Interacting with Hemoglobin: Paracoccidioides spp. Recruits hsp30 on Its Cell Surface for Enhanced Ability to Use This Iron Source
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Strains and Growth Conditions
2.3. Extraction of P. Lutzii Cell Wall Proteins (CWPs)
2.4. Preparation of Complex Samples for NanoUPLC-MSE
2.5. Data Acquisition by NanoUPLC-MSE
2.6. Spectra Processing and Proteomic Analysis
2.7. Expression of the Recombinant HSP30 Protein in Escherichia Coli, Protein Purification, and Polyclonal Antibodies
2.8. Far-Western Blot Analyses
2.9. Flow Cytometry and Immunofluorescence Assays
2.10. Structural Alignment of HSP30 and Human Heme Oxygenase
2.11. Preparation of Three-Dimensional (3D) Structures and Molecular Docking
2.12. Molecular Dynamics Simulations
2.13. Construction of P. Brasiliensis HSP30 Antisense (AsHSP30) Strain
2.14. Characterization of the Knockdown Strain
3. Results
3.1. Hemoglobin Promotes Changes at the P. Lutzii Cell Wall Proteome
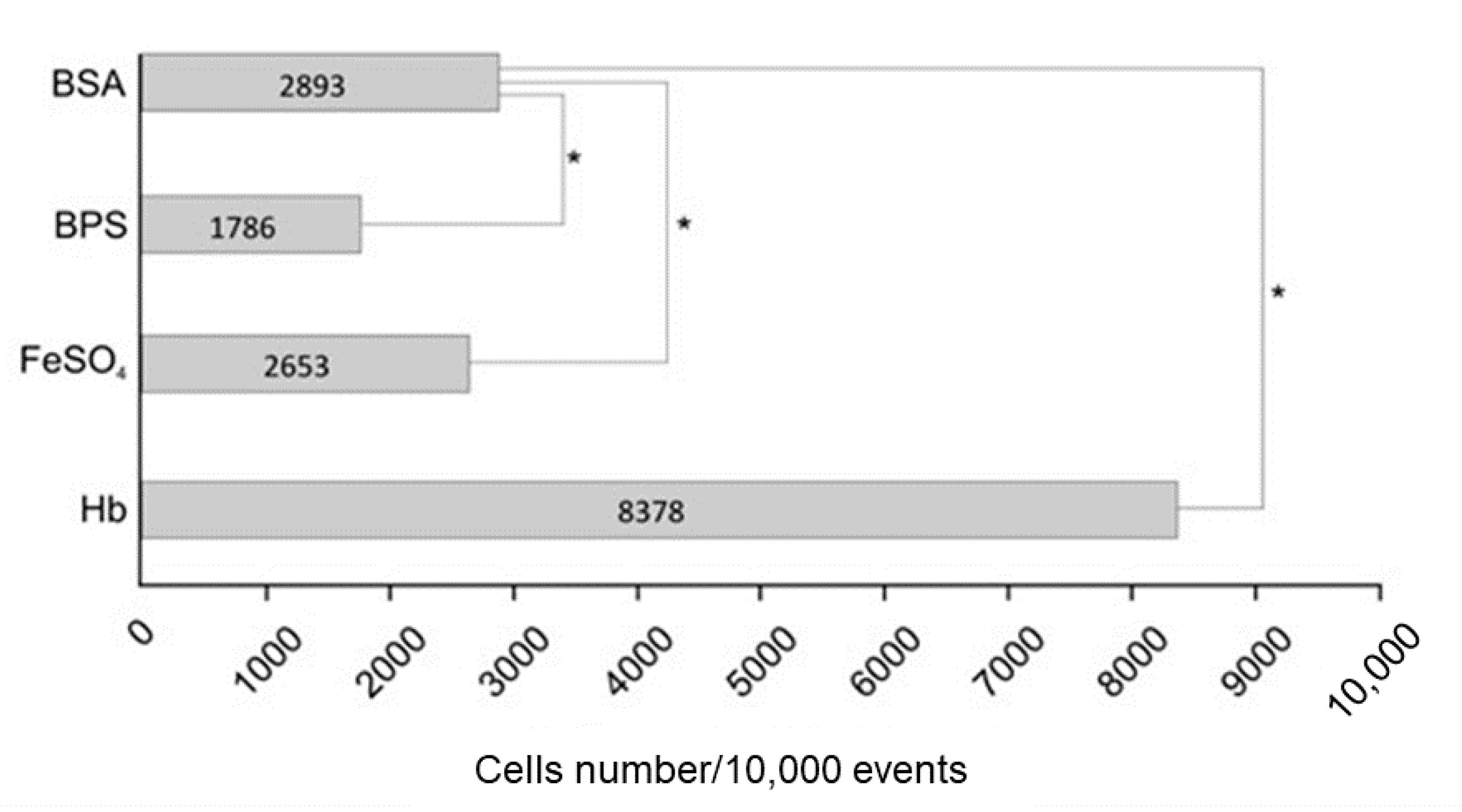
3.2. Hemoglobin Increases HSP30 Expression at Cell Surface
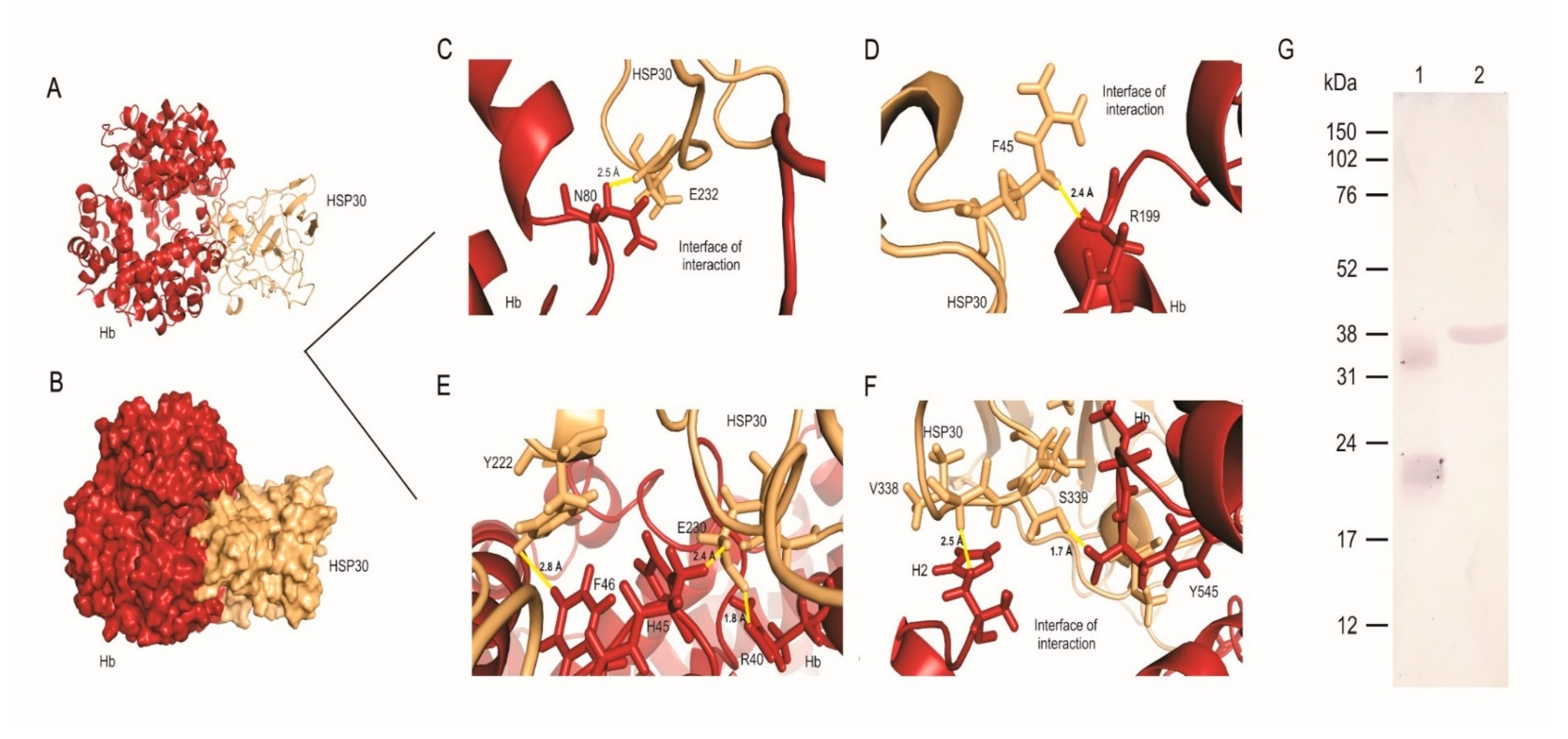
3.3. HSP30 Binds to Hemoglobin at the Fungus Cell Wall by Hydrogen Bonds
3.4. Knockdown of HSP30 Promotes Decreased Cell Growth Post Cultivation in Medium Containing Hemoglobin as Sole Iron Source
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nairz, M.; Schroll, A.; Sonnweber, T.; Weiss, G. The struggle for iron—A metal at the host-pathogen interface. Cell. Microbiol. 2010, 12, 1691–1702. [Google Scholar] [CrossRef] [PubMed]
- Núñez, G.; Sakamoto, K.; Soares, M.P. Innate nutritional immunity. J. Immunol. 2018, 201, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Iron and infection. Int. J. Hematol. 2018, 107, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Nevitt, T. War-Fe-re: Iron at the core of fungal virulence and host immunity. Biometals 2011, 24, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Bairwa, G.; Jung, W.H.; Kronstad, J.W. Iron acquisition in fungal pathogens of humans. Metallomics 2017, 9, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Bailão, E.F.L.C.; Parente, A.F.A.; Parente, J.A.; Silva-Bailão, M.G.; de Castro, K.P.; Kmetzsch, L.; Staats, C.C.; Schrank, A.; Vainstein, M.H.; Borges, C.L.; et al. Metal acquisition and homeostasis in fungi. Curr. Fungal Infect. Rep. 2012, 6, 257–266. [Google Scholar] [CrossRef]
- Caza, M.; Kronstad, J.W. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front. Cell. Infect. Microbiol. 2013, 3, 1–23. [Google Scholar] [CrossRef]
- Roy, U.; Kornitzer, D. Heme-iron acquisition in fungi. Curr. Opin. Microbiol. 2019, 52, 77–83. [Google Scholar] [CrossRef]
- Weissman, Z.; Shemer, R.; Conibear, E.; Kornitzer, D. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol. Microbiol. 2008, 69, 201–217. [Google Scholar] [CrossRef]
- Kuznets, G.; Vigonsky, E.; Weissman, Z.; Lalli, D.; Gildor, T.; Kauffman, S.J.; Turano, P.; Becker, J.; Lewinson, O.; Kornitzer, D. A relay network of extracellular heme-binding proteins drives C. albicans iron acquisition from hemoglobin. PLoS Pathog. 2014, 10, 1–15. [Google Scholar] [CrossRef]
- Weissman, Z.; Kornitzer, D. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol. Microbiol. 2004, 53, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Okamoto-Shibayama, K.; Kikuchi, Y.; Kokubu, E.; Sato, Y.; Ishihara, K. Csa2, a member of the Rbt5 protein family, is involved in the utilization of iron from human hemoglobin during Candida albicans hyphal growth. FEMS Yeast Res. 2014, 14, 674–677. [Google Scholar] [CrossRef] [PubMed]
- Nasser, L.; Weissman, Z.; Pinsky, M.; Amartely, H.; Dvir, H.; Kornitzer, D. Structural basis of haem-iron acquisition by fungal pathogens. Nat. Microbiol. 2016, 1, 16156. [Google Scholar] [CrossRef] [PubMed]
- Cadieux, B.; Lian, T.; Hu, G.; Wang, J.; Biondo, C.; Teti, G.; Liu, V.; Murphy, M.E.P.; Creagh, A.L.; Kronstad, J.W. The mannoprotein cig1 supports iron acquisition from heme and virulence in the pathogenic fungus Cryptococcus neoformans. J. Infect. Dis. 2013, 207, 1339–1347. [Google Scholar] [CrossRef]
- Hu, G.; Caza, M.; Cadieux, B.; Chan, V.; Liu, V.; Kronstad, J. Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence. Infect. Immun. 2013, 81, 292–302. [Google Scholar] [CrossRef]
- Hu, G.; Caza, M.; Cadieux, B.; Bakkeren, E.; Do, E.; Jung, W.H.; Kronstad, J.W. The endosomal sorting complex required for transport machinery influences haem uptake and capsule elaboration in Cryptococcus neoformans. Mol. Microbiol. 2015, 96, 973–992. [Google Scholar] [CrossRef]
- Bairwa, G.; Caza, M.; Horianopoulos, L.; Hu, G.; Kronstad, J. Role of clathrin-mediated endocytosis in the use of heme and hemoglobin by the fungal pathogen Cryptococcus neoformans. Cell. Microbiol. 2019, 21, 1–42. [Google Scholar] [CrossRef]
- Rappleye, C.A.; Goldman, W.E. Defining virulence genes in the dimorphic fungi. Annu. Rev. Microbiol. 2006, 60, 281–303. [Google Scholar] [CrossRef]
- Brummer, E.; Castaneda, E.; Restrepo, A. Paracoccidioidomycosis: An update. Clin. Microbiol. Rev. 1993, 6, 89–117. [Google Scholar] [CrossRef]
- San-blas, G.; Niño-Vega, G.; Iturriaga, T. Paracoccidioides brasiliensis and paracoccidioidomycosis: Molecular approaches to morphogenesis, diagnosis, epidemiology, taxonomy and genetics. Med. Mycol. 2002, 40, 225–242. [Google Scholar] [CrossRef]
- Teixeira, M.M.; Theodoro, R.C.; de Carvalho, M.J.A.; Fernandes, L.; Paes, H.C.; Hahn, R.C.; Mendoza, L.; Bagagli, E.; San-Blas, G.; Felipe, M.S.S. Phylogenetic analysis reveals a high level of speciation in the Paracoccidioides genus. Mol. Phylogenet. Evol. 2009, 52, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Turissini, D.A.; Gomez, O.M.; Teixeira, M.M.; Mcewen, J.G.; Matute, D.R. Species boundaries in the human pathogen Paracoccidioides. Fungal Genet. Biol. 2017, 106, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Restrepo, A.; McEwen, J.G.; Castañeda, E. The habitat of Paracoccidioides brasiliensis: How far from solving the riddle? Med. Mycol. 2001, 39, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Parente, A.F.A.; Bailão, A.M.; Borges, C.L.; Parente, J.A.; Magalhães, A.D.; Ricart, C.A.O.; Soares, C.M.A. Proteomic analysis reveals that iron availability alters the metabolic status of the pathogenic fungus Paracoccidioides brasiliensis. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Silva-Bailão, M.G.; Bailão, E.F.L.C.; Lechner, B.E.; Gauthier, G.M.; Lindner, H.; Bailão, A.M.; Haas, H.; de Almeida Soares, C.M. Hydroxamate production as a high affinity iron acquisition mechanism in Paracoccidioides spp. PLoS ONE 2014, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bailão, E.F.L.C.; de Sousa Lima, P.; Silva-Bailão, M.G.; Bailão, A.M.; da Rocha Fernandes, G.; Kosman, D.J.; de Almeida Soares, C.M. Paracoccidioides spp. ferrous and ferric iron assimilation pathways. Front. Microbiol. 2015, 6, 1–12. [Google Scholar] [CrossRef]
- Bailão, E.F.L.C.; Parente, J.A.; Pigosso, L.L.; de Castro, K.P.; Fonseca, F.L.; Silva-Bailão, M.G.; Báo, S.N.; Bailão, A.M.; Rodrigues, M.L.; Hernandez, O.; et al. Hemoglobin uptake by Paracoccidioides spp. is receptor-mediated. PLoS Negl. Trop. Dis. 2014, 8, 1–20. [Google Scholar] [CrossRef]
- Araújo, D.S.; de Sousa Lima, P.; Baeza, L.C.; Parente, A.F.A.; Bailão, A.B.; Borges, C.L.; de Almeida Soares, C.M. Employing proteomic analysis to compare Paracoccidioides lutzii yeast and mycelium cell wall proteins. BBA Proteins Proteom. 2017, 1865, 1304–1314. [Google Scholar] [CrossRef]
- Borges, C.L.; Pereira, M.; Felipe, M.S.S.; de Faria, F.P.; Gomez, F.J.; Deepe, G.S.; Soares, C.M.A. The antigenic and catalytically active formamidase of Paracoccidioides brasiliensis: Protein characterization, cDNA and gene cloning, heterologous expression and functional analysis of the recombinant protein. Microbes Infect. 2005, 7, 66–77. [Google Scholar] [CrossRef]
- Pereira, L.A.; Báo, S.N.; Barbosa, M.S.; da Silva, M.J.L.; Felipe, M.S.S.; Santana, J.M.; Mendes-Giannini, M.J.S.; de Almeida Soares, C.M. Analysis of the Paracoccidioides brasiliensis triosephosphate isomerase suggests the potential for adhesin function. FEMS Yeast Res. 2007, 7, 1381–1388. [Google Scholar] [CrossRef]
- Nogueira, S.V.; Fonseca, F.L.; Rodrigues, M.L.; Mundodi, V.; Abi-Chacra, E.A.; Winters, M.S.; Alderete, J.F.; de Almeida Soares, C.M. Paracoccidioides brasiliensis enolase is a surface protein that binds plasminogen and mediates interaction of yeast forms with host cells. Infect. Immun. 2010, 78, 4040–4050. [Google Scholar] [CrossRef] [PubMed]
- Bailão, A.M.; Nogueira, S.V.; Bonfim, S.M.R.C.; de Castro, K.P.; da Silva, J.D.F.; Mendes Giannini, M.J.S.; Pereira, M.; de Almeida Soares, C.M. Comparative transcriptome analysis of Paracoccidioides brasiliensis during in vitro adhesion to type I collagen and fibronectin: Identification of potential adhesins. Res. Microbiol. 2012, 163, 182–191. [Google Scholar] [CrossRef] [PubMed]
- De Sousa Lima, P.; Bailão, E.F.L.C.; Silva, M.G.; Castro, N.D.S.; Báo, S.N.; Orlandi, I.; Vai, M.; de Almeida Soares, C.M. Characterization of the Paracoccidioides beta-1,3-glucanosyltransferase family. FEMS Yeast Res. 2012, 12, 685–702. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tomazett, M.V.; Baeza, L.C.; Paccez, J.D.; Parente-Rocha, J.A.; Ribeiro-Dias, F.; de Almeida Soares, C.M. Identification and characterization of Paracoccidioides lutzii proteins interacting with macrophages. Microbes Infect. 2019, 1–11. [Google Scholar] [CrossRef]
- Restrepo, A.; Jiménez, B.E. Growth of Paracoccidioides brasiliensis yeast phase in a chemically defined culture medium. J. Clin. Microbiol. 1980, 12, 279–281. [Google Scholar] [CrossRef]
- Pitarch, A.; Nombela, C.; Gil, C. Cell wall fractionation for yeast and fungal proteomics. Methods Mol. Biol. 2008, 425, 217–239. [Google Scholar] [CrossRef]
- Geromanos, S.J.; Vissers, J.P.C.; Silva, J.C.; Dorschel, C.A.; Li, G.Z.; Gorenstein, M.V.; Bateman, R.H.; Langridge, J.I. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics 2009, 9, 1683–1695. [Google Scholar] [CrossRef]
- Chaves, E.A.C.; Weber, S.S.; Báo, S.N.; Pereira, L.A.; Bailão, A.B.; Borges, C.L.; de Almeida Soares, C.M. Analysis of Paracoccidioides secreted proteins reveals fructose 1,6-bisphosphate aldolase as a plasminogen-binding protein. BMC Microbiol. 2015, 15, 1–14. [Google Scholar] [CrossRef]
- D’Agostino, R.B.; Chase, W.; Belanger, A. The appropriateness of some common procedures for testing the equality of two independent binomial populations. Am. Stat. 1988, 42, 198–202. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Mitchell, J.C. KFC2: A knowledge-based hot spot prediction method based on interface solvation, atomic density, and plasticity features. Proteins Struct. Funct. Bioinform. 2011, 79, 2671–2683. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef]
- Eichenberger, A.P.; Allison, J.R.; Dolenc, J.; Geerke, D.P.; Horta, B.A.C.; Meier, K.; Oostenbrink, C.; Schmid, N.; Steiner, D.; Wang, D.; et al. GROMOS++ software for the analysis of biomolecular simulation trajectories. J. Chem. Theory Comput. 2011, 7, 3379–3390. [Google Scholar] [CrossRef]
- Parente-rocha, J.A.; Flávia, A.; Parente, A.; Baeza, L.C.; Bail, M.; Taborda, C.P.; Borges, C.L.; De, C.M. Macrophage interaction with Paracoccidioides brasiliensis yeast cells modulates fungal metabolism and generates a response to oxidative stress. PLoS ONE 2015, 1–18. [Google Scholar] [CrossRef]
- Silva, M.G.; de Curcio, J.S.; Silva-Bailão, M.G.; Lima, R.M.; Tomazett, M.V.; de Souza, A.F.; Cruz-Leite, V.R.M.; Sbaraini, N.; Bailão, A.M.; Rodrigues, F.; et al. Molecular characterization of siderophore biosynthesis in Paracoccidioides brasiliensis. IMA Fungus 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Rappleye, C.A.; Engle, J.T.; Goldman, W.E. RNA interference in Histoplasma capsulatum demonstrates a role for α-(1,3)-glucan in virulence. Mol. Microbiol. 2004, 53, 153–165. [Google Scholar] [CrossRef]
- De Groot, M.J.A.; Bundock, P.; Hooykaas, P.J.J.; Beijersbergen, A.G.M. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 1998, 16, 839–842. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Dias-Melicio, L.A.; Calvi, S.A.; Peraçoli, M.T.S.; Soares, Â.M.V.D.C. Inhibitory effect of deferoxamine on Paracoccidioides brasiliensis survival in human monocytes: Reversal by holotransferrin not by apotransferrin. Rev. Inst. Med. Trop. Sao Paulo 2005, 47, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Dias-melicio, L.A.; Moreira, A.P.; Calvi, S.A.; Maria, A.; De Campos, V. Chloroquine inhibits Paracoccidioides brasiliensis survival within human monocytes by limiting the availability of intracellular iron. Microbiol. Immunol. 2006, 50, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Sorgo, A.G.; Brul, S.; de Koster, C.G.; de Koning, L.J.; Klis, F.M. Iron restriction-induced adaptations in the wall proteome of Candida albicans. Microbiology 2013, 159, 1673–1682. [Google Scholar] [CrossRef]
- Pitarch, A.; Sánchez, M.; Nombela, C.; Gil, C. Sequential fractionation and two-dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol. Cell. Proteom. 2002, 1, 967–982. [Google Scholar] [CrossRef]
- Longo, L.V.G.; da Cunha, J.P.C.; Sobreira, T.J.P.; Puccia, R. Proteome of cell wall-extracts from pathogenic Paracoccidioides brasiliensis: Comparison among morphological phases, isolates, and reported fungal extracellular vesicle proteins. EuPA Open Proteom. 2014, 3, 216–228. [Google Scholar] [CrossRef]
- Nombela, C.; Gil, C.; Chaffin, W.L. Non-conventional protein secretion in yeast. Trends Microbiol. 2006, 14, 15–21. [Google Scholar] [CrossRef]
- Karkowska-Kuleta, J.; Kozik, A. Cell wall proteome of pathogenic fungi. Acta Biochim. Pol. 2015, 62, 339–351. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Jensen, L.J.; Blom, N.; von Heijne, G.; Brunak, S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 2004, 17, 349–356. [Google Scholar] [CrossRef]
- Marcos, C.M.; de Oliveira, H.C.; da Silva, J.D.F.; Assato, P.A.; Yamazaki, D.S.; da Silva, R.A.M.; Santos, C.T.; Santos-Filho, N.A.; Portuondo, D.L.; Mendes-Giannini, M.J.S.; et al. Identification and characterisation of elongation factor Tu, a novel protein involved in Paracoccidioides brasiliensis-host interaction. FEMS Yeast Res. 2016, 16. [Google Scholar] [CrossRef]
- Gonzalez, A.; Hernandez, O. New insights into a complex fungal pathogen: The case of Paracoccidioides spp. Yeast 2016, 33, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Ewing, J.F.; Maines, M.D. Histochemical localization of heme oxygenase-2 protein and mRNA expression in rat brain. Brain Res. Protoc. 1997, 1, 165–174. [Google Scholar] [CrossRef]
- Bidmon, H.J.; Emde, B.; Oermann, E.; Kubitz, R.; Witte, O.W.; Zilles, K. Heme oxygenase-1 (HSP-32) and heme oxygenase-2 induction in neurons and glial cells of cerebral regions and its relation to iron accumulation after focal cortical photothrombosis. Exp. Neurol. 2001, 168, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Maines, M.D.; Trakshel, G.M.; Kutty, R.K. Characterization of two constitutive forms of rat liver microsomal heme oxygenase. J. Biol. Chem. 1986, 261, 411–419. [Google Scholar]
- Maines, M.D. Heme oxygenase: Function, multiplicity, regulatory mechanisms, and clinical applications. FASEB J. 1988, 2, 2557–2568. [Google Scholar] [CrossRef]
- Lathrop, J.T.; Timko, M.P. Regulation by heme of mitochondrial protein transport through a conserved amino acid motif. Science 1993, 259, 522–525. [Google Scholar] [CrossRef]
- Zhang, L.; Guarente, L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 1995, 14, 313–320. [Google Scholar] [CrossRef]
- Brewitz, H.H.; Goradia, N.; Schubert, E.; Galler, K.; Kühl, T.; Syllwasschy, B.; Popp, J.; Neugebauer, U.; Hagelueken, G.; Schiemann, O.; et al. Heme interacts with histidine- and tyrosine-based protein motifs and inhibits enzymatic activity of chloramphenicol acetyltransferase from Escherichia coli. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 1343–1353. [Google Scholar] [CrossRef]
- Almeida, A.J.; Carmona, J.A.; Cunha, C.; Carvalho, A.; Rappleye, C.A.; Goldman, W.E.; Hooykaas, P.J.; Leão, C.; Ludovico, P.; Rodrigues, F. Towards a molecular genetic system for the pathogenic fungus Paracoccidioides brasiliensis. Fungal Genet. Biol. 2007, 44, 1387–1398. [Google Scholar] [CrossRef]
- Almeida, A.J.; Cunha, C.; Carmona, J.A.; Sampaio-Marques, B.; Carvalho, A.; Malavazi, I.; Steensma, H.Y.; Johnson, D.I.; Leão, C.; Logarinho, E.; et al. Cdc42p controls yeast-cell shape and virulence of Paracoccidioides brasiliensis. Fungal Genet. Biol. 2009, 46, 919–926. [Google Scholar] [CrossRef]
- Menino, J.F.; Saraiva, M.; Gomes-Rezende, J.; Sturme, M.; Pedrosa, J.; Castro, A.G.; Ludovico, P.; Goldman, G.H.; Rodrigues, F.P. brasiliensis Virulence is affected by SconC, the negative regulator of inorganic sulfur assimilation. PLoS ONE 2013, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Torres, I.; Hernandez, O.; Tamayo, D.; Muñoz, J.F.; García, A.M.; Gómez, B.L.; Restrepo, A.; McEwen, J.G. Paracoccidioides brasiliensis PbP27 gene: Knockdown procedures and functional characterization. FEMS Yeast Res. 2014, 14, 270–280. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Torres, I.; Hernandez, O.; Tamayo, D.; Muñoz, J.F.; Leitão, N.P.; García, A.M.; Restrepo, A.; Puccia, R.; McEwen, J.G. Inhibition of PbGP43 expression may suggest that gp43 is a virulence factor in Paracoccidioides brasiliensis. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Tamayo, D.; Muñoz, J.F.; Torres, I.; Almeida, A.J.; Restrepo, A.; McEwen, J.G.; Hernández, O. Involvement of the 90kDa heat shock protein during adaptation of Paracoccidioides brasiliensis to different environmental conditions. Fungal Genet. Biol. 2013, 51, 34–41. [Google Scholar] [CrossRef]
- Goes, T.; Bailão, E.F.L.C.; Correa, C.R.; Bozzi, A.; Santos, L.I.; Gomes, D.A.; Soares, C.M.A.; Goes, A.M. New developments of RNAi in Paracoccidioides brasiliensis: Prospects for high-throughput, genome-wide, functional genomics. PLoS Negl. Trop. Dis. 2014, 8. [Google Scholar] [CrossRef]
- Fernandes, F.F.; Oliveira, A.F.; Landgraf, T.N.; Cunha, C.; Carvalho, A.; Vendruscolo, P.E.; Gonçales, R.A.; Almeida, F.; da Silva, T.A.; Rodrigues, F.; et al. Impact of paracoccin gene silencing on Paracoccidioides brasiliensis virulence. MBio 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Nora, L.C.; Gonçales, R.A.; Martins-Santana, L.; Ferreira, B.H.; Rodrigues, F.; Silva-Rocha, R. Synthetic and minimalist vectors for Agrobacterium tumefaciens-mediated transformation of fungi. Genet. Mol. Biol. 2019, 42, 395–398. [Google Scholar] [CrossRef]
- Oliveira, L.N.; Gonçales, R.A.; Silva, M.G.; Lima, R.M.; Tomazett, M.V.; de Curcio, J.S.; Paccez, J.D.; Cruz-Leite, V.R.M.; Rodrigues, F.; de Sousa Lima, P.; et al. Characterization of a heme-protein responsive to hypoxia in Paracoccidioides brasiliensis. Fungal Genet. Biol. 2020, 144, 1–11. [Google Scholar] [CrossRef]
- Ryter, S.W.; Kvam, E.; Tyrrel, R.M. Heme oxygenase activity. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2000; Volume 99, pp. 369–391. [Google Scholar]
- Vile, G.F.; Basu-Modak, S.; Waltner, C.; Tyrrell, R.M. Heme oxygenase 1 mediates an adaptive response to oxidative stress in human skin fibroblasts. Proc. Natl. Acad. Sci. USA 1994, 91, 2607–2610. [Google Scholar] [CrossRef]
- Ascenzi, P.; Bocedi, A.; Visca, P.; Altruda, F.; Tolosano, E.; Beringhelli, T.; Fasano, M. Hemoglobin and heme scavenging. IUBMB Life 2005, 57, 749–759. [Google Scholar] [CrossRef]
- Dantas, A.S.; Andrade, R.V.; de Carvalho, M.J.; Felipe, M.S.S.; Campos, É.G. Oxidative stress response in Paracoccidioides brasiliensis: Assessing catalase and cytochrome c peroxidase. Mycol. Res. 2008, 112, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Navarathna, D.H.M.L.P.; Roberts, D.D. Candida albicans heme oxygenase and its product CO contribute to pathogenesis of candidemia and alter systemic chemokine and cytokine expression. Free Radic. Biol. Med. 2010, 49, 1561–1573. [Google Scholar] [CrossRef] [PubMed]
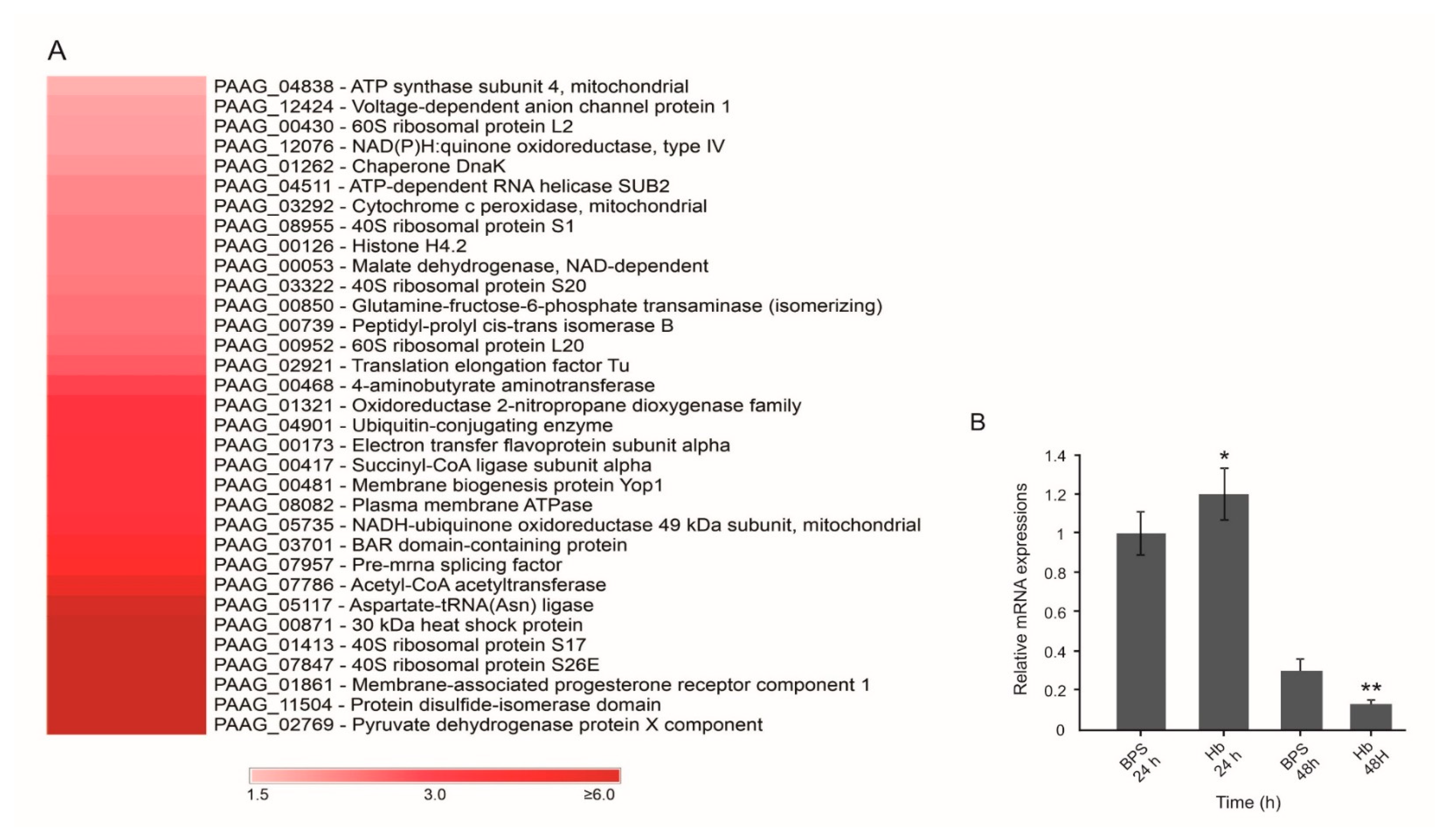


| Accession a | Description b | Score c | Expression LevelsRatio (Hb/BPS) d | SignalP e | SecretomeP f |
|---|---|---|---|---|---|
| PAAG_00871 | 30 kDa heat shock protein (HSP30) • | 754.8 | * | - | 0.786 |
| PAAG_08955 | 40S ribosomal protein S1 | 672.2 | 1.62 | - | 0.720 |
| PAAG_01413 | 40S ribosomal protein S17 | 1850.1 | * | - | 0.738 |
| PAAG_03322 | 40S ribosomal protein S20 | 1303.5 | 1.63 | - | 0.750 |
| PAAG_07847 | 40S ribosomal protein S26E | 409.6 | * | - | 0.613 |
| PAAG_00468 | 4-aminobutyrate aminotransferase | 977.2 | 1.75 | - | 0.601 |
| PAAG_00430 | 60S ribosomal protein L2 | 360.1 | 1.55 | - | 0.853 |
| PAAG_00952 | 60S ribosomal protein L20 | 1022.6 | 1.67 | - | 0.712 |
| PAAG_07786 | Acetyl-CoA acetyltransferase | 1043.1 | 4.10 | - | 0.655 |
| PAAG_05117 | Aspartate-tRNA (Asn) ligase | 565.1 | 5.53 | - | 0.609 |
| PAAG_04838 | ATP synthase subunit 4, mitochondrial | 663.4 | 1.51 | - | 0.781 |
| PAAG_04511 | ATP-dependent RNA helicase SUB2 | 2259.1 | 1.60 | - | 0.722 |
| PAAG_03701 | BAR domain-containing protein | 844.8 | 2.48 | - | 0.614 |
| PAAG_01262 | Chaperone DnaK | 2982.3 | 1.57 | 0.864 | - |
| PAAG_03292 | Cytochrome c peroxidase, mitochondrial • | 3045.4 | 1.60 | - | 0.809 |
| PAAG_00173 | Electron transfer flavoprotein subunit alpha | 465.5 | 1.88 | - | 0.642 |
| PAAG_00850 | Glutamine-fructose-6-phosphate transaminase (isomerizing) | 1305.8 | 1.65 | - | 0.693 |
| PAAG_00126 | Histone H4.2 | 4337.0 | 1.62 | 0.792 | - |
| PAAG_00053 | Malate dehydrogenase, NAD-dependent | 1037.2 | 1.62 | - | 0.651 |
| PAAG_00481 | Membrane biogenesis protein Yop1 | 922.3 | 1.93 | - | 0.902 |
| PAAG_01861 | Membrane-associated progesterone receptor component 1 | 443.0 | * | - | 0.735 |
| PAAG_12076 | NAD(P)H:quinone oxidoreductase, type IV • | 1770.3 | 1.55 | 0.718 | - |
| PAAG_05735 | NADH-ubiquinone oxidoreductase 49 kDa subunit, mitochondrial | 668.4 | 2.05 | - | 0.675 |
| PAAG_01321 | Oxidoreductase 2-nitropropane dioxygenase family • | 2254.9 | 1.79 | - | 0.707 |
| PAAG_00739 | Peptidyl-prolyl cis-trans isomerase B | 583.7 | 1.65 | 0.641 | - |
| PAAG_08082 | Plasma membrane ATPase | 738.9 | 1.93 | - | 0.712 |
| PAAG_07957 | Pre-mRNA splicing factor • | 539.9 | 2.69 | - | 0.801 |
| PAAG_11504 | Protein disulfide-isomerase domain • | 364.5 | * | - | 0.783 |
| PAAG_02769 | Pyruvate dehydrogenase protein X component • | 325.5 | * | - | 0.685 |
| PAAG_00417 | Succinyl-CoA ligase subunit alpha | 1400.3 | 1.88 | - | 0.624 |
| PAAG_02921 | Translation elongation factor Tu | 990.0 | 1.70 | - | 0.773 |
| PAAG_04901 | Ubiquitin-conjugating enzyme | 555.9 | 1.79 | - | 0.883 |
| PAAG_12424 | Voltage-dependent anion channel protein 1 | 2827.7 | 1.54 | - | 0.761 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Souza, A.F.; Tomazett, M.V.; Freitas e Silva, K.S.; de Curcio, J.S.; Pereira, C.A.; Baeza, L.C.; Paccez, J.D.; Gonçales, R.A.; Rodrigues, F.; Pereira, M.; et al. Interacting with Hemoglobin: Paracoccidioides spp. Recruits hsp30 on Its Cell Surface for Enhanced Ability to Use This Iron Source. J. Fungi 2021, 7, 21. https://doi.org/10.3390/jof7010021
de Souza AF, Tomazett MV, Freitas e Silva KS, de Curcio JS, Pereira CA, Baeza LC, Paccez JD, Gonçales RA, Rodrigues F, Pereira M, et al. Interacting with Hemoglobin: Paracoccidioides spp. Recruits hsp30 on Its Cell Surface for Enhanced Ability to Use This Iron Source. Journal of Fungi. 2021; 7(1):21. https://doi.org/10.3390/jof7010021
Chicago/Turabian Stylede Souza, Aparecido Ferreira, Mariana Vieira Tomazett, Kleber Santiago Freitas e Silva, Juliana Santana de Curcio, Christie Ataides Pereira, Lilian Cristiane Baeza, Juliano Domiraci Paccez, Relber Aguiar Gonçales, Fernando Rodrigues, Maristela Pereira, and et al. 2021. "Interacting with Hemoglobin: Paracoccidioides spp. Recruits hsp30 on Its Cell Surface for Enhanced Ability to Use This Iron Source" Journal of Fungi 7, no. 1: 21. https://doi.org/10.3390/jof7010021
APA Stylede Souza, A. F., Tomazett, M. V., Freitas e Silva, K. S., de Curcio, J. S., Pereira, C. A., Baeza, L. C., Paccez, J. D., Gonçales, R. A., Rodrigues, F., Pereira, M., & de Almeida Soares, C. M. (2021). Interacting with Hemoglobin: Paracoccidioides spp. Recruits hsp30 on Its Cell Surface for Enhanced Ability to Use This Iron Source. Journal of Fungi, 7(1), 21. https://doi.org/10.3390/jof7010021






