Biosensors and Diagnostics for Fungal Detection
Abstract
1. Introduction
2. Conventional Diagnostic Tools
3. Galactomannan Detection
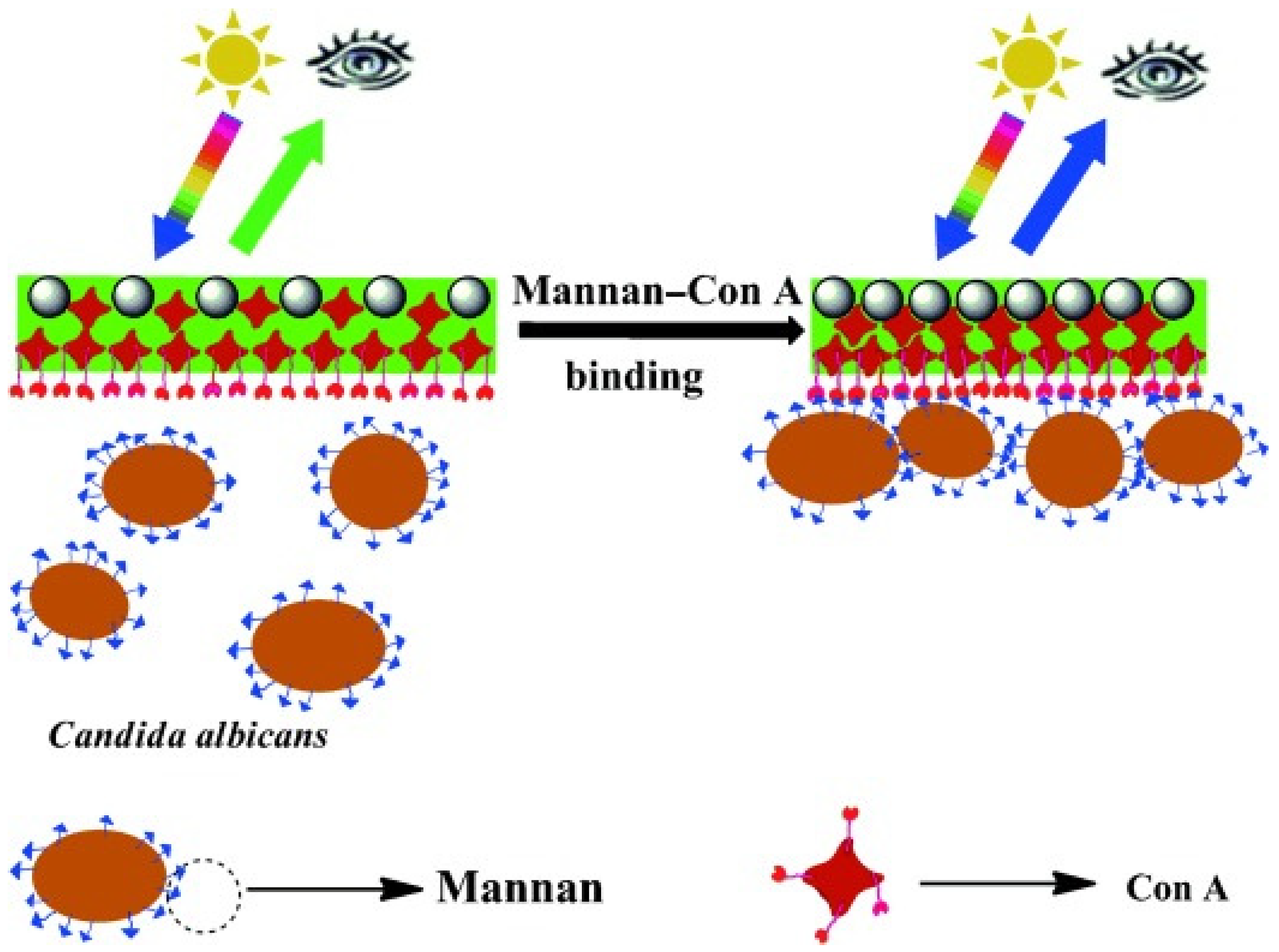
4. Mannan
5. β-(1,3)-D-Glucan
6. Cryptococcal Capsular Polysaccharide
7. Point-of-Care Tests (POCT) in Fungal Diagnosis
8. Nucleic Acids Based Diagnostics
9. Biosensors for Fungal Detection
10. Working Principal of Biosensors
10.1. Electrochemical Biosensors
10.2. Optical Biosensors
10.3. Piezoelectric Biosensors
10.4. Thermal biosensors
11. Emerging Diagnostic Methods
12. Current and Future Fungal Biosensors and Biomarkers
13. Pattern Recognition Receptors
14. Siderophores
15. Mycotoxins
16. Biosensor Immobilization Techniques
16.1. Chemical Immobilization
16.2. Physical Immobilization
17. Characterization of Immobilised Biomolecules
18. Conclusions and Future Perspective
Funding
Conflicts of Interest
References
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Zhan, P.; Liu, W. The changing face of dermatophytic infections worldwide. Mycopathologia 2017, 182, 77–86. [Google Scholar] [CrossRef]
- Kullberg, B.J.; Arendrup, M.C. Invasive candidiasis. N. Engl. J. Med. 2015, 373, 1445–1456. [Google Scholar] [CrossRef]
- Denning, D.W.; Kneale, M.; Sobel, J.D.; Rautemaa-Richardson, R. Global burden of recurrent vulvovaginal candidiasis: A systematic review. Lancet Infect. Dis. 2018, 18, e339–e347. [Google Scholar] [CrossRef]
- Von Eiff, M.; Roos, N.; Schulten, R.; Hesse, M.; Zühlsdorf, M.; Van de Loo, J. Pulmonary aspergillosis: Early diagnosis improves survival. Respiration 1995, 62, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Morrell, M.; Fraser, V.J.; Kollef, M.H. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: A potential risk factor for hospital mortality. Antimicrob. Agents Chemother. 2005, 49, 3640–3645. [Google Scholar] [CrossRef]
- Head, M.G.; Fitchett, J.R.; Atun, R.; May, R.C. Systematic analysis of funding awarded for mycology research to institutions in the UK, 1997–2010. BMJ Open 2014, 4, e004129. [Google Scholar] [CrossRef] [PubMed]
- White, P.L.; Barnes, R.A. Molecular diagnosis of fungal diseases. In Oxford Textbook of Medical Mycology; Kibbler, C.C., Barton, R.C., Gow, N.A.R., Howell, S., MacCallum, D.M., Manuel, R., Eds.; Oxford University Press: Hong Kong, China, 2018; pp. 313–326. [Google Scholar]
- Patel, R. MALDI-TOF MS for the diagnosis of infectious diseases. Clin. Chem. 2015, 61, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Ashbee, H.R. General Approaches for direct detection and identification of fungi. In Manual of Clinical Microbiology; ASM Press: Washington, DC, USA, 2015; pp. 1965–1983. [Google Scholar]
- Kauffman, C.A.; Fisher, J.F.; Sobel, J.D.; Newman, C.A. Candida urinary tract infections—Diagnosis. Clin. Infect. Dis. 2011, 52, 452–456. [Google Scholar] [CrossRef]
- D’Haese, J.; Theunissen, K.; Vermeulen, E.; Schoemans, H.; De Vlieger, G.; Lammertijn, L.; Meersseman, P.; Meersseman, W.; Lagrou, K.; Maertens, J. Detection of galactomannan in bronchoalveolar lavage fluid samples of patients at risk for invasive pulmonary aspergillosis: Analytical and clinical validity. J. Clin. Microbiol. 2012, 50, 1258–1263. [Google Scholar] [CrossRef]
- Vyzantiadis, T.A.A.; Johnson, E.M.; Kibbler, C.C. From the patient to the clinical mycology laboratory: How can we optimise microscopy and culture methods for mould identification? J. Clin. Pathol. 2012, 65, 475–483. [Google Scholar] [CrossRef] [PubMed]
- McGowan, K.L. Specimen collection, transport, and processing: Mycology. In Manual of Clinical Microbiology; ASM Press: Washington, DC, USA, 2015; pp. 1944–1954. [Google Scholar]
- Borman, A.M.; Johnson, E.M. Genomics and proteomics as compared to conventional phenotypic approaches for the identification of the agents of invasive fungal infections topical collection on advances in diagnosis of invasive fungal infections. Curr. Fungal Infect. Rep. 2013, 7, 235–243. [Google Scholar] [CrossRef]
- Borman, A.M.; Fraser, M.; Johnson, E.M. CHROMagarTM Candida plus: A novel chromogenic agar that permits the rapid identification of Candida auris. Med. Mycol. 2020, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kozel, T.R.; Wickes, B.L. Fungal diagnostics. Cold Spring Harb. Perspect. Med. 2014, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Azar, M.M.; Hage, C.A. Laboratory diagnostics for histoplasmosis. J. Clin. Microbiol. 2017, 55, 1612–1620. [Google Scholar] [CrossRef]
- Donnelly, J.P.; Chen, S.C.; Kauffman, C.A.; Steinbach, W.J.; Baddley, J.W.; Verweij, P.E.; Clancy, C.J.; Wingard, J.R.; Lockhart, S.R.; Groll, A.H.; et al. Revision and update of the consensus definitions of invasive fungal disease from the European organization for research and treatment of cancer and the mycoses study group education and research consortium. Clin. Infect. Dis. 2020, 71, 1367–1376. [Google Scholar] [CrossRef]
- Rüchel, R.; Schaffrinski, M. Versatile fluorescent staining of fungi in clinical specimens by using the optical brightener blankophor. J. Clin. Microbiol. 1999, 37, 2694–2696. [Google Scholar] [CrossRef]
- Hamer, E.; Moore, C.; Denning, D. Comparison of two fluorescent whiteners, calcoflour and blankophor, for the detection of fungal elements in clinical specimens in the diagnostic laboratory. Clin. Microbiol. Infect. 2006, 12, 178–196. [Google Scholar] [CrossRef]
- Yao, Y.; Shi, L.; Zhang, C.; Sun, H.; Wu, L. Application of fungal fluorescent staining in oral candidiasis: Diagnostic analysis of 228 specimens. BMC Microbiol. 2019, 19, 1–5. [Google Scholar] [CrossRef]
- Kakoschke, T.K.; Kleinemeier, C.; Langenmayer, M.C.; Ebel, F. Tape mount immunostaining: A versatile method for immunofluorescence analysis of fungi. Future Microbiol. 2019, 14, 275–282. [Google Scholar] [CrossRef]
- Lindsley, M.D.; Snyder, J.W.; Atlas, R.M.; Larocco, M.T. Reagents, stains, and media: Mycology. In Manual of Clinical Microbiology; ASM Press: Washington, DC, USA, 2015; pp. 1955–1964. [Google Scholar]
- Bhavasar, R.; Goje, S.; Takalkar, A.; Ganvir, S.; Hazaray, V.; Gosavi, S. Detection of candida by calcoflour white. Acta Cytol. 2010, 54, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Schelenz, S.; Barnes, R.A.; Barton, R.C.; Cleverley, J.R.; Lucas, S.B.; Kibbler, C.C.; Denning, D.W. British society for medical mycology best practice recommendations for the diagnosis of serious fungal diseases. Lancet Infect. Dis. 2015, 15, 461–474. [Google Scholar] [CrossRef]
- Coovadia, Y.; Mahomed, S.; Dorasamy, A.; Chang, C. A comparative evaluation of the gram stain and India ink stain for the rapid diagnosis of cryptococcal meningitis in HIV infected patients in Durban. S. Afr. J. Infect. Dis. 2015, 30, 61–63. [Google Scholar] [CrossRef]
- Liu, L.; Yuan, Y.; Zhang, J.; Lei, H.; Wang, Q.; Liu, J.; Du, X.; Ni, G.; Liu, Y. Automatic identification of fungi under complex microscopic fecal images. J. Biomed. Opt. 2015, 20, 076004. [Google Scholar] [CrossRef] [PubMed]
- Abbott, J.J.; Hamacher, K.L.; Ahmed, I. In situ hybridization in cutaneous deep fungal infections: A valuable diagnostic adjunct to fungal morphology and tissue cultures. J. Cutan. Pathol. 2006, 33, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Guschin, A.; Tang, Q.; Dörffel, Y.; Verstraelen, H.; Tertychnyy, A.; Khayrullina, G.; Luo, X.; Sobel, J.D.; Jiang, X. Vulvovaginal candidiasis: Histologic lesions are primarily polymicrobial and invasive and do not contain biofilms. Am. J. Obstet. Gynecol. 2019, 220, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Harris, D.M.; Hata, D.J. Rapid identification of bacteria and candida using pna-fish from blood and peritoneal fluid cultures: A retrospective clinical study. Ann. Clin. Microbiol. Antimicrob. 2013, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Hall, L.; Le Febre, K.M.; Deml, S.M.; Wohlfiel, S.L.; Wengenack, N.L. Evaluation of the yeast traffic light PNA FISH probes for identification of Candida species from positive blood cultures. J. Clin. Microbiol. 2012, 50, 1446–1448. [Google Scholar] [CrossRef] [PubMed]
- Cooper, R.M.; Leslie, D.C.; Domansky, K.; Jain, A.; Yung, C.; Cho, M.; Workman, S.; Super, M.; Ingber, D.E. A microdevice for rapid optical detection of magnetically captured rare blood pathogens. Lab Chip 2014, 14, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Guarner, J.; Brandt, M.E. Histopathologic diagnosis of fungal infections in the 21st century. Clin. Microbiol. Rev. 2011, 24, 247–280. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Arora, S.K.; Rajwanshi, A.; Nijhawan, R.; Srinivasan, R. Histoplasmosis: Cytodiagnosis and review of literature with special emphasis on differential diagnosis on cytomorphology. Cytopathology 2010, 21, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Frickmann, H.; Loderstaedt, U.; Racz, P.; Tenner-Racz, K.; Eggert, P.; Haeupler, A.; Bialek, R.; Hagen, R.M. Detection of tropical fungi in formalin-fixed, paraffin-embedded tissue: Still an indication for microscopy in times of sequence-based diagnosis? Biomed. Res. Int. 2015, 2015, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, I.S.; Sanche, S.; Wiederhold, N.P.; Patterson, T.F.; Sigler, L. Emergomyces canadensis, a dimporphic fungus causing fatal systemic human diease in North America. Emerg. Infect. Dis. 2018, 24, 758–761. [Google Scholar] [CrossRef] [PubMed]
- Frater, J.L.; Hall, G.S.; Procop, G.W. Histologic features of zygomycosis: Emphasis on perineural invasion and fungal morphology. Arch. Pathol. Lab. Med. 2001, 125, 375–378. [Google Scholar] [PubMed]
- Cornely, O.A.; Arikan-Akdagli, S.; Dannaoui, E.; Groll, A.H.; Lagrou, K.; Chakrabarti, A.; Lanternier, F.; Pagano, L.; Skiada, A.; Akova, M.; et al. ESCMID and ECMM joint clinical guidelines for the diagnosis and management of mucormycosis 2013. Clin. Microbiol. Infect. 2014, 20, 5–26. [Google Scholar] [CrossRef] [PubMed]
- Challa, S.; Uppin, S.G.; Uppin, M.S.; Pamidimukkala, U.; Vemu, L. Diagnosis of filamentous fungi on tissue sections by immunohistochemistry using anti-aspergillus antibody. Med. Mycol. 2015, 53, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Park, Y.S.; Sung, H.; Song, J.S.; Lee, S.O.; Choi, S.H.; Kim, Y.S.; Woo, J.H.; Kim, S.H. Using immunohistochemistry to assess the accuracy of histomorphologic diagnosis of aspergillosis and mucormycosis. Clin. Infect. Dis. 2015, 61, 1664–1670. [Google Scholar]
- Schuetz, A.N.; Cohen, C. Aspergillus immunohistochemistry of culture-proven. Appl. Immunohistochem. Mol. Morphol. 2009, 17, 524–529. [Google Scholar] [CrossRef]
- Li, X.S.; Zhu, H.X.; Fan, H.X.; Zhu, L.; Wang, H.X.; Song, Y.L. Pulmonary fungal infections after bone marrow transplantation: The value of high-resolution computed tomography in predicting their etiology. Chin. Med. J. (Engl.) 2011, 124, 3249–3254. [Google Scholar]
- Legouge, C.; Caillot, D.; Chrétien, M.L.; Lafon, I.; Ferrant, E.; Audia, S.; Pagès, P.B.; Roques, M.; Estivalet, L.; Martin, L.; et al. The reversed halo sign: Pathognomonic pattern of pulmonary mucormycosis in leukemic patients with neutropenia? Clin. Infect. Dis. 2014, 58, 672–678. [Google Scholar] [CrossRef]
- Maturu, V.N.; Agarwal, R. Reversed halo sign: A systematic review. Respir. Care 2014, 59, 1440–1449. [Google Scholar] [CrossRef] [PubMed]
- Escuissato, D.L.; Gasparetto, E.L.; Marchiori, E.; De Melo Rocha, G.; Inoue, C.; Pasquini, R.; Müller, N.L. Pulmonary infections after bone marrow transplantation: High-resolution CT findings in 111 patients. Am. J. Roentgenol. 2005, 185, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Bruno, C.; Minniti, S.; Vassanelli, A.; Pozzi-Mucelli, R. Comparison of CT features of aspergillus and bacterial pneumonia in severely neutropenic patients. J. Thorac. Imaging 2007, 22, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Starkey, J.; Moritani, T.; Kirby, P. MRI of CNS fungal infections: Review of aspergillosis to histoplasmosis and everything in between. Clin. Neuroradiol. 2014, 24, 217–230. [Google Scholar] [CrossRef]
- Wine, Y.; Horton, A.; Ippolito, G.; Georgiou, G. Serology in the 21st century: The molecular-level analysis of the serum antibody repertoire. Curr. Opin. Microbiol. 2015, 35, 89–97. [Google Scholar] [CrossRef]
- Richardson, M.D.; Page, I.D. Aspergillus serology: Have we arrived yet? Med. Mycol. 2017, 55, 48–55. [Google Scholar] [CrossRef]
- Kamikawa, C.M.; Mendes, R.P.; Vicentini, A.P. Standardization and validation of Dot-ELISA assay for paracoccidioides brasiliensis antibody detection. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 1–7. [Google Scholar] [CrossRef]
- Heldt, S.; Hoenigl, M. Lateral flow assays for the diagnosis of invasive aspergillosis: Current status. Curr. Fungal Infect. Rep. 2017, 11, 45–51. [Google Scholar] [CrossRef]
- Meersseman, W.; Lagrou, K.; Maertens, J.; Wilmer, A.; Hermans, G.; Vanderschueren, S.; Spriet, I.; Verbeken, E.; Van Wijngaerden, E. Galactomannan in bronchoalveolar lavage fluid: A tool for diagnosing aspergillosis in intensive care unit patients. Am. J. Respir. Crit. Care Med. 2008, 177, 27–34. [Google Scholar] [CrossRef]
- Mikulska, M.; Calandra, T.; Sanguinetti, M.; Poulain, D.; Viscoli, C. The use of mannan antigen and anti-mannan antibodies in the diagnosis of invasive candidiasis: Recommendations from the third European conference on infections in Leukemia. Crit. Care 2010, 14, 1–14. [Google Scholar] [CrossRef]
- Karageorgopoulos, D.E.; Vouloumanou, E.K.; Ntziora, F.; Michalopoulos, A.; Rafailidis, P.I.; Falagas, M.E. β-D-glucan assay for the diagnosis of invasive fungal infections: A meta-analysis. Clin. Infect. Dis. 2011, 52, 750–770. [Google Scholar] [CrossRef] [PubMed]
- De Vlieger, G.; Lagrou, K.; Maertens, J.; Verbeken, E.; Meersseman, W.; Van Wijngaerden, E. Beta-D-glucan detection as a diagnostic test for invasive aspergillosis in immunocompromised critically ill patients with symptoms of respiratory infection: An autopsy-based study. J. Clin. Microbiol. 2011, 49, 3783–3787. [Google Scholar] [CrossRef] [PubMed]
- Amich, J.; Mokhtari, Z.; Strobel, M.; Vialetto, E.; Sheta, D.; Yu, Y.; Hartweg, J.; Kalleda, N.; Jarick, K.J.; Brede, C.; et al. Three-dimensional light sheet fluorescence microscopy of lungs to dissect local host immune-aspergillus fumigatus interactions. MBio 2020, 11, e02752-19. [Google Scholar] [CrossRef] [PubMed]
- Fukutomi, Y.; Tanimoto, H.; Yasueda, H.; Taniguchi, M. Serological diagnosis of allergic bronchopulmonary mycosis: Progress and challenges. Allergol. Int. 2016, 65, 30–36. [Google Scholar] [CrossRef]
- Farrell, P.M.; Govan, J.R.W. Pseudomonas seroloqy: Confusion, controversy, and challenges. Thorax 2006, 61, 645–647. [Google Scholar] [CrossRef][Green Version]
- Oliveira, L.; Almeida-Paes, R.; Pizzini, C.; Gutierrez-Galhardo, M.; Freitas, D.; Zancope-Oliveira, R. Diagnostic performance of mycologic and serologic methods in a cohort of patients with suspected sporotrichosis. Rev. Iberoam. Micol. 2019, 36, 61–65. [Google Scholar] [CrossRef]
- Singh, S.; Kaur, H.; Choudhary, H.; Sethi, S.; Malhotra, P.; Gupta, K.; Rudramurthy, S.; Chakrabarti, A. Evaluation of biomarkers: Galactomannan and 1,3-beta-D-glucan assay for the diagnosis of invasive fungal infections in immunocompromised patients from a tertiary care centre. Indian J. Med. Microbiol. 2018, 36, 557–563. [Google Scholar] [CrossRef]
- Wong, S.S.W.; Krylov, V.B.; Argunov, D.A.; Karelin, A.A.; Bouchara, J.-P.; Fontaine, T.; Latgé, J.-P.; Nifantiev, N.E. Potential of chemically synthesized oligosaccharides to define the carbohydrate moieties of the fungal cell wall responsible for the human immune response, using aspergillus fumigatus galactomannan as a model. mSphere 2020, 5, e00688-19. [Google Scholar] [CrossRef]
- Bassetti, M.; Giacobbe, D.R.; Grecchi, C.; Rebuffi, C.; Zuccaro, V.; Scudeller, L.; Akova, M.; Alastruey-Izquierdo, A.; Arikan-Akdagli, S.; Azoulay, E.; et al. Performance of existing definitions and tests for the diagnosis of invasive aspergillosis in critically ill, adult patients: A systematic review with qualitative evidence synthesis. J. Infect. 2020, 81, 131–146. [Google Scholar] [CrossRef]
- Safavieh, M.; Coarsey, C.; Esiobu, N.; Memic, A.; Vyas, J.M.; Shafiee, H.; Asghar, W. Advances in Candida detection platforms for clinical and point-of-care applications. Crit. Rev. Biotechnol. 2017, 37, 441–458. [Google Scholar] [CrossRef]
- Theel, E.S.; Doern, C.D. β-D-Glucan testing is important for diagnosis of invasive fungal infections. J. Clin. Microbiol. 2013, 51, 3478–3483. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R.; Latgé, J.P.; Munro, C.A. The fungal cell wall: Structure, biosynthesis, and function. In The Fungal Kingdom; Heitman, J., Gow, N.A.R., Howlett, B., Crous, P., Stukenbroch, E., James, T., Eds.; ASM Press: Washington, DC, USA, 2017; pp. 267–292. [Google Scholar]
- Odabasi, Z.; Paetznick, V.L.; Rodriguez, J.R.; Chen, E.; McGinnis, M.R.; Ostrosky-Zeichner, L. Differences in beta-glucan levels in culture supernatants of a variety of fungi. Med. Mycol. 2006, 44, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Morjaria, S.; Frame, J.; Franco-Garcia, A.; Geyer, A.; Kamboj, M.; Esther Babady, N. Clinical performance of (1,3) Beta-D glucan for the diagnosis of pneumocystis pneumonia (PCP) in cancer patients tested with PCP polymerase chain reaction. Clin. Infect. Dis. 2019, 69, 1303–1309. [Google Scholar] [CrossRef] [PubMed]
- Gerbst, A.G.; Grachev, A.A.; Yashunsky, D.V.; Tsvetkov, Y.E.; Shashkov, A.S.; Nifantiev, N.E. Theoretical and experimental conformational studies of oligoglucosides structurally related to fragments of fungal cell wall β-(1→3)-D-glucan. J. Carbohydr. Chem. 2013, 32, 205–221. [Google Scholar] [CrossRef]
- Matveev, A.L.; Krylov, V.B.; Khlusevich, Y.A.; Baykov, I.K.; Yashunsky, D.V.; Emelyanova, L.A.; Tsvetkov, Y.E.; Karelin, A.A.; Bardashova, A.V.; Wong, S.S.W.; et al. Novel mouse monoclonal antibodies specifically recognizing β-(1,3)-D-glucan antigen. PLoS ONE 2019, 14, e0215535. [Google Scholar] [CrossRef]
- Jaye, D.L.; Waites, K.B.; Parker, B.; Bragg, S.L.; Moser, S.A. Comparison of two rapid latex agglutination tests for detection of cryptococcal capsular polysaccharide. Am. J. Clin. Pathol. 1998, 109, 634–641. [Google Scholar] [CrossRef][Green Version]
- Lindsley, M.D.; Mekha, N.; Baggett, H.C.; Surinthong, Y.; Autthateinchai, R.; Sawatwong, P.; Harris, J.R.; Park, B.J.; Chiller, T.; Arunmozhi Balajee, S.; et al. Evaluation of a newly developed lateral flow immunoassay for the diagnosis of cryptococcosis. Clin. Infect. Dis. 2011, 53, 321–325. [Google Scholar] [CrossRef]
- Williams, D.A.; Kiiza, T.; Kwizera, R.; Kiggundu, R.; Velamakanni, S.; Meya, D.B.; Rhein, J.; Boulware, D.R. Evaluation of fingerstick cryptococcal antigen lateral flow assay in HIV-infected persons: A diagnostic accuracy study. Clin. Infect. Dis. 2015, 61, 464–467. [Google Scholar] [CrossRef]
- Peeling, R.W.; Holmes, K.K.; Mabey, D.; Ronald, A. Rapid tests for sexually transmitted infections (STIs): The way forward. Sex. Transm. Infect. 2006, 82, v1–v6. [Google Scholar] [CrossRef]
- Prattes, J.; Heldt, S.; Eigl, S.; Hoenigl, M. Point of care testing for the diagnosis of fungal infections: Are we there yet? Curr. Fungal Infect. Rep. 2016, 10, 43–50. [Google Scholar] [CrossRef]
- Thornton, C.R. Development of an immunochromatographic lateral-flow device for rapid serodiagnosis of invasive aspergillosis. Clin. Vaccine Immunol. 2008, 15, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- White, P.L.; Parr, C.; Thornton, C.; Barnes, R.A. Evaluation of real-time PCR, galactomannan enzyme-linked immunosorbent assay (ELISA), and a novel lateral-flow device for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 2013, 51, 1510–1516. [Google Scholar] [CrossRef] [PubMed]
- Takazono, T.; Ito, Y.; Tashiro, M.; Nishimura, K.; Saijo, T.; Yamamoto, K.; Imamura, Y.; Miyazaki, T.; Yanagihara, K.; Mukae, H.; et al. Evaluation of aspergillus-specific lateral-flow device test using serum and bronchoalveolar lavage fluid for diagnosis of chronic pulmonary aspergillosis. J. Clin. Microbiol. 2019, 57, e00095-19. [Google Scholar] [CrossRef]
- Dufresne, S.F.; Datta, K.; Li, X.; Dadachova, E.; Staab, J.F.; Patterson, T.F.; Feldmesser, M.; Marr, K.A. Detection of urinary excreted fungal galactomannan-like antigens for diagnosis of invasive aspergillosis. PLoS ONE 2012, 7, e42736. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Shoji, H.; Takuma, T.; Niki, Y. Clinical viability of Fungitell, a new (1-3)- β-D-Glucan measurement kit, fr diagnosis of invasive fungal infection, and comparison with other kits available in Japan. J. Infect. Chemother. 2011, 4, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Petinataud, D.; Berger, S.; Contet-audonneau, N.; Machouart, M. Molecular diagnosis of onychomycosis. J. Mycol. Med. 2014, 24, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Badiee, P.; Gandomi, B.; Sabz, G.; Khodami, B.; Choopanizadeh, M.; Jafarian, H. Evaluation of nested PCR in diagnosis of fungal rhinosinusitis. Iran. J. Microbiol. 2015, 7, 62–66. [Google Scholar]
- Wagner, K.; Springer, B.; Pires, V.P.; Keller, P.M. Molecular detection of fungal pathogens in clinical specimens by 18S rDNA high-throughput screening in comparison to ITS PCR and culture. Sci. Rep. 2018, 8, 1–7. [Google Scholar] [CrossRef]
- Lewis White, P.; Loeffler, J.; Barnes, R.A.; Peter Donnelly, J. Towards a standard for aspergillus PCR—Requirements, process and results. Infectio 2012, 16, 64–72. [Google Scholar] [CrossRef][Green Version]
- Hage, C.A.; Carmona, E.M.; Epelbaum, O.; Evans, S.E.; Gabe, L.M.; Haydour, Q.; Knox, K.S.; Kolls, J.K.; Murad, M.H.; Wengenack, N.L.; et al. Microbiological laboratory testing in the diagnosis of fungal infections in pulmonary and critical care practice: An official American thoracic society clinical practice guideline. Am. J. Respir. Crit. Care Med. 2019, 200, 535–550. [Google Scholar] [CrossRef]
- Gaviria, M.; Rivera, V.; Muñoz-Cadavid, C.; Cano, L.E.; Naranjo, T.W. Validation and clinical application of a nested PCR for paracoccidioidomycosis diagnosis in clinical samples from Colombian patients. Braz. J. Infect. Dis. 2015, 19, 376–383. [Google Scholar] [CrossRef] [PubMed]
- Haghani, I.; Amirinia, F.; Nowroozpoor Dailami, K.; Shokohi, T. Detection of fungi by conventional methods and semi-nested PCR in patients with presumed fungal keratitis. Curr. Med. Mycol. 2015, 1, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Rath, P.M.; Steinmann, J. Overview of commercially available PCR assays for the detection of Aspergillus spp. DNA in patient samples. Front. Microbiol. 2018, 9, 740. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Beer, K.; Toda, M. Azole-resistant aspergillus fumigatus: What you need to know. Clin. Microbiol. Newsl. 2020, 42, 1–6. [Google Scholar] [CrossRef]
- Sturaro, L.L.; Gonoi, T.; Busso-Lopes, A.F.; Tararam, C.A.; Levy, C.E.; Lyra, L.; Trabasso, P.; Schreiber, A.Z.; Kamei, K.; Moretti, M.L. Visible DNA microarray system as an adjunctive molecular test in identification of pathogenic fungi directly from a blood culture bottle. J. Clin. Microbiol. 2018, 56, 1–9. [Google Scholar] [CrossRef]
- Ricna, D.; Lengerova, M.; Bezdicek, M.; Kocmanova, I.; Drgona, L.; Weinbergerova, B.; Mayer, J.; Racil, Z. Detection and identification of fungi in bronchoalveolar lavage fluid from immunocompromised patients using panfungal PCR. Folia Microbiol. 2018, 64, 421–428. [Google Scholar] [CrossRef]
- Luo, G.; Mitchell, T.G. Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J. Clin. Microbiol. 2002, 40, 2860–2865. [Google Scholar] [CrossRef]
- Fuchs, S.; Lass-Flörl, C.; Posch, W. Diagnostic performance of a novel multiplex PCR assay for candidemia among ICU patients. J. Fungi 2019, 5, 86. [Google Scholar] [CrossRef]
- Scotter, J.M.; Campbell, P.; Anderson, T.P.; Murdoch, D.R.; Chambers, S.T.; Patton, W.N. Comparison of PCR-ELISA and galactomannan detection for the diagnosis of invasive aspergillosis. Pathology 2005, 37, 246–253. [Google Scholar] [CrossRef]
- Sue, M.J.; Yeap, S.K.; Omar, A.R.; Tan, S.W. Application of PCR-ELISA in molecular diagnosis. Biomed. Res. Int. 2014, 2014, 653014. [Google Scholar] [CrossRef]
- Salehi, E.; Hedayati, M.T.; Zoll, J.; Rafati, H.; Ghasemi, M.; Doroudinia, A.; Abastabar, M.; Tolooe, A.; Snelders, E.; Van Der Lee, H.A.; et al. Discrimination of aspergillosis, mucormycosis, fusariosis, and scedosporiosis in formalin-fixed paraffin-embedded tissue specimens by use of multiple real-time quantitative PCR assays. J. Clin. Microbiol. 2016, 54, 2798–2803. [Google Scholar] [CrossRef] [PubMed]
- Bidartondo, M. Preserving accuracy in GenBank. Science 2008, 319, 1616. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Clancy, C.J.; Ostrosky-Zeichner, L.; Garey, K.W.; Alangaden, G.J.; Vazquez, J.A.; Groeger, J.S.; Judson, M.A.; Vinagre, Y.M.; Heard, S.O.; et al. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: A clinical trial. Clin. Infect. Dis. 2015, 60, 892–899. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.F.; Drake, S.K.; Calhoun, L.B.; Henderson, C.M.; Zelazny, A.M. Development of a clinically comprehensive database and a simple procedure for identification of molds from solid media by matrix-assisted laser desorption ionization-Time of flight mass spectrometry. J. Clin. Microbiol. 2013, 51, 828–834. [Google Scholar] [CrossRef] [PubMed]
- Hendrickx, M. MALDI-TOF MS and filamentous fungal identification: A success story? Curr. Fungal Infect. Rep. 2017, 11, 60–65. [Google Scholar] [CrossRef]
- Becker, P.; Normand, A.C.; Vanantwerpen, G.; Vanrobaeys, M.; Haesendonck, R.; Vercammen, F.; Stubbe, D.; Piarroux, R.; Hendrickx, M. Identification of fungal isolates by MALDI-TOF mass spectrometry in veterinary practice: Validation of a web application. J. Vet. Diagn. Investig. 2019, 31, 471–474. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhang, Q.; Xu, C.; Shi, W. MALDI-TOF MS for the rapid identification and drug susceptibility testing of filamentous fungi. Exp. Ther. Med. 2019, 18, 4865–4873. [Google Scholar] [CrossRef]
- Barker, A.P.; Horan, J.L.; Susan Slechta, E.; Alexander, B.D.; Hanson, K.E. Complexities associated with the molecular and proteomic identification of Paecilomyces species in the clinical mycology laboratory. Med. Mycol. 2014, 52, 535–543. [Google Scholar] [CrossRef][Green Version]
- Triest, D.; Stubbe, D.; De Cremer, K.; Piérard, D.; Normand, A.C.; Piarroux, R.; Detandt, M.; Hendrickx, M. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of molds of the fusarium genus. J. Clin. Microbiol. 2015, 53, 465–476. [Google Scholar] [CrossRef]
- Chalupová, J.; Raus, M.; Sedlářová, M.; Šebela, M. Identification of fungal microorganisms by MALDI-TOF mass spectrometry. Biotechnol. Adv. 2014, 32, 230–241. [Google Scholar] [CrossRef]
- Sanguinetti, M.; Posteraro, B. Identification of mycobacteria by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. J. Clin. Microbiol. 2017, 55, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Normand, A.C.; Gabriel, F.; Riat, A.; Cassagne, C.; Bourgeois, N.; Huguenin, A.; Chauvin, P.; De Geyter, D.; Bexkens, M.; Rubio, E.; et al. Optimization of MALDI-ToF mass spectrometry for yeast identification: A multicenter study. Med. Mycol. 2020, 58, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Florio, W.; Tavanti, A.; Ghelardi, E.; Lupetti, A. MALDI-TOF MS applications to the detection of antifungal resistance: State of the art and future perspectives. Front. Microbiol. 2018, 9, 2577. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.F.; Walchak, R.C.; Miller, H.B.; Slechta, E.S.; Kamboj, K.; Riebe, K.; Robertson, A.E.; Gilbreath, J.J.; Mitchell, K.F.; Wallace, M.A.; et al. Multicenter study demonstrates standardization requirements for mold identification by MALDI-TOF MS. Front. Microbiol. 2019, 10, 2098. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; He, Y.; Maier, T.; Quinn, C.; Shi, G.; Li, H.; Stratton, C.W.; Kostrzewa, M.; Tang, Y.W. Improved identification of yeast species directly from positive blood culture media by combining sepsityper specimen processing and microflex analysis with the matrix-assisted laser desorption ionization biotyper system. J. Clin. Microbiol. 2011, 49, 2528–2532. [Google Scholar] [CrossRef] [PubMed]
- Spanu, T.; Posteraro, B.; Fiori, B.; D’Inzeo, T.; Campoli, S.; Ruggeri, A.; Tumbarello, M.; Canu, G.; Trecarichi, E.M.; Parisi, G.; et al. Direct MALDI-TOF mass spectrometry assay of blood culture broths for rapid identification of Candida species causing bloodstream infections: An observational study in two large microbiology laboratories. J. Clin. Microbiol. 2012, 50, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Yang, Q.; Kudinha, T.; Sun, L.; Zhang, R.; Liu, C.; Yu, S.; Xiao, M.; Kong, F.; Zhao, Y.; et al. An improved in-house MALDI-TOF MS protocol for direct cost-effective identification of pathogens from blood cultures. Front. Microbiol. 2017, 8, 1824. [Google Scholar] [CrossRef]
- Thvenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification (Technical Report). Pure Appl. Chem. 1999, 71, 2333–2348. [Google Scholar] [CrossRef]
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef]
- Cosnier, S. Electrochemical Biosensors, 1st ed.; Jenny Standford Publishing: Singapore, 2015; ISBN 9789814411479. [Google Scholar]
- Sahin, B.; Kaya, T. Electrochemical amperometric biosensor applications of nanostructured metal oxides: A review. Mater. Res. Express 2019, 6, 042003. [Google Scholar] [CrossRef]
- Hussain, K.K.; Gurudatt, N.G.; Akthar, M.H.; Seo, K.D.; Park, D.S.; Shim, Y.B. Nano-biosensor for the in vitro lactate detection using bi-functionalized conducting polymer/N, S-doped carbon; the effect of αCHC inhibitor on lactate level in cancer cell lines. Biosens. Bioelectron. 2020, 155, 112094. [Google Scholar] [CrossRef] [PubMed]
- Hussain, K.K.; Moon, J.M.; Park, D.S.; Shim, Y.B. Electrochemical detection of hemoglobin: A review. Electroanalysis 2017, 29, 1–11. [Google Scholar] [CrossRef]
- Ding, J.; Qin, W. Recent advances in potentiometric biosensors. TrAC Trends Anal. Chem. 2020, 124, 115803. [Google Scholar] [CrossRef]
- Kaisti, M. Detection principles of biological and chemical FET sensors. Biosens. Bioelectron. 2017, 98, 437–448. [Google Scholar] [CrossRef]
- Yoshinobu, T.; Miyamoto, K.I.; Werner, C.F.; Poghossian, A.; Wagner, T.; Schöning, M.J. Light-addressable potentiometric sensors for quantitative spatial imaging of chemical species. Annu. Rev. Anal. Chem. 2017, 10, 225–246. [Google Scholar] [CrossRef]
- Bahadir, E.B.; Sezgintürk, M.K. A review on impedimetric biosensors. Artif. Cells Nanomed. Biotechnol. 2016, 44, 248–262. [Google Scholar] [CrossRef]
- Kwasny, D.; Tehrani, S.E.; Almeida, C.; Schjødt, I.; Dimaki, M.; Svendsen, W.E. Direct detection of Candida albicans with a membrane based electrochemical impedance spectroscopy sensor. Sensors 2018, 18, 2214. [Google Scholar] [CrossRef]
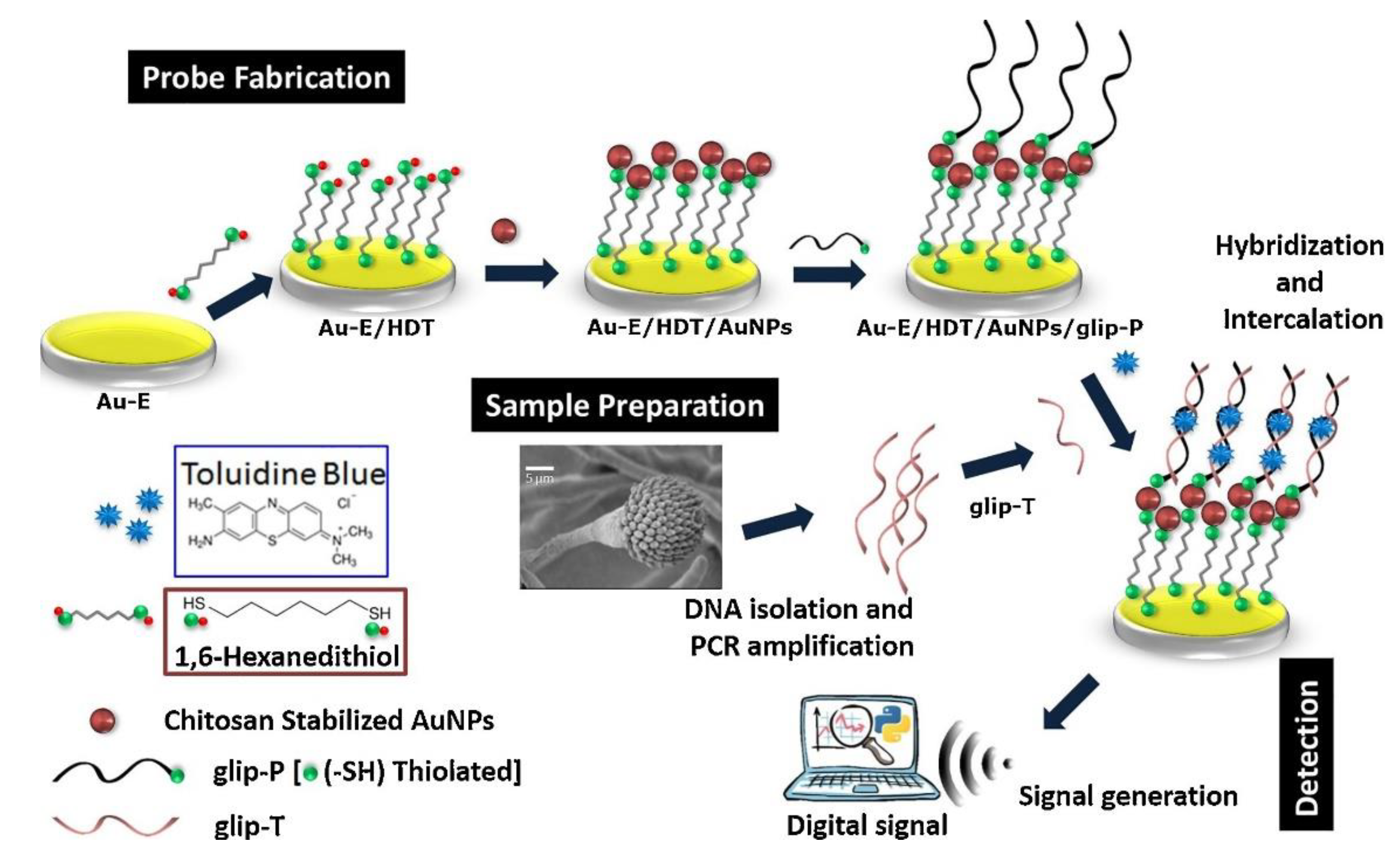
- Bhatnagar, I.; Mahato, K.; Ealla, K.K.R.; Asthana, A.; Chandra, P. Chitosan stabilized gold nanoparticle mediated self-assembled gliP nanobiosensor for diagnosis of invasive aspergillosis. Int. J. Biol. Macromol. 2018, 110, 449–456. [Google Scholar] [CrossRef]
- Damborský, P.; Švitel, J.; Katrlík, J. Optical biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar]
- Asghar, W.; Sher, M.; Khan, N.S.; Vyas, J.M.; Demirci, U. Microfluidic chip for detection of fungal infections. ACS Omega 2019, 4, 7474–7481. [Google Scholar] [CrossRef]
- Chen, C.; Wang, J. Optical biosensors: An exhaustive and comprehensive review. Analyst 2020, 145, 1605–1628. [Google Scholar] [CrossRef] [PubMed]
- Huertas, C.S.; Calvo-Lozano, O.; Mitchell, A.; Lechuga, L.M. Advanced evanescent-wave optical biosensors for the detection of nucleic acids: An analytic perspective. Front. Chem. 2019, 7, 724. [Google Scholar] [CrossRef] [PubMed]
- Altintas, Z. Optical biosensors and applications to drug discovery for cancer cases. In Biosensors and Nanotechnology: Applications in Health Care Diagnostics; Wiley: Hoboken, NJ, USA, 2018; ISBN 9781119065036. [Google Scholar]
- Cai, Z.; Kwak, D.H.; Punihaole, D.; Hong, Z.; Velankar, S.S.; Liu, X.; Asher, S.A. A photonic crystal protein hydrogel sensor for Candida albicans. Angew. Chemie Int. Ed. 2015, 55, 13036–13040. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; White, I.M.; Shopova, S.I.; Zhu, H.; Suter, J.D.; Sun, Y. Sensitive optical biosensors for unlabeled targets: A review. Anal. Chim. Acta 2008, 620, 8–26. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, F.; Deshui, Y. Optical Whispering Gallery Modes for Biosensing; Springer: Berlin, Germany, 2020. [Google Scholar]
- Vincent, S.; Subramanian, S.; Vollmer, F. Optoplasmonic characterisation of reversible disulfide interactions at single thiol sites in the attomolar regime. Nat. Commun. 2020, 11, 2043. [Google Scholar] [CrossRef] [PubMed]
- Pohanka, M. Overview of piezoelectric biosensors, immunosensors and DNA sensors and their applications. Materials 2018, 11, 448. [Google Scholar] [CrossRef]
- Chorsi, M.T.; Curry, E.J.; Chorsi, H.T.; Das, R.; Baroody, J.; Purohit, P.K.; Ilies, H.; Nguyen, T.D. Piezoelectric biomaterials for sensors and actuators. Adv. Mater. 2019, 31, 1–15. [Google Scholar] [CrossRef]
- Kopparthy, V.L.; Tangutooru, S.M.; Guilbeau, E.J. Label free detection of l-glutamate using microfluidic based thermal biosensor. Bioengineering 2015, 2, 1–14. [Google Scholar] [CrossRef]
- Jarvis, J.N.; Percival, A.; Bauman, S.; Pelfrey, J.; Meintjes, G.; Williams, G.N.; Longley, N.; Harrison, T.S.; Kozel, T.R. Evaluation of a novel point-of-care cryptococcal antigen test on serum, plasma, and urine from patients with HIV-associated cryptococcal meningitis. Clin. Infect. Dis. 2011, 53, 1019–1023. [Google Scholar] [CrossRef]
- Martins, J.F.S.; Castilho, M.L.; Cardoso, M.A.G.; Carreiro, A.P.; Martin, A.A.; Raniero, L. Identification of paracoccidioides brasiliensis by gold nanoprobes. In Proceedings of the Biomedical Vibrational Spectroscopy V: Advances in Research and Industry, San Francisco, CA, USA, 31 January 2012. [Google Scholar]
- De Oliveira Lima, E.; Navarro, L.C.; Morishita, K.N.; Kamikawa, C.M.; Rodrigues, R.G.M.; Dabaja, M.Z.; De Oliveira, D.N.; Delafiori, J.; Dias-Audibert, F.L.; da Silva Ribeiro, M.; et al. Metabolomics and machine learning approaches combined in pursuit for more accurate paracoccidioidomycosis diagnoses. mSystems 2020, 5, e00258. [Google Scholar]
- Rickerts, V.; Khot, P.D.; Myerson, D.; Ko, D.L.; Lambrecht, E.; Fredricks, D.N. Comparison of quantitative real time PCR with sequencing and ribosomal RNA-FISH for the identification of fungi in formalin fixed, paraffin-embedded tissue specimens. BMC Infect. Dis. 2011, 11, 202. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, R.M.; Da Silva Neto, J.R.; Santos, C.S.; Frickmann, H.; Poppert, S.; Cruz, K.S.; Koshikene, D.; De Souza, J.V.B. Evaluation of fluorescence in situ hybridisation (FISH) for the detection of fungi directly from blood cultures and cerebrospinal fluid from patients with suspected invasive mycoses. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, C.; Gicquel, A.; Sendid, B.; Meyer, J.; Accoceberry, I.; François, N.; Morio, F.; Desoubeaux, G.; Chandenier, J.; Kauffmann-Lacroix, C.; et al. Evaluation of two matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) systems for the identification of Candida species. Clin. Microbiol. Infect. 2014, 20, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Sutarlie, L.; Ow, S.Y.; Su, X. Nanomaterials-based biosensors for detection of microorganisms and microbial toxins. Biotechnol. J. 2017, 12, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Nayl, A.A.; Abd-Elhamid, A.I.; El-Moghazy, A.Y.; Hussin, M.; Abu-Saied, M.A.; El-Shanshory, A.A.; Soliman, H.M.A. The nanomaterials and recent progress in biosensing systems: A review. Trends Environ. Anal. Chem. 2020, 26, e00087. [Google Scholar] [CrossRef]
- Zhou, W.; Le, J.; Chen, Y.; Cai, Y.; Hong, Z.; Chai, Y. Recent advances in microfluidic devices for bacteria and fungus research. TrAC Trends Anal. Chem. 2019, 112, 175–195. [Google Scholar] [CrossRef]
- Busser, F.D.; Coelho, V.C.; de Abreu Fonseca, C.; Del Negro, G.M.B.; Shikanai-Yasuda, M.A.; Lopes, M.H.; Magri, M.M.C.; de Freitas, V.L.T. A real time PCR strategy for the detection and quantification of Candida albicans in human blood. Rev. Inst. Med. Trop. Sao Paulo 2020, 62, e9. [Google Scholar] [CrossRef]
- Rigby, S.; Procop, G.W.; Haase, G.; Wilson, D.; Hall, G.; Kurtzman, C.; Oliveira, K.; Von Oy, S.; Hyldig-Nielsen, J.J.; Coull, J.; et al. Fluorescence in situ hybridization with peptide nucleic acid probes for rapid identification of Candida albicans directly from blood culture bottles. J. Clin. Microbiol. 2002, 40, 2182–2186. [Google Scholar] [CrossRef]
- Yoo, S.M.; Kang, T.; Kang, H.; Lee, H.; Kang, M.; Lee, S.Y.; Kim, B. Combining a nanowire SERRS sensor and a target recycling reaction for ultrasensitive and multiplex identification of pathogenic fungi. Small 2011, 7, 3371–3376. [Google Scholar] [CrossRef]
- Naja, G.; Hrapovic, S.; Male, K.; Bouvrette, P.; Luong, J.H.T. Rapid detection of microorganisms with nanoparticles and electron microscopy. Microsc. Res. Tech. 2008, 71, 742–748. [Google Scholar] [CrossRef]
- Neely, L.A.; Audeh, M.; Phung, N.A.; Min, M.; Suchocki, A.; Plourde, D.; Blanco, M.; Demas, V.; Skewis, L.R.; Anagnostou, T.; et al. T2 magnetic resonance enables nanoparticle-mediated rapid detection of candidemia in whole blood. Sci. Transl. Med. 2013, 5, 182ra54. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.C.; Horn, T.; Zayats, M.; Rizk, G.; Major, S.; Zhu, H.; Russell, J.; Xu, Z.; Rothman, R.E.; Celedon, A. Ultra-sensitive and rapid detection of nucleic acids and microorganisms in body fluids using single-molecule tethering. Nat. Commun. 2020, 11, 4774. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.I.; Jang, S.C.; Chung, J.; Choi, W.K.; Hong, C.; Ahn, G.R.; Kim, S.H.; Lee, B.Y.; Chung, W.J. Colorimetric allergenic fungal spore detection using peptide-modified gold nanoparticles. Sens. Actuators B Chem. 2020, 327, 128894. [Google Scholar] [CrossRef]
- Villamizar, R.A.; Maroto, A.; Rius, F.X. Improved detection of Candida albicans with carbon nanotube field-effect transistors. Sens. Actuators B Chem. 2009, 136, 451–457. [Google Scholar] [CrossRef]
- Huppler, A.R.; Fisher, B.T.; Lehrnbecher, T.; Walsh, T.J.; Steinbach, W.J. Role of molecular biomarkers in the diagnosis of invasive fungal diseases in children. J. Pediatric Infect. Dis. Soc. 2017, 6, 32–44. [Google Scholar] [CrossRef]
- Erwig, L.P.; Gow, N.A.R. Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 2016, 14, 163–176. [Google Scholar] [CrossRef]
- Dambuza, I.; Wagener, J.; Brown, G.; Gow, N.A.R. Immunology of fungal diseases. In Oxford Textbook of Medical Mycology: A Handbook for Scientists and Clinicians; Kibbler, C.C., Gow, N.A.R., Barton, R.C., Howell, R., MacCallum, D.M., Eds.; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Vendele, I.; Willment, J.A.; Silva, L.M.; Palma, A.S.; Chai, W.; Liu, Y.; Feizi, T.; Spyrou, M.; Stappers, M.H.T.; Brown, G.D.; et al. Mannan detecting C-type lectin receptor probes recognise immune epitopes with diverse chemical, spatial and phylogenetic heterogeneity in fungal cell walls. PLoS Pathog. 2020, 16, e1007927. [Google Scholar] [CrossRef]
- Kerscher, B.; Willment, J.A.; Brown, G.D. The Dectin-2 family of C-type lectin-like receptors: An update. Int. Immunol. 2013, 25, 271–277. [Google Scholar] [CrossRef]
- Hussain, K.K.; Vollmer, F.; Gow, N.A.R. Sandwich fungal biosensor based on human receptor immobilized on to conductive polymer/AuZnO nanocomposite. in preparation.
- Kong, H.; Cheng, W.; Wei, H.; Yuan, Y.; Yang, Z.; Zhang, X. An overview of recent progress in siderophore-antibiotic conjugates. Eur. J. Med. Chem. 2019, 182, 271–277. [Google Scholar] [CrossRef]
- Chauhan, R.; Singh, J.; Sachdev, T.; Basu, T.; Malhotra, B.D. Recent advances in mycotoxins detection. Biosens. Bioelectron. 2016, 81, 532–545. [Google Scholar] [CrossRef]
- Vidal, A.; Mengelers, M.; Yang, S.; De Saeger, S.; De Boevre, M. Mycotoxin biomarkers of exposure: A comprehensive review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1127–1155. [Google Scholar] [CrossRef]
- Goud, K.Y.; Reddy, K.K.; Satyanarayana, M.; Kummari, S.; Gobi, K.V. A review on recent developments in optical and electrochemical aptamer-based assays for mycotoxins using advanced nanomaterials. Microchim. Acta 2020, 187, 29. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, W.; Hong, T. Novel approaches for biomolecule immobilization in microscale systems. Analyst 2019. [Google Scholar] [CrossRef] [PubMed]
- Mohamad, N.R.; Marzuki, N.H.C.; Buang, N.A.; Huyop, F.; Wahab, R.A. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 2015, 29, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.H.; Kim, M. An overview of techniques in enzyme immobilization. Appl. Sci. Converg. Technol. 2017, 26, 157–163. [Google Scholar] [CrossRef]
- Dwevedi, A. Basics of enzyme immobilization. In Enzyme Immobilization; Springer Nature: Cham, Switzerland, 2016. [Google Scholar]
- Odabasi, Z.; Mattiuzzi, G.; Estey, E.; Kantarjian, H.; Saeki, F.; Ridge, R.J.; Ketchum, P.A.; Finkelman, M.A.; Rex, J.H.; Ostrosky-Zeichner, L. β-D-glucan as a diagnostic adjunct for invasive fungal infections: Validation, cutoff development, and performance in patients with acute myelogenous leukemia and myelodysplastic syndrome. Clin. Infect. Dis. 2004, 39, 199–205. [Google Scholar] [CrossRef]


| Type of Fungus | Assay Time | LOD or Sensitivity | References |
|---|---|---|---|
| Aspergillus spp. | 15 min | 37 ng/mL | [78,134] |
| Cryptococcus neoformans | 10 min | >5 ng/mL | [137] |
| Cryptococcus spp. | 5–15 min | 100% (serum) 70.7–92% (urine) | [72] |
| Fungal Species | Assay Time | Material | Sample | LOD or Sensitivity | Reference |
|---|---|---|---|---|---|
| Paracoccidioides brasiliensis | - | Au-NPs | Fungal DNA | >4 mg mL | [138] |
| Candida albicans | 1 h | CNT | Fungal solution | 50 CFU/mL | [153] |
| Candida spp. | 30 min | Au-NPs | Waste water effluent | - | [149] |
| Candida albicans | 2.5 h | PNA | Blood culture | 100% | [147] |
| A. fumigatus, C. glabrata, C. krusei, Cryptococcus neoformans | A few hours | Au-nanowire | Fungal DNA | 100 fM | [148] |
| Candida spp. | <3 h | Nanoparticles | Whole blood | 1 CFU/mL | [150] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
K. Hussain, K.; Malavia, D.; M. Johnson, E.; Littlechild, J.; Winlove, C.P.; Vollmer, F.; Gow, N.A.R. Biosensors and Diagnostics for Fungal Detection. J. Fungi 2020, 6, 349. https://doi.org/10.3390/jof6040349
K. Hussain K, Malavia D, M. Johnson E, Littlechild J, Winlove CP, Vollmer F, Gow NAR. Biosensors and Diagnostics for Fungal Detection. Journal of Fungi. 2020; 6(4):349. https://doi.org/10.3390/jof6040349
Chicago/Turabian StyleK. Hussain, Khalil, Dhara Malavia, Elizabeth M. Johnson, Jennifer Littlechild, C. Peter Winlove, Frank Vollmer, and Neil A. R. Gow. 2020. "Biosensors and Diagnostics for Fungal Detection" Journal of Fungi 6, no. 4: 349. https://doi.org/10.3390/jof6040349
APA StyleK. Hussain, K., Malavia, D., M. Johnson, E., Littlechild, J., Winlove, C. P., Vollmer, F., & Gow, N. A. R. (2020). Biosensors and Diagnostics for Fungal Detection. Journal of Fungi, 6(4), 349. https://doi.org/10.3390/jof6040349







