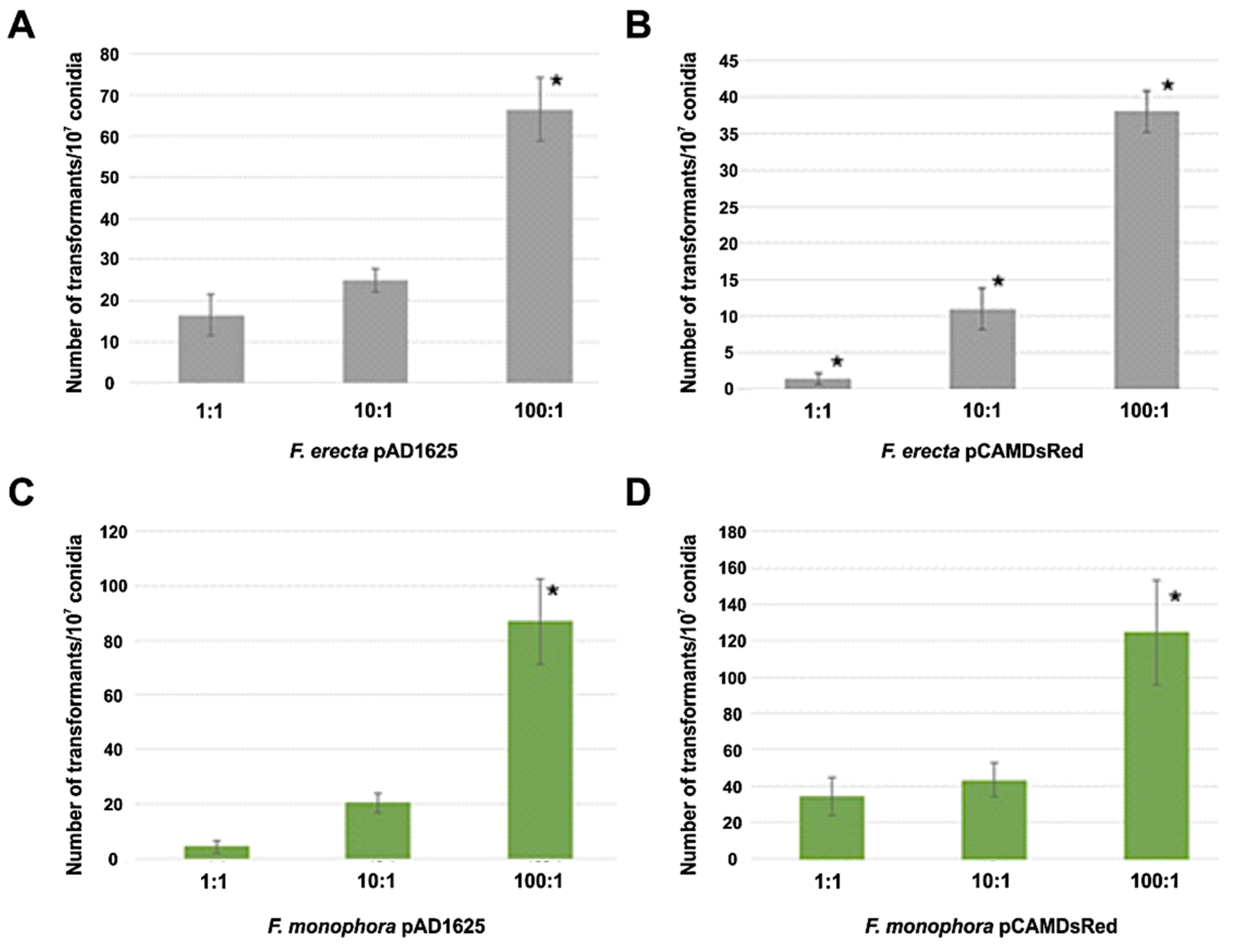
Clinical F. monophora and environmental F. erecta were transformed by Agrobacterium with the pAD1625 and pCAMDsRed plasmids. Figure 1 shows the number of transformants per 107 conidia for both plasmids and both species tested after 72 h of co-cultivation at ratios bacteria:conidia of 1:1, 10:1, and 100:1. The AMT with EHA105 harboring pAD1625 with F. erecta provided the highest number of transformants (statistically significant) with respect to the pCAMDsRed plasmid. However, for the F. monophora, the number of transformants with pCAMDsRed showed a statistical increase compared to pAD1625. The optimal of transformation efficiency was obtained with the ratio bacteria: conidia of 100:1 (Figure 1). All transformants tested were mitotically stable for resistance to hygromycin B at a concentration of 100 µg/mL after five passages in non-selective medium. The transformants conserved the parental morphology of velvet-melanized colonies on SGA plates.
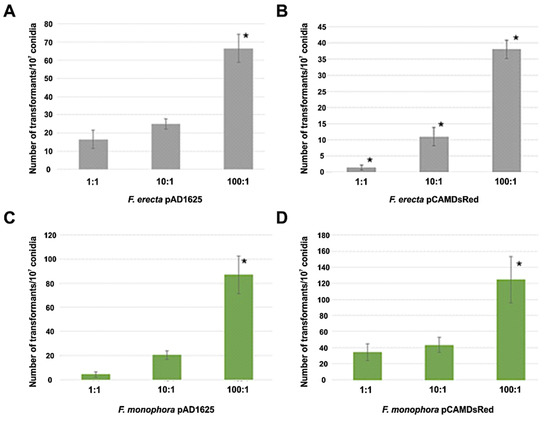
Figure 1.
Number of transformants per 107 conidia obtained by AMT. Comparison of transformation efficiency between 1:1, 10:1 and 100:1 (bacteria:conidia) ratios of Fonsecaea erecta with pAD1625 (A) and pCAMDsRed (B), and Fonsecaea monophora with pAD1625 (C) and pCAMDsRed (D) plasmids. The statistical tests applied were one-way ANOVA and Tukey’s post-test. * indicates significant statistical difference with 95% confidence.
3.3. PCR and DNA Hybridization
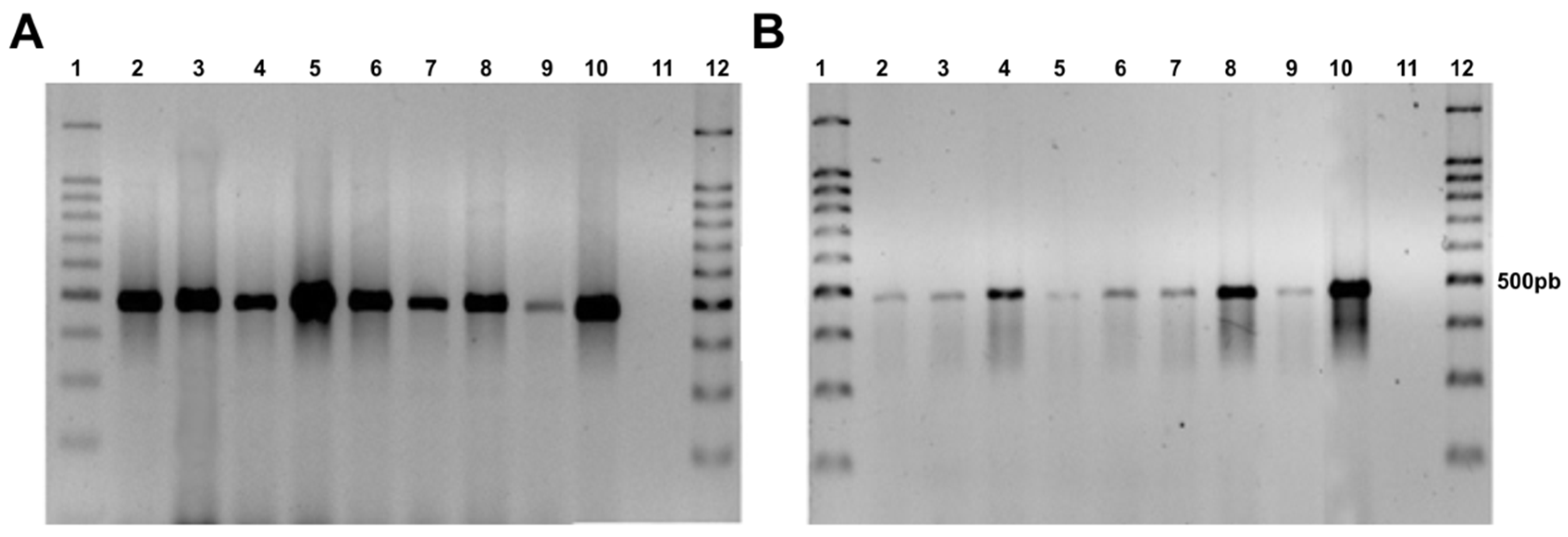
The 500 bp fragment of the hph gene was amplified by PCR, demonstrating the presence of the hygromycin B resistance gene in the genome of the F. erecta and F. monophora transformants, while the wild-type strain did not show any amplification (Figure 2).
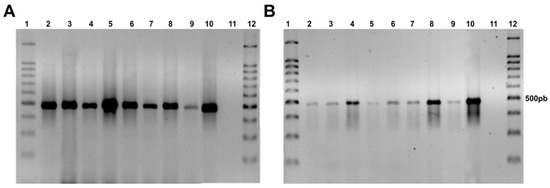
Figure 2.
Agarose gel electrophoresis showing presence of the hph gene in transformed colonies with pAD1625 (A) and pCAMDsRed (B). Fonsecaea erecta (2–5), Fonsecaea monophora (6–9), Fonsecaea pedrosoi gGFP as a positive control (10), F. monophora wild type (11), and 100 bp ladder (1,12).
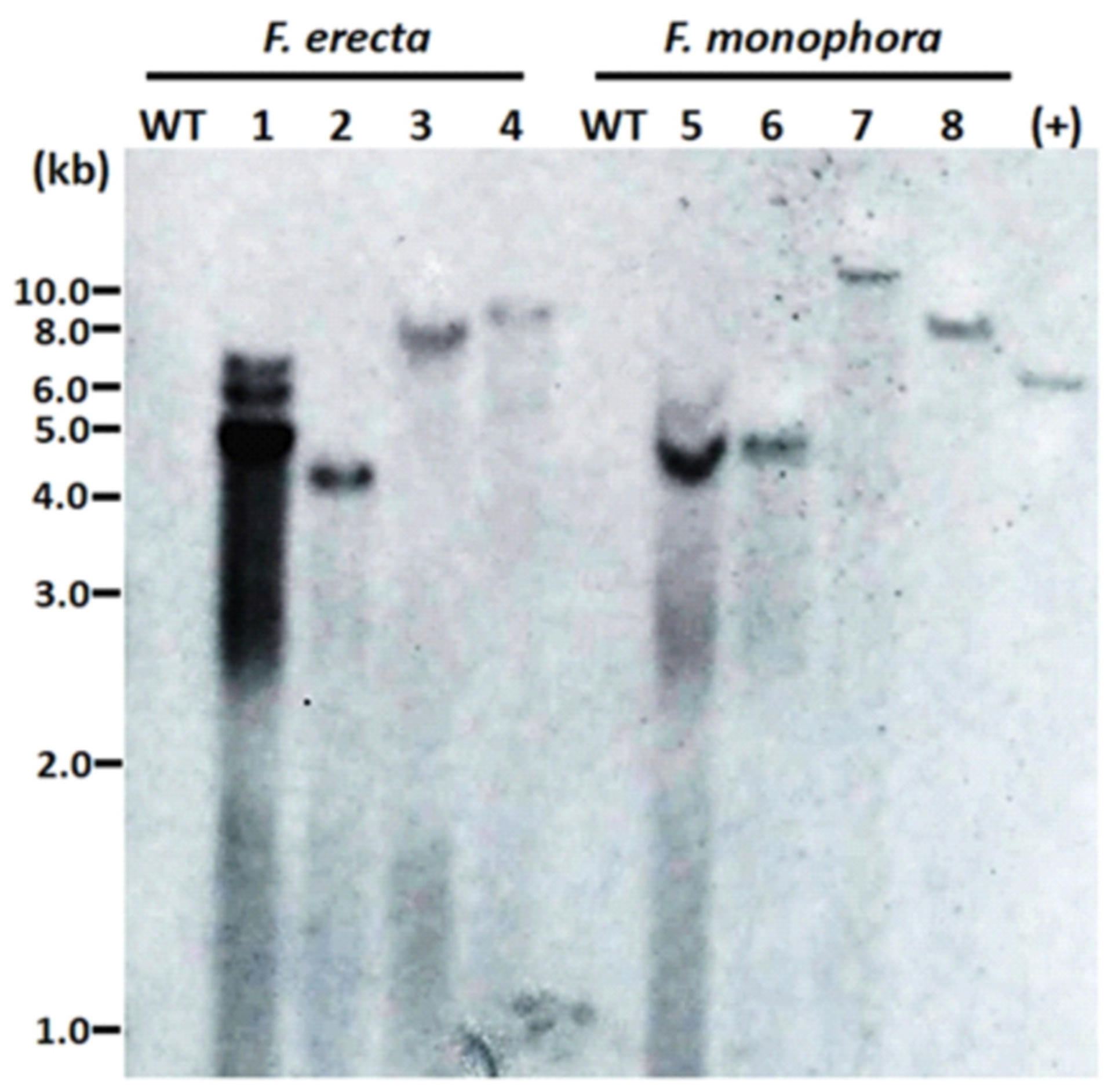
The number of T-DNA copies inserted in the genome of the transformants was evaluated by DNA hybridization. Different sites of integration were observed due to size differences of hybridized fragments. In the majority of transformants, we observed single bands, confirming the insertion of a single copy of T-DNA. In one of the transformants, three bands were observed (Figure 3).
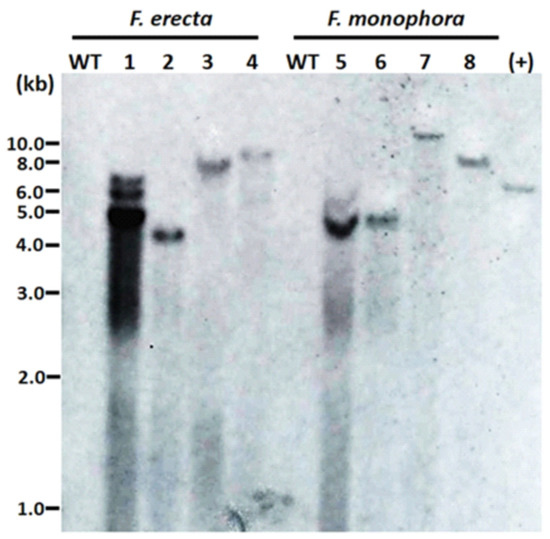
Figure 3.
Integrations of T-DNA containing the hph gene into the genome of the Fonsecaea erecta and Fonsecaea monophora after Agrobacterium-mediated transformation with pAD1625 (1, 2, 5, 6) or pCAMdsRED (3, 4, 7, 8). A total of 38 μg of genomic DNA was digested with BglII, and 0.6 kb digoxigenin-labelled hph was used as probe. The genomic DNA from the wild-type strain of F. erecta (CBS 125763) and F. monophora (CBS 269.37) were used as the untransformed negative control (WT), while the HindIII linearized pAN7.1 was the positive control.
3.4. Reporter Fluorescence Emission by Fonsecaea Transformants
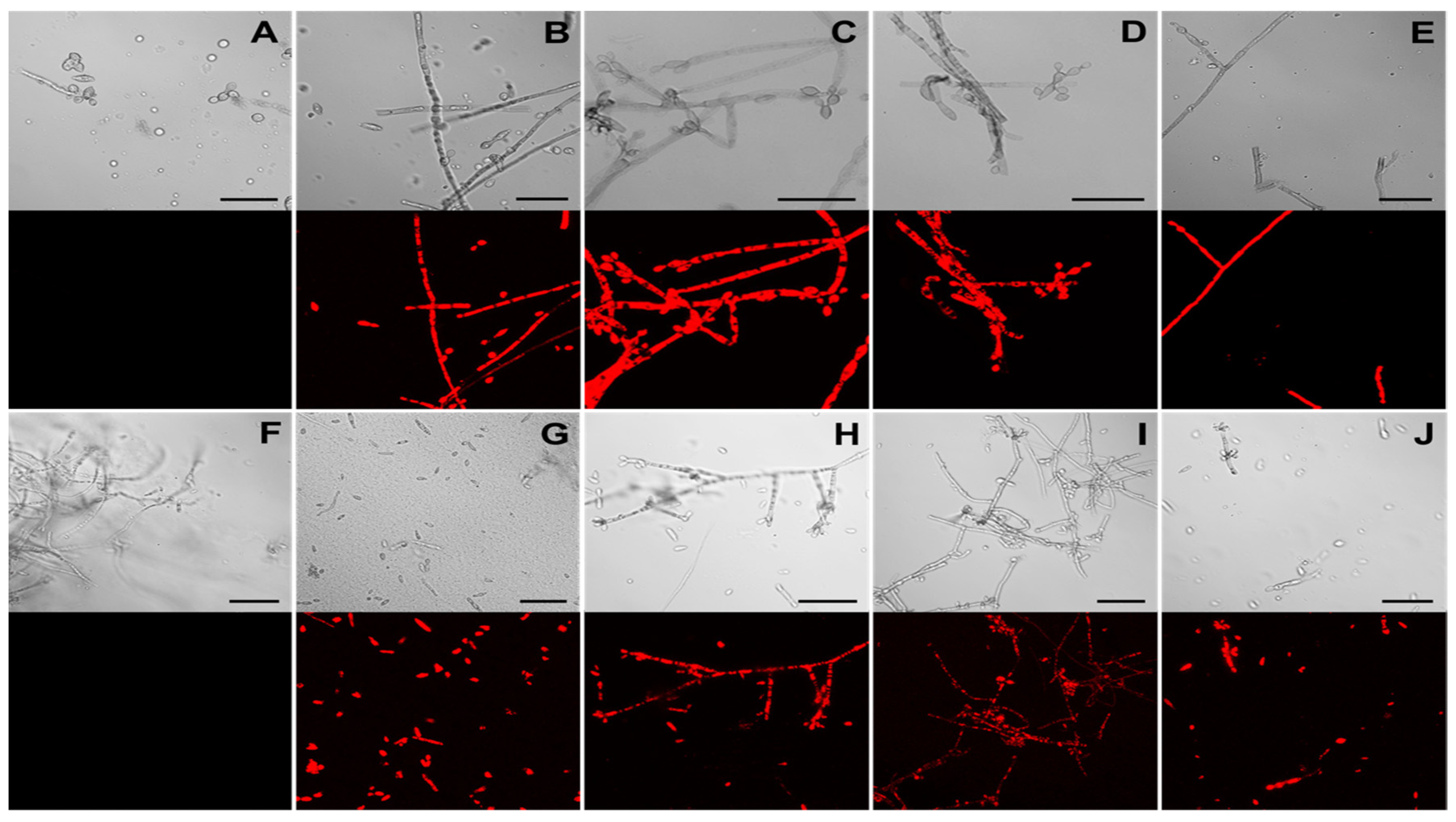
The presence of the functional DsRed protein in F. erecta and F. monophora indicating the pCAMDsRed transformed colonies were observed using fluorescent microscopy. Transformed strains exhibited red fluorescence in the cytoplasm of hyphae and conidia, while no red emission was detected in the wild-type strains (Figure 4A,B).
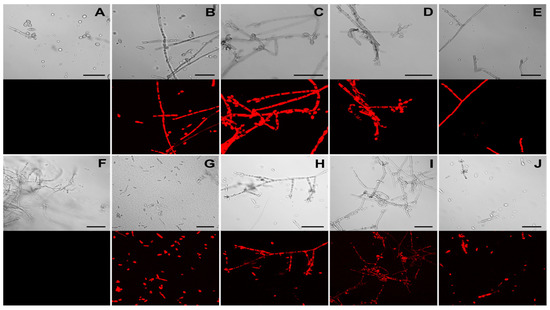
Figure 4.
Detection of DsRed fluorescence in Fonsecaea erecta (A–E) and Fonsecaea monophora (F–J) by confocal microscopy. Images of wild-type (A,F) and transformed strains F. erecta 1-pCAMDsRed (B), 2-pCAMDsRed (C), 3-pCAMDsRed (D) and 4-pCAMDsRed (E); F. monophora 1-pCAMDsRed (G), 2-pCAMDsRed (H), 3-pCAMDsRed (I) and 4-pCAMDsRed (J) are shown. The upper panels depict DIC (Differential Interference Contrast) images and the red emitted fluorescence panels are on the bottom.
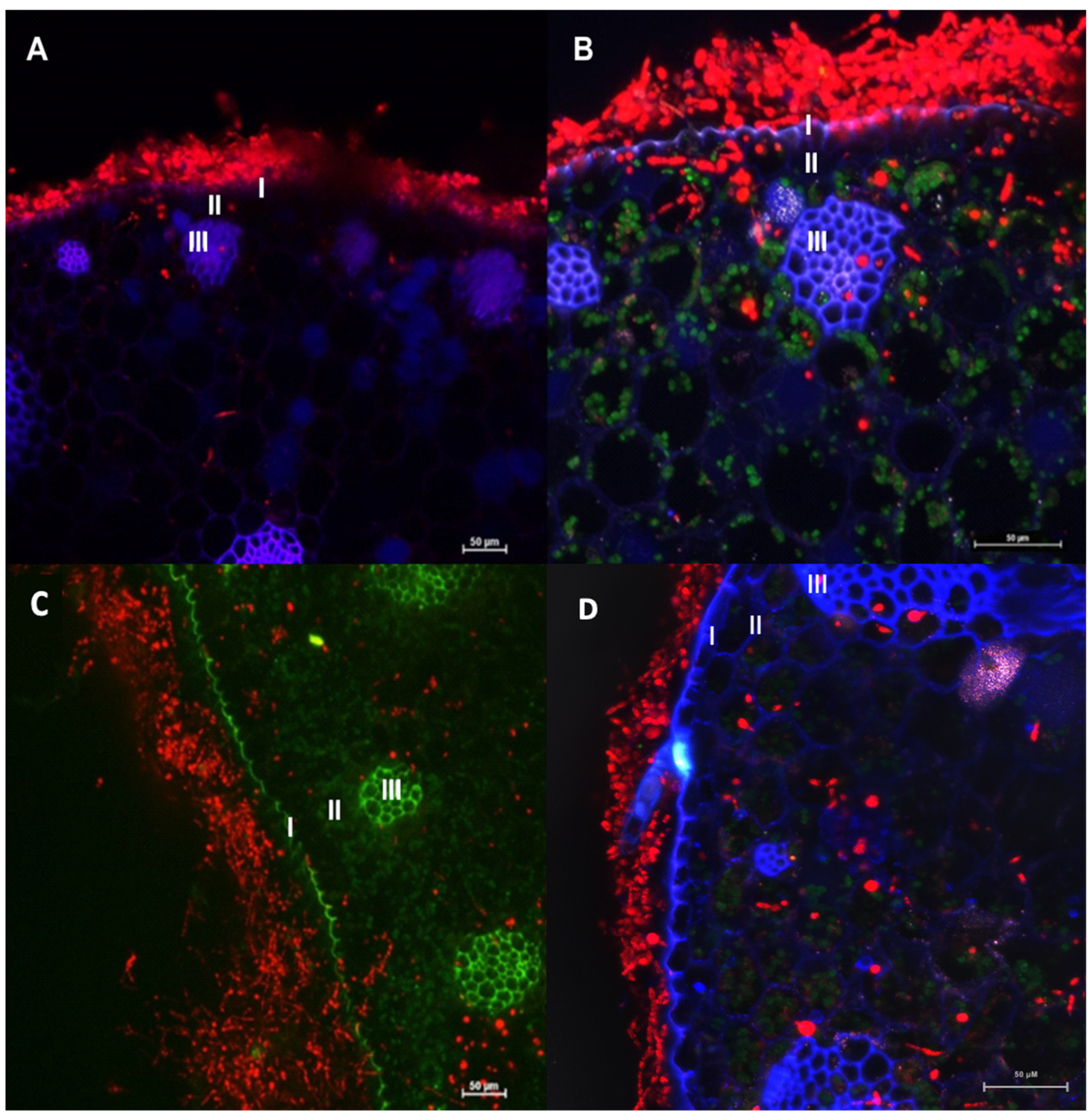
The transformants of F. erecta (4-pCAMDsRed) and F. monophora (4-pCAMDsRed) characterized by red fluorescent were inoculated into the model plant B. gasipaes, with the wild-type strains as control. Fonsecaea erecta and F. monophora expressed red fluorescent proteins while colonizing the intercellular spaces (Figure 5).
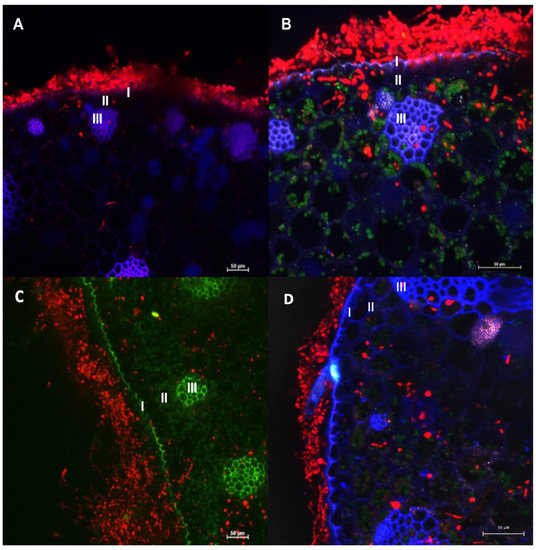
Figure 5.
Confocal microscopy images of Bactris gasipaes plants inoculated with transformed Fonsecaea erecta (A,B) and Fonsecaea monophora (C,D) expressing DsRed gene (red fluorescence). Blue and green are autofluorescence of plant. It is possible to identify epidermis (I), parenchyma (II) and vascular tissue (III). Green autofluorescence reveals the chloroplasts in B and D.
Moreover, conidia, germinated cells and hyphae were observed on the surface of the epidermis, and in parenchyma and vascular tissue of the plant in Figure 5. Both clinical and environmental strains demonstrated ability to germinate and invade the plant tissues.
4. Discussion
Chromoblastomycosis (CMB) is a neglected tropical disease (NTD) caused by dematiaceous fungi and characterized by the presence of muriform cells in tissue, in contrast to phaeohyphomycosis that presents with hyphae. The onset of infection is traumatic inoculation of the fungi from an environmental source [1]. Melanized fungi can be isolated from a wide diversity of natural sources, but the environmental habitat of the clinical species has rarely been revealed. Clinical Fonsecaea species have occasionally been detected on plant material [16,36]. Fonsecaea monophora and F. pedrosoi are generally isolated from clinical samples, while F. erecta and F. minima are saprobes which have as yet not been found in CBM lesions [16].
The present study demonstrated the feasibility of an Agrobacterium tumefaciens-mediated genetic transformation system for F. erecta and F. monophora. AMT is a powerful tool as it has high transformation frequencies, does not require special equipment, and the transformed cells usually receive a single copy of DNA [26]. The variables used for AMT in the present paper were modified according to Florencio et al. [31], who described a high frequency of transformants using 108 rather than 106 conidia, and a co-cultivation period of 72 instead of 48 h. We used 107 conidia and 72 h of co-cultivation. In addition, we employed A. tumefaciens EHA105 as this clone was constructed as a high-virulent strain and was proven to perform excellently in producing high numbers of transformants [37].
Fonsecaea erecta and F. monophora were completely inhibited by hygromycin B at concentrations up to 50 µg/mL of the antibiotic, confirming data of Florencio et al. [31] for F. pedrosoi. This confirms that hygromycin B resistance is an excellent dominant selective marker for genetic transformation of these fungi. A close relationship between conidia ratios and the frequency of Fonsecaea transformants was found. Changing the ratio Agrobacterium:conidia for both Fonsecaea species carrying any of the plasmids (pAD1625 and pCAMDsRed) from 1:1 to 100:1 led to an increase of 4–25 fold in transformation efficiency. The 100:1 ratio generated the highest number of transformants. Florencio et al. [31] used another conidial density, but they also reported an increase in transformation efficiency when a higher conidial ratio was used. Abuodeh et al. [30] reported similar results with Coccidioides immitis and Xiao et al. [38] noted that the transformation also increased in the ratio of 100:1 to transform F. monophora conidia using A. tumefaciens strains AGL-1, although they observed that in EHA105 strains, the highest transformation efficiency was a 1:1 ratio.
The F. erecta and F. monophora transformants were stable after five rounds of growth. Several investigations described AMT transformants to be mitotically very stable [39,40]. The marker gene (hph) allowed proper selection of transformed strains. Confirmation of hph integration into the genome was performed using PCR and DNA hybridization. The evaluated transformants were amplified using hph specific primers, and Southern blot showed that just a single transformant had three randomly inserted copies of hph, while the remaining transformants presented only one hybridized fragment. Michielse et al. [41] also reported that AMT of T-DNA usually inserts a single copy into the fungal genome, with a higher level of mitotic stability of the transformants. Other previous published works also used the same strategy with probes specific to the inserted gene, that in this case was the HPH [27,31,42].
The reporter genes codifying fluorescent proteins can be employed as diagnostic tools in determining the presence of fungal cells in plant hosts [26]. Markers such as GFP and DsRed are widely used because they do not require cofactors or substrates [43], have low toxicity and can be visualized in vivo in individual cells and in cell populations [44]. The DsRed protein (pCAMDsRed) was microscopically detected in hyphae and conidia of all evaluated Fonsecaea transformants at a high intensity emission. Eckert et al. [29] noted higher fluorescence emission of DsRed protein than GFP protein in mycelium of Leptosphaeria spp. and Oculimacula spp.; this may be attributed to the strength and compatibility of the promoter (PgpdA) controlling the reporter genes.
Bactris gasipaes is a palm native to the tropical forests reported as the habitat of melanized fungi [45]. Based on this fact, and wondering how Fonsecaea species can use the plant as a natural reservoir, we inoculated B. gasipaes with F. erecta and F. monophora transformed conidial cells expressing red fluorescent protein. The inoculum in vitro was applied around the root according to Fornari et al. [35], and both clinical and environmental strains were observed in the root on the location of application of the inoculum and also in the tissues of the stem indicating the ability of invasion, colonization and adaptation of these species to living plant tissue.
Judging from the expression of transformed fungal cells of both species inside the plant tissues, it was observed that transformed strains were able to invade the epidermis, parenchyma and vascular and grow inside tissue of B. gasipaes (Figure 5). However, in this study, we observed that the clinical species F. monophora was also able to grow in the vascular tissue. Chromoblastomycosis is characterized by the presence of muriform cells inside host tissues. In these studies, as well as in Fornari et al. [36], the cells were not observed inside the plant tissues. However, de Hoog et al. [45] demonstrated that in Cladophialophora both the clinical species, C. carrionii, and the environmental species, C. yegresii could produce muriform cells upon their artificial inoculation into cactus plants.
Since the A. tumefaciens-mediated transformation technique is used for random insertional mutagenesis, it is important to assess whether the biological function has been maintained. We observed that the transformed isolates kept their ability to invade plant tissues since they remained able to penetrate the epidermis, reaching the cortical region and clearly growing in the intercellular spaces that were colonized by the fungus. Therefore, the clinical and environmental Fonsecaea strains obtained in this study could be used for research in the future.
Our results highlight that both species, F. monophora and F. erecta, have the ability to penetrate plant epidermis and deeper tissue of our model plant, where we found the transformants expressing DsRed in the area that generates the thorns, which have been the hypothesized transmission route of Fonsecaea from the environment to the human host. Our study proposes a new protocol allowing observation of Fonsecaea sp. inside plants without histological staining. This approach enables evaluation of the fungi in living tissue. This presents perspectives to understand conidial germination, penetration, colonization and nutrient acquisition of the environmental form of Fonsecaea, and may clarify the route of infection of human skin, possibly explaining the infection route of agents of chromoblastomycosis.
Author Contributions
Conceptualization, R.R.G., S.d.H. and V.A.V.; Methodology, C.I.F.V., R.R.G, L.F., C.S.F., A.B., S.d.H., V.A.V.; Software, C.I.F.V., R.R.G., L.F., C.S.F., A.B., V.A.V.; Validation, C.I.F., R.R.G., L.F., C.S.F., A.B., E.d.S.T., S.d.H. and V.A.V.; Formal Analysis, C.I.F.V., R.R.G., L.F., C.S.F., E.d.S.T., S.d.H. and V.A.V.; Investigation, C.I.F.V., R.R.G., L.F., C.S.F., S.d.H. and V.A.V.; Resources, C.I.F.V., R.R.G., L.F., C.S.F., S.d.H. and V.A.V.; Data Curation, C.I.F.V., R.R.G., L.F., C.S.F., S.d.H. and V.A.V.; Writing—Original Draft Preparation, C.I.F.V., R.R.G. and V.A.V.; Writing—Review and Editing, C.I.F.V., R.R.G., M.E.G., E.S.T., S.d.H. and V.A.V.; Visualization, R.R.G., L.F., M.E.G., E.d.S.T., S.d.H. and V.A.V.; Supervision, R.R.G., L.F., S.d.H. and V.A.V.; Project Administration, S.d.H. and V.A.V.; Funding Acquisition, L.F. and V.A.V. All authors contributed to the overall data interpretation, provided intellectual input. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Coordination for the Improvement of Higher Education Personnel (CAPES), Brazil; National Council for Scientific and Technological Development (CNPq), Brazil (grant number: 400011/2016-6; and 312811/2018-7), Federal District Research Support Foundation (FAP-DF), Brazil (grant number: 00193.00000180/2019-91), and the institutional Program of Internationalization CAPES/Print, Brazil (grant number 8887.311835/2018-00-AUXPE-2796/2018).
Conflicts of Interest
Authors declare no potential conflicts of interest.
References
- Queiroz-Telles, F.; De Hoog, S.; Santos, D.W.C.L.; Salgado, C.G.; Vicente, V.A.; Bonifaz, A.; Roilides, E.; Xi, L.; Azevedo, C.D.M.P.E.S.; Da Silva, M.B.; et al. Chromoblastomycosis. Clin. Microbiol. Rev. 2016, 30, 233–276. [Google Scholar] [CrossRef]
- Kurien, G.; Sugumar, K.; Chandran, V. Chromoblastomycosis (Chromomycosis). StatPearls [Internet]; 2020. Available online: www.ncbi.nlm.nih.gov/books/NBK470253/ (accessed on 14 September 2020).
- Smith, G.; Chen, A.F.; Weiss, E. Chromoblastomycosis infection from a house plant. Cutis 2017, 100, E13–E14. [Google Scholar]
- Subhadarshani, S.; Yadav, D. Dermoscopy of chromoblastomycosis. Dermatol. Pract. Concept. 2017, 7, 23–24. [Google Scholar] [CrossRef] [PubMed]
- Ventura-Flores, R.; Failoc-Rojas, V.E.; Silva-Díaz, H. Cromoblastomicosis: Características clínicas y microbiológicas de una enfermedad desatendida. Rev. Chil. Infectología 2017, 34, 404–407. [Google Scholar] [CrossRef] [PubMed]
- Gomes, R.R.; Vicente, V.A.; De Azevedo, C.M.P.S.; Salgado, C.G.; Da Silva, M.B.; Queiroz-Telles, F.; Marques, S.G.; Santos, D.W.C.L.; De Andrade, T.S.; Takagi, E.H.; et al. Molecular Epidemiology of Agents of Human Chromoblastomycosis in Brazil with the Description of Two Novel Species. PLoS Negl. Trop. Dis. 2016, 10, e0005102. [Google Scholar] [CrossRef] [PubMed]
- De Azevedo, C.M.P.S.; Gomes, R.R.; Vicente, V.A.; Santos, D.W.C.L.; Marques, S.G.; Nascimento, M.M.F.D.; Andrade, C.E.W.; Silva, R.R.; Queiroz-Telles, F.; De Hoog, G.S. Fonsecaea pugnacius, a Novel Agent of Disseminated Chromoblastomycosis. J. Clin. Microbiol. 2015, 53, 2674–2685. [Google Scholar] [CrossRef]
- Chowdhary, A.; Perfect, J.; De Hoog, G.S. Black Molds and Melanized Yeasts Pathogenic to Humans. Cold Spring Harb. Perspect. Med. 2014, 5, a019570. [Google Scholar] [CrossRef]
- Garnica, M.; Nucci, M.; Queiroz-Telles, F. Difficult mycoses of the skin: Advances in the epidemiology and management of eumycetoma, phaeohyphomycosis and chromoblastomycosis. Curr. Opin. Infect. Dis. 2009, 22, 559–563. [Google Scholar] [CrossRef]
- Vicente, V.A.; Orélis-Ribeiro, R.; Najafzadeh, M.; Sun, J.; Guerra, R.S.; Miesch, S.; Ostrensky, A.; Meis, J.F.; Klaassen, C.H.; De Hoog, G.; et al. Black yeast-like fungi associated with Lethargic Crab Disease (LCD) in the mangrove-land crab, Ucides cordatus (Ocypodidae). Vet. Microbiol. 2012, 158, 109–122. [Google Scholar] [CrossRef]
- Najafzadeh, M.; Vicente, V.; Sun, J.; Meis, J.; De Hoog, G.S. Fonsecaea multimorphosa sp. nov, a new species of Chaetothyriales isolated from a feline cerebral abscess. Fungal Biol. 2011, 115, 1066–1076. [Google Scholar] [CrossRef]
- Najafzadeh, M.J.; Sun, J.; Vicente, V.; Xi, L.; Ende, A.H.G.G.V.D.; De Hoog, G.S. Fonsecaea nubicasp. nov, a new agent of human chromoblastomycosis revealed using molecular data. Med. Mycol. 2010, 48, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Najafzadeh, M.J.; Rezusta, A.; Cameo, M.I.; Zubiri, M.L.; Yus, M.C.; Badali, H.; Revillo, M.J.; De Hoog, G.S. Successful treatment of chromoblastomycosis of 36 years duration caused byFonsecaea monophora. Med. Mycol. 2010, 48, 390–393. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Najafzadeh, M.J.; Gueidan, C.; Badali, H.; Ende, A.H.G.G.V.D.; Xi, L.; De Hoog, G.S. Genetic diversity and species delimitation in the opportunistic genusFonsecaea. Med. Mycol. 2009, 47, 17–25. [Google Scholar] [CrossRef] [PubMed]
- De Hoog, G.S.; Attili-Angelis, D.; Vicente, V.A.; Ende, A.H.G.G.V.D.; Queiroz-Telles, F. Molecular ecology and pathogenic potential ofFonsecaeaspecies. Med. Mycol. 2004, 42, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Vicente, V.A.; Najafzadeh, M.J.; Sun, J.; Gomes, R.R.; Robl, D.; Marques, S.G.; Azevedo, C.M.P.S.; De Hoog, G.S. Environmental siblings of black agents of human chromoblastomycosis. Fungal Divers. 2013, 65, 47–63. [Google Scholar] [CrossRef]
- Vicente, V.; Attili-Angelis, D.; Pie, M.; Queiroz-Telles, F.; Cruz, L.; Najafzadeh, M.; De Hoog, G.; Zhao, J.; Pizzirani-Kleiner, A. Environmental isolation of black yeast-like fungi involved in human infection. Stud. Mycol. 2008, 61, 137–144. [Google Scholar] [CrossRef]
- Vicente, V.A.; De Angelis, D.A.; Filho, F.Q.-T.; Pizzirani-Kleiner, A.A. Isolation of herpotrichiellacious fungi from the environment. Braz. J. Microbiol. 2001, 32, 47–51. [Google Scholar] [CrossRef]
- Vicente, V.A.; Weiss, V.A.; Bombassaro, A.; Moreno, L.F.; Costa, F.F.; Raittz, R.T.; Leão, A.C.; Gomes, R.R.; Bocca, A.L.; Fornari, G.; et al. Comparative Genomics of Sibling Species of Fonsecaea Associated with Human Chromoblastomycosis. Front. Microbiol. 2017, 8, 1924. [Google Scholar] [CrossRef]
- Seyedmousavi, S.; Guillot, J.; De Hoog, G.S. Phaeohyphomycoses, Emerging Opportunistic Diseases in Animals. Clin. Microbiol. Rev. 2013, 26, 19–35. [Google Scholar] [CrossRef]
- Badali, H.; Gueidan, C.; Najafzadeh, M.J.; Bonifaz, A.; Ende, A.G.V.D.; De Hoog, G.S. Biodiversity of the genus Cladophialophora. Stud. Mycol. 2008, 61, 175–191. [Google Scholar] [CrossRef]
- De Hoog, G.S.; Zeng, J.S.; Harrak, M.J.; Sutton, D.A. Exophiala xenobiotica sp. nov., an opportunistic black yeast inhabiting environments rich in hydrocarbons. Antonie Van Leeuwenhoek 2006, 90, 257–268. [Google Scholar] [CrossRef]
- Koo, S.; Klompas, M.; Marty, F.F. Fonsecaea monophoracerebral phaeohyphomycosis: Case report of successful surgical excision and voriconazole treatment and review. Med. Mycol. 2010, 48, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Surash, S.; Tyagi, A.; De Hoog, G.S.; Zeng, J.-S.; Barton, R.C.; Hobson, R.P. Cerebral phaeohyphomycosis caused byFonsecaea monophora. Med. Mycol. 2005, 43, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Mullins, E.; Kang, S. Transformation: A tool for studying fungal pathogens of plants. Cell. Mol. Life Sci. 2001, 58, 2043–2052. [Google Scholar] [CrossRef] [PubMed]
- Michielse, C.B.; Hooykaas, P.J.J.; Hondel, C.A.M.J.J.V.D.; Ram, A.F.J. Agrobacterium-mediated transformation of the filamentous fungus Aspergillus awamori. Nat. Protoc. 2008, 3, 1671–1678. [Google Scholar] [CrossRef]
- De Groot, M.J.; Bundock, P.; Hooykaas, P.J.; Beijersbergen, A.G. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 1998, 16, 839–842. [Google Scholar] [CrossRef] [PubMed]
- Eckert, M.; Maguire, K.; Urban, M.; Foster, S.J.; Fitt, B.; Lucas, J.; Hammond-Kosack, K.E. Agrobacterium tumefaciens-mediated transformation of Leptosphaeria spp. and Oculimacula spp. with the reef coral gene DsRed and the jellyfish gene gfp. FEMS Microbiol. Lett. 2005, 253, 67–74. [Google Scholar] [CrossRef]
- Dos Santos, P.J.C.; Savi, D.C.; Gomes, R.R.; Goulin, E.H.; Senkiv, C.D.C.; Tanaka, F.A.O.; Almeida, Á.M.R.; Galli-Terasawa, L.; Kava, V.; Glienke, C. Diaporthe endophytica and D. terebinthifolii from medicinal plants for biological control of Phyllosticta citricarpa. Microbiol. Res. 2016, 186, 153–160. [Google Scholar] [CrossRef]
- Abuodeh, R.O.; Orbach, M.J.; Mandel, M.A.; Das, A.; Galgiani, J.N. Genetic Transformation ofCoccidioides immitisFacilitated byAgrobacterium tumefaciens. J. Infect. Dis. 2000, 181, 2106–2110. [Google Scholar] [CrossRef]
- Florencio, C.S.; Brandão, F.; Teixeira, M.D.M.; Bocca, A.L.; Felipe, M.S.S.; Vicente, V.A.; Fernandes, L. Genetic manipulation of Fonsecaea pedrosoi using particles bombardment and Agrobacterium mediated transformation. Microbiol. Res. 2018, 207, 269–279. [Google Scholar] [CrossRef]
- Sebastianes, F.L.S.; Lacava, P.T.; Fávaro, L.C.L.; Rodrigues, M.B.C.; Araújo, W.L.; Azevedo, J.L.; Pizzirani-Kleiner, A.A. Genetic transformation of Diaporthe phaseolorum, an endophytic fungus found in mangrove forests, mediated by Agrobacterium tumefaciens. Curr. Genet. 2012, 58, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Malonek, S.; Meinhardt, F. Agrobacterium tumefaciens-mediated genetic transformation of the phytopathogenic ascomycete Calonectria morganii. Curr. Genet. 2001, 40, 152–155. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Fornari, G.; Gomes, R.R.; Degenhardt-Goldbach, J.; Dos Santos, S.S.; De Almeida, S.R.; Dos Santos, G.D.; Muro, M.D.; Bona, C.; Scola, R.H.; Trindade, E.S.; et al. A Model for Trans-Kingdom Pathogenicity in Fonsecaea Agents of Human Chromoblastomycosis. Front. Microbiol. 2018, 9, 2211. [Google Scholar] [CrossRef]
- Salgado, C.G.; Da Silva, J.P.; Diniz, J.A.P.; Da Silva, M.B.; Da Costa, P.F.; Teixeira, C.; Salgado, U.I. Isolation of Fonsecaea pedrosoi from thorns of Mimosa pudica, a probable natural source of chromoblastomycosis. Rev. Do Inst. De Med. Trop. De São Paulo 2004, 46, 33–36. [Google Scholar] [CrossRef][Green Version]
- Hood, E.E.; Gelvin, S.B.; Melchers, L.S.; Hoekema, A. NewAgrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 1993, 2, 208–218. [Google Scholar] [CrossRef]
- Xiao, X.; Li, Y.; Qin, J.; He, Y.; Cai, W.; Chen, Z.; Xi, L.; Zhang, J. An optimized Agrobacterium tumefaciens-mediated transformation system for random insertional mutagenesis in Fonsecaea monophora. J. Microbiol. Methods 2020, 170, 105838. [Google Scholar] [CrossRef]
- Rho, H.S.; Kang, S.; Lee, Y.H. Agrobacterium tumefaciens-mediated transformation of the plant pathogenic fungus, Magnaporthe grisea. Mol. Cells 2001, 12, 407–411. [Google Scholar]
- Figueiredo, J.A.G.; Goulin, E.; Tanaka, F.; Stringari, D.; Kava, V.; Galli-Terasawa, L.V.; Staats, C.C.; Schrank, A.; Glienke, C. Agrobacterium tumefaciens-mediated transformation of Guignardia citricarpa. J. Microbiol. Methods 2010, 80, 143–147. [Google Scholar] [CrossRef]
- Michielse, C.B.; Hooykaas, P.J.J.; Hondel, C.A.M.J.J.V.D.; Ram, A. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 2005, 48, 1–17. [Google Scholar] [CrossRef]
- Nahálková, J.; Fatehi, J. Red fluorescent protein (DsRed2) as a novel reporter inFusarium oxysporumf. sp.lycopersici. FEMS Microbiol. Lett. 2003, 225, 305–309. [Google Scholar] [CrossRef]
- Lorang, J.M.; Tuori, R.P.; Martinez, J.P.; Sawyer, T.L.; Redman, R.S.; Rollins, J.A.; Wolpert, T.J.; Johnson, K.B.; Rodriguez, R.J.; Dickman, M.B.; et al. Green Fluorescent Protein Is Lighting Up Fungal Biology. Appl. Environ. Microbiol. 2001, 67, 1987–1994. [Google Scholar] [CrossRef] [PubMed]
- Caligiorne, R.B.; Licinio, P.; Dupont, J.; De Hoog, G.S. Internal Transcribed Spacer rRNA Gene-Based Phylogenetic Reconstruction Using Algorithms with Local and Global Sequence Alignment for Black Yeasts and Their Relatives. J. Clin. Microbiol. 2005, 43, 2816–2823. [Google Scholar] [CrossRef] [PubMed]
- De Hoog, G.; Nishikaku, A.; Fernandez-Zeppenfeldt, G.; Padín-González, C.; Burger, E.; Badali, H.; Richard-Yegres, N.; Ende, A.G.V.D. Molecular analysis and pathogenicity of the Cladophialophora carrionii complex, with the description of a novel species. Stud. Mycol. 2007, 58, 219–234. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).