A New Double-Stranded RNA Mycovirus in Cryphonectria naterciae Is Able to Cross the Species Barrier and Is Deleterious to a New Host
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Isolates
2.2. Viral Detection and RNA-Seq
2.3. Specific Viral Primer and RT-PCR
2.4. Genome Terminal Sequences and Phylogeny
2.5. SSR-PCR of Cryphonectria naterciae
2.6. Horizontal CnFGV1 Transmission and Isogenic Strains
2.7. Effects on the Fungal Fitness
2.8. Effect of CnFGV1 Infection on the Fungus–Plant Interaction
3. Results
3.1. dsRNA Represents a Non-Segmented Genome of a Novel Virus Encoding the Conserved RdRp Domain
3.2. ORFs Were Phylogenetically Placed within the Proposed Family Fusagraviridae
3.3. CnFGV1 Was Able to Infect New Hosts beyond the Incompatibility and Species Barrier
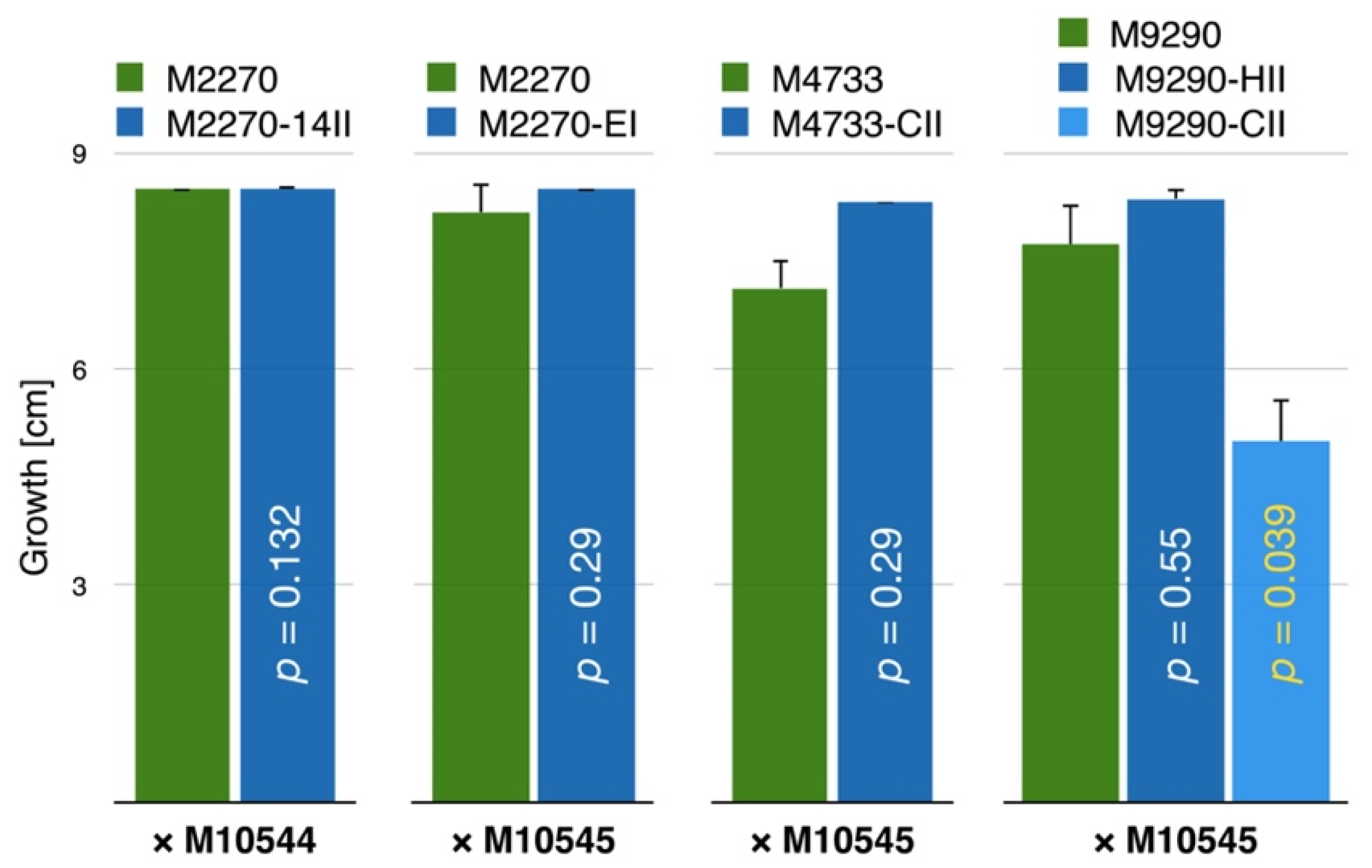
3.4. CnFGV1 Reduced the Growth Rate of Newly Infected Hosts
3.5. CnFGV1 Was Vertically Transmitted at 100% to the Next Generation through Asexual Spores, but Not So in New Host Species
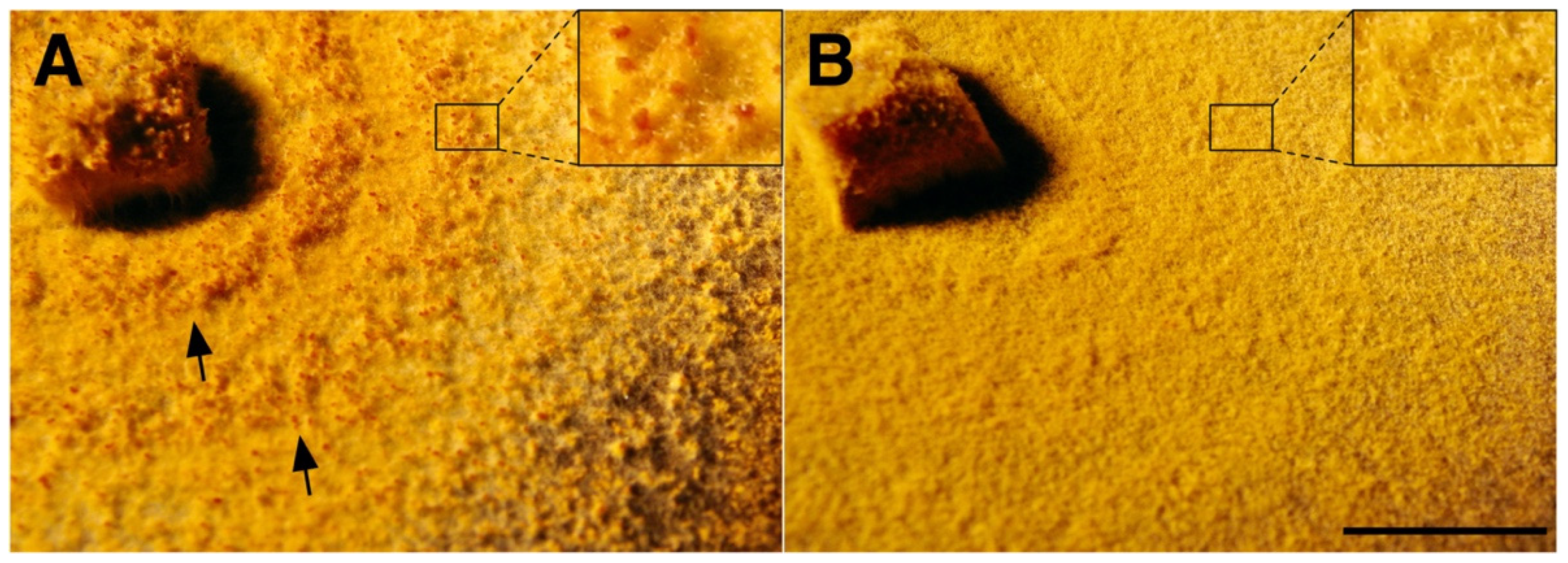
3.6. In Planta Tests Show That CnFGV1 Has No Major Effect on the C. naterciae–Tree Interaction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koonin, E.V.; Dolja, V.V. Metaviromics: A tectonic shift in understanding virus evolution. Virus Res. 2018, 246, A1–A3. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Silas, S.; Wang, Y.; Wu, S.; Bocek, M.; Kazlauskas, D.; Krupovic, M.; Fire, A.; Dolja, V.V.; Koonin, E.V. Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome. Nat. Microbiol. 2020, 5, 1262–1270. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Kondo, H.; Hisano, S.; Chiba, S.; Maruyama, K.; Andika, I.B.; Toyoda, K.; Fujimori, F.; Suzuki, N. Sequence and phylogenetic analyses of novel totivirus-like double-stranded RNAs from field-collected powdery mildew fungi. Virus Res. 2016, 213, 353–364. [Google Scholar] [CrossRef] [Green Version]
- Kondo, H.; Fujita, M.; Hisano, H.; Hyodo, K.; Andika, I.B.; Suzuki, N. Virome Analysis of aphid populations that infest the barley field: The discovery of two novel groups of Nege/Kita-like viruses and other novel RNA viruses. Front. Microbiol. 2020, 11, 509. [Google Scholar] [CrossRef]
- Roossinck, M.J.; Bazán, E.R. Symbiosis: Viruses as intimate partners. Ann. Rev. Virol. 2017, 4, 123–139. [Google Scholar] [CrossRef]
- Ali, A. Chapter 17—Fungal viruses: An unlikely ally. In Applied Plant Virology; Awasthi, L.P., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 229–236. [Google Scholar]
- Myers, J.M.; Bonds, A.E.; Clemons, R.A.; Thapa, N.A.; Simmons, D.R.; Carter-House, D.; Ortanez, J.; Liu, P.; Miralles-Durán, A.; Desirò, A.; et al. Survey of early-diverging lineages of fungi reveals abundant and diverse mycoviruses. mBio 2020, 11, e02027-20. [Google Scholar] [CrossRef]
- Marzano, S.-Y.L.; Nelson, B.D.; Ajayi-Oyetunde, O.; Bradley, C.A.; Hughes, T.J.; Hartman, G.L.; Eastburn, D.M.; Domier, L.L. Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J. Virol. 2016, 90, 6846. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Chen, X.; Li, P.; Qiu, D.; Guo, L. Complete genome sequence of a Fusarium graminearum double-stranded RNA virus in a newly proposed Family, Alternaviridae. Genome Announc. 2018, 6, e00064-18. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Cheng, J.; Fu, Y.; Jiang, D.; Xie, J. Molecular characterization of a novel positive-sense, single-stranded RNA mycovirus infecting the plant pathogenic fungus Sclerotinia sclerotiorum. Viruses 2015, 7, 2470. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Zheng, D.; Cheng, J.; Chen, T.; Fu, Y.; Jiang, D.; Xie, J. Characterization of a novel Sclerotinia sclerotiorum RNA virus as the prototype of a new proposed family within the order Tymovirales. Virus Res. 2016, 219, 92–99. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, J.; Zhang, H.; Qiu, D.; Guo, L. Two novel relative double-stranded RNA mycoviruses infecting Fusarium poae strain SX63. Int. J. Mol. Sci. 2016, 17, 641. [Google Scholar] [CrossRef] [Green Version]
- Wolf, Y.I.; Kazlauskas, D.; Iranzo, J.; Lucía-Sanz, A.; Kuhn, J.H.; Krupovic, M.; Dolja, V.V.; Koonin, E.V. Origins and evolution of the global RNA virome. mBio 2018, 9, e02329-18. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Li, B.; Fu, Y.; Jiang, D.; Ghabrial, S.A.; Li, G.; Peng, Y.; Xie, J.; Cheng, J.; Huang, J.; et al. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. USA 2010, 107, 8387–8392. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Wang, S.; Zhang, L.; Qiu, D.; Zhou, X.; Guo, L. A tripartite ssDNA mycovirus from a plant pathogenic fungus is infectious as cloned DNA and purified virions. Sci. Adv. 2020, 6, eaay9634. [Google Scholar] [CrossRef] [Green Version]
- Feng, C.; Feng, J.; Wang, Z.; Pedersen, C.; Wang, X.; Saleem, H.; Domier, L.; Marzano Shin-Yi, L.; Simon Anne, E. Identification of the viral determinant of hypovirulence and host range in Sclerotiniaceae of a genomovirus reconstructed from the plant metagenome. J. Virol. 2021, 95, e00264-21. [Google Scholar] [CrossRef]
- Gryzenhout, M.; Wingfield, B.D.; Wingfield, M.J. Taxonomy, Phylogeny, and Ecology of Bark-Inhabiting and Tree-Pathogenic Fungi in the Cryphonectriaceae; American Phytopathological Society (APS Press): St. Paul, MN, USA, 2009. [Google Scholar]
- Jiang, N.; Fan, X.; Tian, C.; Crous, P.W. Reevaluating Cryphonectriaceae and allied families in Diaporthales. Mycologia 2020, 112, 267–292. [Google Scholar] [CrossRef]
- Cornejo, C.; Hauser, A.; Beenken, L.; Cech, T.; Rigling, D. Cryphonectria carpinicola sp. nov. associated with hornbeam decline in Europe. Fungal Biol. 2021, 125, 347–356. [Google Scholar] [CrossRef]
- Bragança, H.; Rigling, D.; Diogo, E.; Capelo, J.; Phillips, A.; Tenreiro, R. Cryphonectria naterciae: A new species in the Cryphonectria–Endothia complex and diagnostic molecular markers based on microsatellite-primed PCR. Fungal Biol. 2011, 115, 852–861. [Google Scholar] [CrossRef]
- Smahi, H.; Belhoucine-Guezouli, L.; Bouhraoua, R.T.; Franceschini, A.; Linaldeddu, B.T. First report of branch canker and dieback caused by Cryphonectria naterciae on Quercus suber in Algeria. Plant Dis. 2017, 102, 251. [Google Scholar] [CrossRef]
- Heiniger, U.; Rigling, D. Biological control of chestnut blight in Europe. Annu. Rev. Phytopathol. 1994, 32, 581–599. [Google Scholar] [CrossRef]
- Prospero, S.; Rigling, D. Chestnut blight. In Infectious Forest Diseases; Gonthier, P., Nicolotti, G., Eds.; CABI International Wallingford: Oxfordshire, UK, 2013; pp. 318–339. [Google Scholar]
- Shapira, R.; Choi, G.H.; Nuss, D.L. Virus-like genetic organization and expression strategy for a double-stranded RNA genetic element associated with biological control of chestnut blight. EMBO J. 1991, 10, 731–739. [Google Scholar] [CrossRef]
- Choi, G.H.; Shapira, R.; Nuss, D.L. Cotranslational autoproteolysis involved in gene expression from a double-stranded RNA genetic element associated with hypovirulence of the chestnut blight fungus. Proc. Natl. Acad. Sci. USA 1991, 88, 1167–1171. [Google Scholar] [CrossRef] [Green Version]
- Ghabrial, S.A.; Suzuki, N. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 2009, 47, 353–384. [Google Scholar] [CrossRef]
- Rigling, D.; Prospero, S. Cryphonectria parasitica, the causal agent of chestnut blight: Invasion history, population biology and disease control. Mol. Plant Pathol. 2018, 19, 7–20. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.N.; Jin, H.Y.; Kwak, M.J.; Khaine, I.; You, H.N.; Lee, T.Y.; Ahn, T.H.; Woo, S.Y. Why does Quercus suber species decline in Mediterranean areas? J. Asia-Pac. Biodiver. 2017, 10, 337–341. [Google Scholar] [CrossRef]
- Ghabrial, S.A.; Castón, J.R.; Jiang, D.; Nibert, M.L.; Suzuki, N. 50-plus years of fungal viruses. Virology 2015, 479–480, 356–368. [Google Scholar] [CrossRef] [Green Version]
- Choi, G.H.; Dawe, A.L.; Churbanov, A.; Smith, M.L.; Milgroom, M.G.; Nuss, D.L. Molecular characterization of vegetative incompatibility genes that restrict hypovirus transmission in the chestnut blight fungus Cryphonectria parasitica. Genetics 2012, 190, 113–127. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.-X.; Spiering, M.J.; Dawe, A.L.; Nuss, D.L. Vegetative incompatibility loci with dedicated roles in allorecognition restrict mycovirus transmission in chestnut blight fungus. Genetics 2014, 197, 701–714. [Google Scholar] [CrossRef] [Green Version]
- Paoletti, M. Vegetative incompatibility in fungi: From recognition to cell death, whatever does the trick. Fungal Biol. Rev. 2016, 30, 152–162. [Google Scholar] [CrossRef]
- Biella, S.; Smith, M.L.; Aist, J.R.; Cortesi, P.; Milgroom, M.G. Programmed cell death correlates with virus transmission in a filamentous fungus. Proc. R. Soc. Lond. B Biol. 2002, 269, 2269. [Google Scholar] [CrossRef]
- Liu, Y.C.; Linder-Basso, D.; Hillman, B.I.; Kaneko, S.; Milgroom, M.G. Evidence for interspecies transmission of viruses in natural populations of filamentous fungi in the genus Cryphonectria. Mol. Ecol. 2003, 12, 1619–1628. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Dynek, J.N.; Hillman, B.I.; Milgroom, M.G. Diversity of viruses in Cryphonectria parasitica and C. nitschkei in Japan and China, and partial characterization of a new chrysovirus species. Mycol. Res. 2007, 111, 433–442. [Google Scholar] [CrossRef]
- Shahi, S.; Chiba, S.; Kondo, H.; Suzuki, N. Cryphonectria nitschkei chrysovirus 1 with unique molecular features and a very narrow host range. Virology 2021, 554, 55–65. [Google Scholar] [CrossRef]
- Stauber, L.; Prospero, S.; Croll, D.; Mitchell Aaron, P. Comparative genomics analyses of lifestyle transitions at the origin of an invasive fungal pathogen in the genus Cryphonectria. mSphere 2020, 5, e00737-20. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Dennert, F.; Rigling, D.; Meyer, J.B.; Schefer, C.; Augustiny, E.; Prospero, S. Testing the pathogenic potential of Cryphonectria parasitica and related species on three common European Fagaceae. Front. For. Glob. Chang. 2020, 3, 52. [Google Scholar] [CrossRef]
- Hoegger, P.J.; Rigling, D.; Holdenrieder, O.; Heiniger, U. Genetic structure of newly established populations of Cryphonectria parasitica. Mycol. Res. 2000, 104, 1108–1116. [Google Scholar] [CrossRef]
- Hoegger, P.J.; Rigling, D.; Holdenrieder, O.; Heiniger, U. Cryphonectria radicalis: Rediscovery of a lost fungus. Mycologia 2002, 94, 105–115. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Urayama, S.-i.; Katoh, Y.; Fukuhara, T.; Arie, T.; Moriyama, H.; Teraoka, T. Rapid detection of Magnaporthe oryzae chrysovirus 1-A from fungal colonies on agar plates and lesions of rice blast. J. Gen. Plant Pathol. 2015, 81, 97–102. [Google Scholar] [CrossRef]
- Suzuki, N.; Supyani, S.; Maruyama, K.; Hillman, B.I. Complete genome sequence of Mycoreovirus-1/Cp9B21, a member of a novel genus within the family Reoviridae, isolated from the chestnut blight fungus Cryphonectria parasitica. J. Gen. Virol. 2004, 85, 3437–3448. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Huson, D.H. SplitsTree: Analyzing and visualizing evolutionary data. Bioinformatics 1998, 14, 68–73. [Google Scholar] [CrossRef]
- Schoebel, C.N.; Brodbeck, S.; Buehler, D.; Cornejo, C.; Gajurel, J.; Hartikainen, H.; Keller, D.; Leys, M.; Říčanová, Š.; Segelbacher, G.; et al. Lessons learned from microsatellite development for nonmodel organisms using 454 pyrosequencing. J. Evol. Biol. 2013, 26, 600–611. [Google Scholar] [CrossRef]
- Stauber, L.; Badet, T.; Feurtey, A.; Prospero, S.; Croll, D. Emergence and diversification of a highly invasive chestnut pathogen lineage across southeastern Europe. eLife 2021, 10, e56279. [Google Scholar] [CrossRef]
- Faircloth, B.C. msatcommander: Detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol. Ecol. Res. 2008, 8, 92–94. [Google Scholar] [CrossRef]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press, Inc.: New York, NY, USA, 1990. [Google Scholar]
- Zhang, T.; Zeng, X.; Cai, X.; Liu, H.; Zeng, Z. Molecular characterization of a novel double-stranded RNA mycovirus of Trichoderma asperellum strain JLM45-3. Arch. Virol. 2018, 163, 3433–3437. [Google Scholar] [CrossRef]
- Wang, J.; Ni, Y.; Liu, X.; Zhao, H.; Xiao, Y.; Xiao, X.; Li, S.; Liu, H. Divergent RNA viruses in Macrophomina phaseolina exhibit potential as virocontrol agents. Virus Evol. 2021, 7, veaa095. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Zhang, M.; Hong, N.; Xiao, F.; Fu, M.; Xiang, J.; Wang, G. Identification and characterization of a novel hepta-segmented dsRNA virus from the phytopathogenic fungus Colletotrichum fructicola. Front. Microbiol. 2018, 9, 754. [Google Scholar] [CrossRef] [PubMed]
- Marvelli, R.A.; Hobbs, H.A.; Li, S.; McCoppin, N.K.; Domier, L.L.; Hartman, G.L.; Eastburn, D.M. Identification of novel double-stranded RNA mycoviruses of Fusarium virguliforme and evidence of their effects on virulence. Arch. Virol. 2014, 159, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Na, B.; Kim, D.-H. Characterization of a novel dsRNA mycovirus of Trichoderma atroviride NFCF377 reveals a member of “Fusagraviridae” with changes in antifungal activity of the host fungus. J. Microbiol. 2020, 58, 1046–1053. [Google Scholar] [CrossRef]
- Williams, S.H.; Levy, A.; Yates, R.A.; Somaweera, N.; Neville, P.J.; Nicholson, J.; Lindsay, M.D.A.; Mackenzie, J.S.; Jain, K.; Imrie, A.; et al. The diversity and distribution of viruses associated with Culex annulirostris mosquitoes from the Kimberley Region of Western Australia. Viruses 2020, 12, 717. [Google Scholar] [CrossRef] [PubMed]
- Arjona-Lopez, J.M.; Telengech, P.; Jamal, A.; Hisano, S.; Kondo, H.; Yelin, M.D.; Arjona-Girona, I.; Kanematsu, S.; Lopez-Herrera, C.J.; Suzuki, N. Novel, diverse RNA viruses from Mediterranean isolates of the phytopathogenic fungus, Rosellinia necatrix: Insights into evolutionary biology of fungal viruses. Environ. Microbiol. 2018, 20, 1464–1483. [Google Scholar] [CrossRef] [PubMed]
- Kotta-Loizou, I.; Coutts, R.H.A. Studies on the virome of the entomopathogenic fungus Beauveria bassiana reveal novel dsRNA elements and mild hypervirulence. PLoS Pathog. 2017, 13, e1006183. [Google Scholar] [CrossRef] [Green Version]
- Mahillon, M.; Decroës, A.; Caulier, S.; Tiendrebeogo, A.; Legrève, A.; Bragard, C. Genomic and biological characterization of a novel partitivirus infecting Fusarium equiseti. Virus Res. 2021, 297, 198386. [Google Scholar] [CrossRef]
- Márquez, L.M.; Redman, R.S.; Rodriguez, R.J.; Roossinck, M.J. A virus in a fungus in a plant: Three-way symbiosis required for thermal tolerance. Science 2007, 315, 513. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Choi, G.H.; Nuss, D.L. Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 1994, 264, 1762–1764. [Google Scholar] [CrossRef] [PubMed]
- Longdon, B.; Day, J.P.; Alves, J.M.; Smith, S.C.L.; Houslay, T.M.; McGonigle, J.E.; Tagliaferri, L.; Jiggins, F.M. Host shifts result in parallel genetic changes when viruses evolve in closely related species. PLoS Pathog. 2018, 14, e1006951. [Google Scholar] [CrossRef] [Green Version]
- McLeish, M.J.; Fraile, A.; García-Arenal, F. Evolution of plant–virus interactions: Host range and virus emergence. Curr. Opin. Virol. 2019, 34, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Rigling, D.; Robin, C.; Prospero, S. Mycovirus-mediated biological control. In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: Oxford, UK, 2021; pp. 468–477. [Google Scholar]
- Kwon, S.-J.; Lim, W.-S.; Park, S.-H.; Park, M.-R.; Kim, K.-H. Molecular characterization of a dsRNA mycovirus, Fusarium graminearum Virus-DK21, which is phylogenetically related to hypoviruses but has a genome organization and gene expression strategy resembling those of plant potex-like viruses. Mol. Cells 2009, 28, 73–74. [Google Scholar] [CrossRef]
- Darissa, O.; Adam, G.; Schäfer, W. A dsRNA mycovirus causes hypovirulence of Fusarium graminearum to wheat and maize. Eur. J. Plant Pathol. 2012, 134, 181–189. [Google Scholar] [CrossRef]
- Campo, S.; Gilbert, K.B.; Carrington, J.C. Small RNA-based antiviral defense in the phytopathogenic fungus Colletotrichum higginsianum. PLoS Pathog. 2016, 12, e1005640. [Google Scholar] [CrossRef] [Green Version]
- Olivé, M.; Campo, S. The dsRNA mycovirus ChNRV1 causes mild hypervirulence in the fungal phytopathogen Colletotrichum higginsianum. Arch. Microbiol. 2021, 203, 241–249. [Google Scholar] [CrossRef]

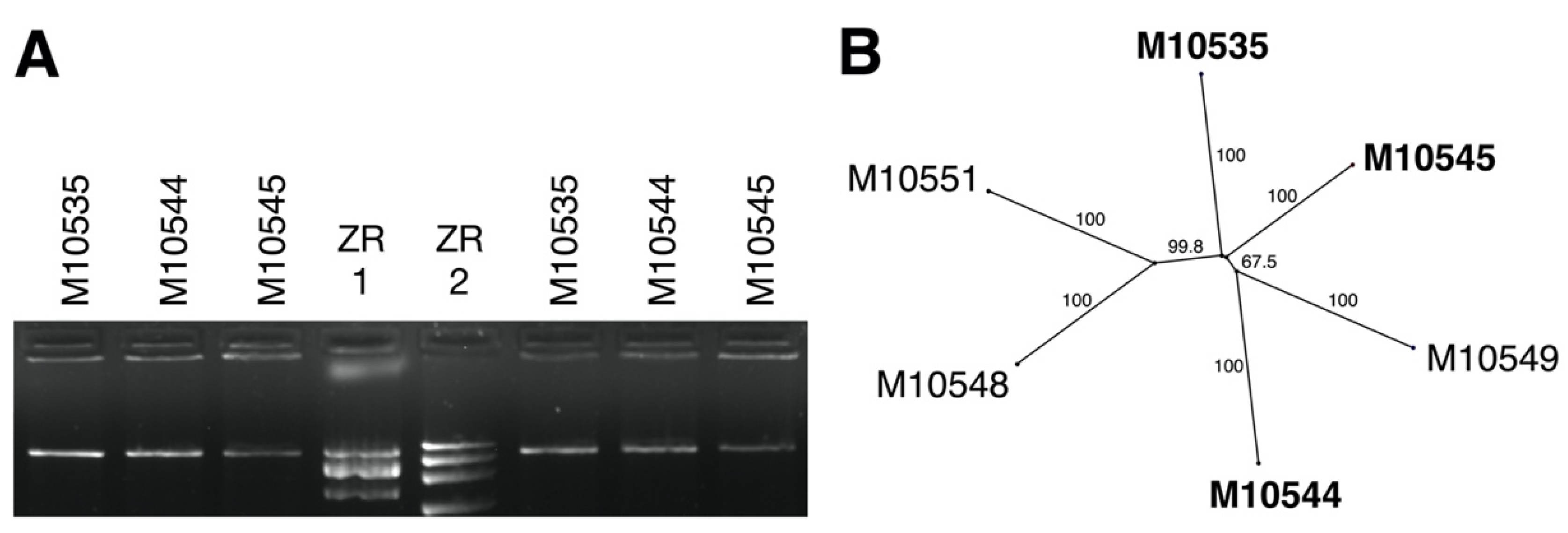
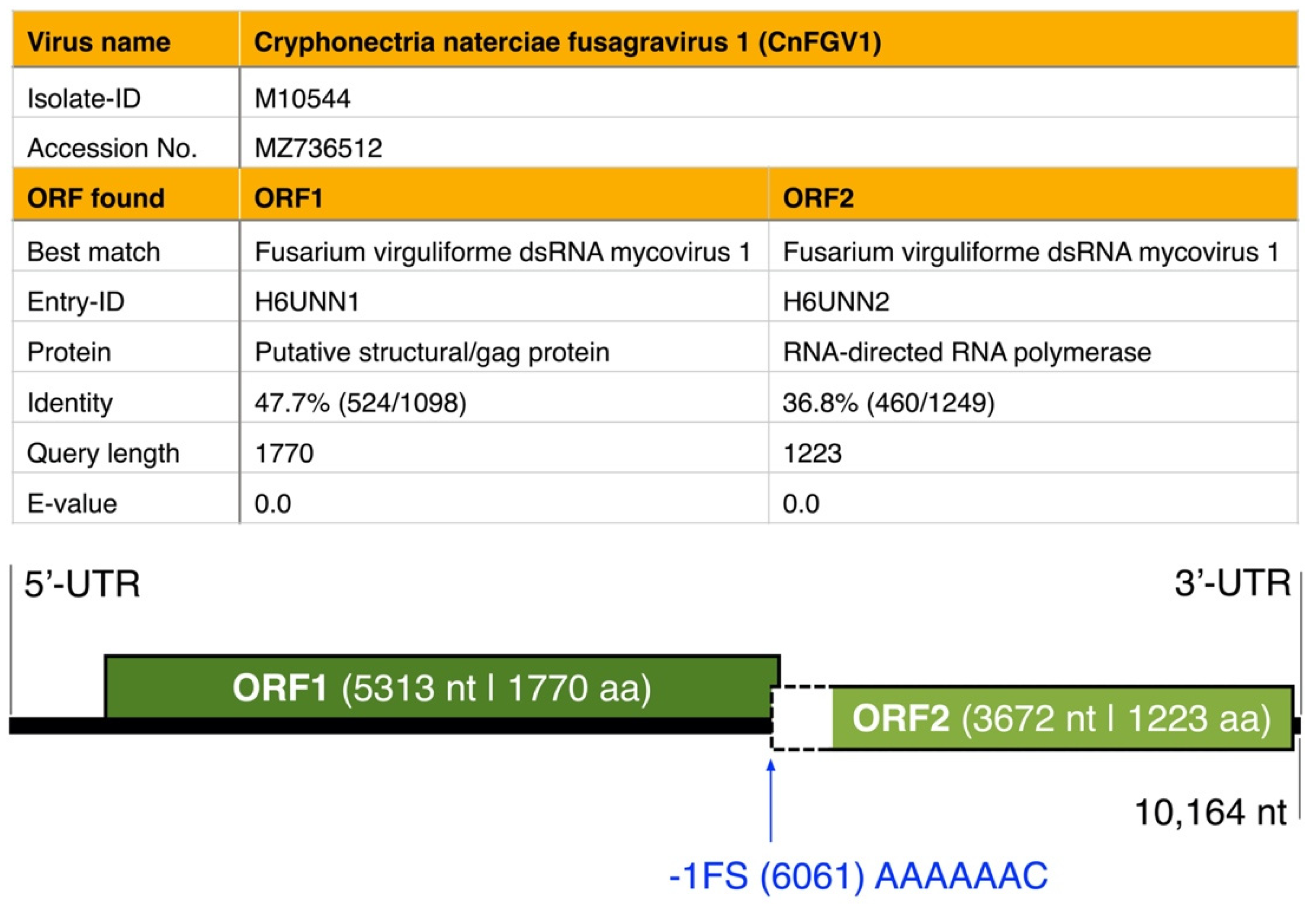
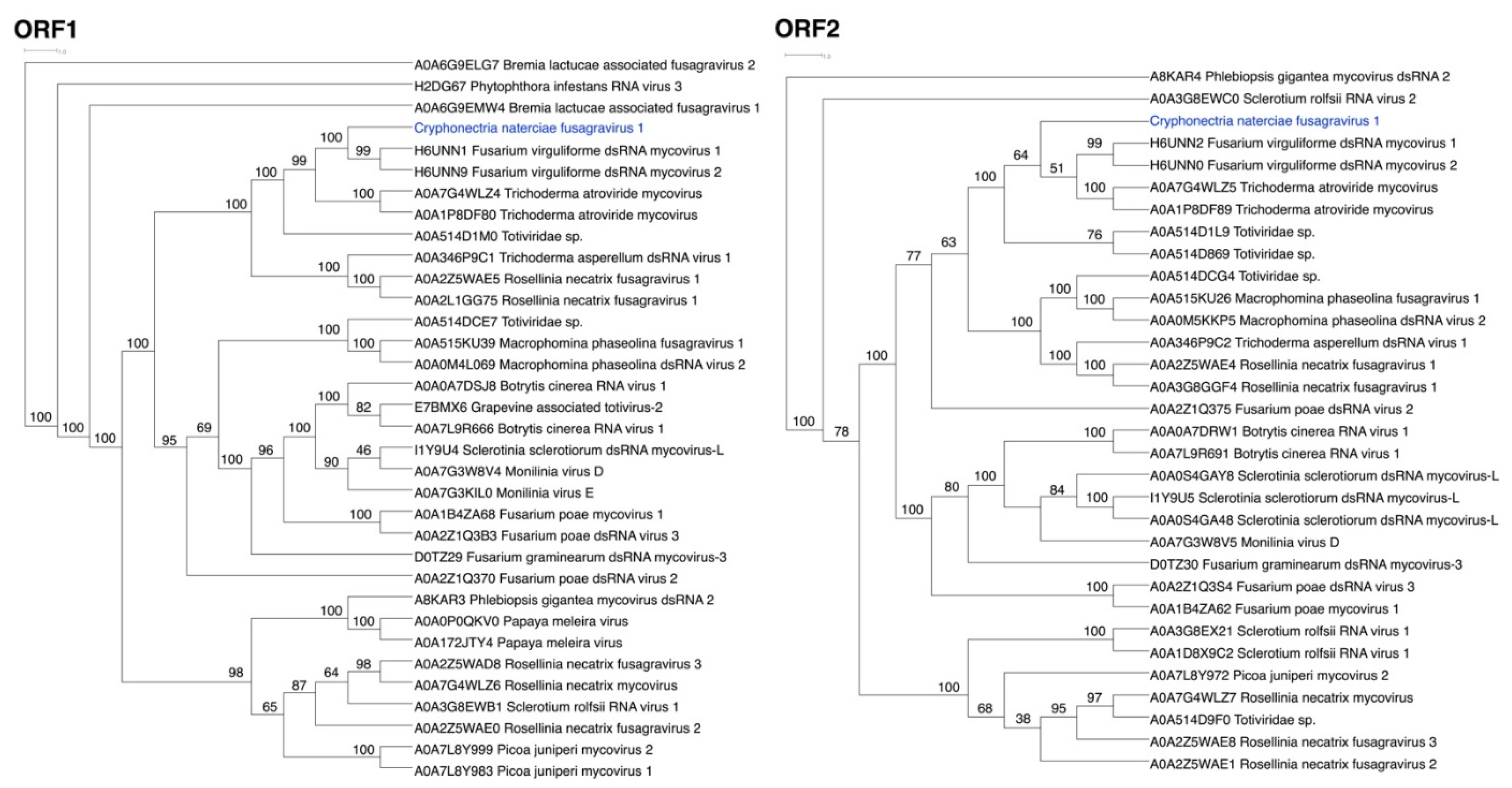
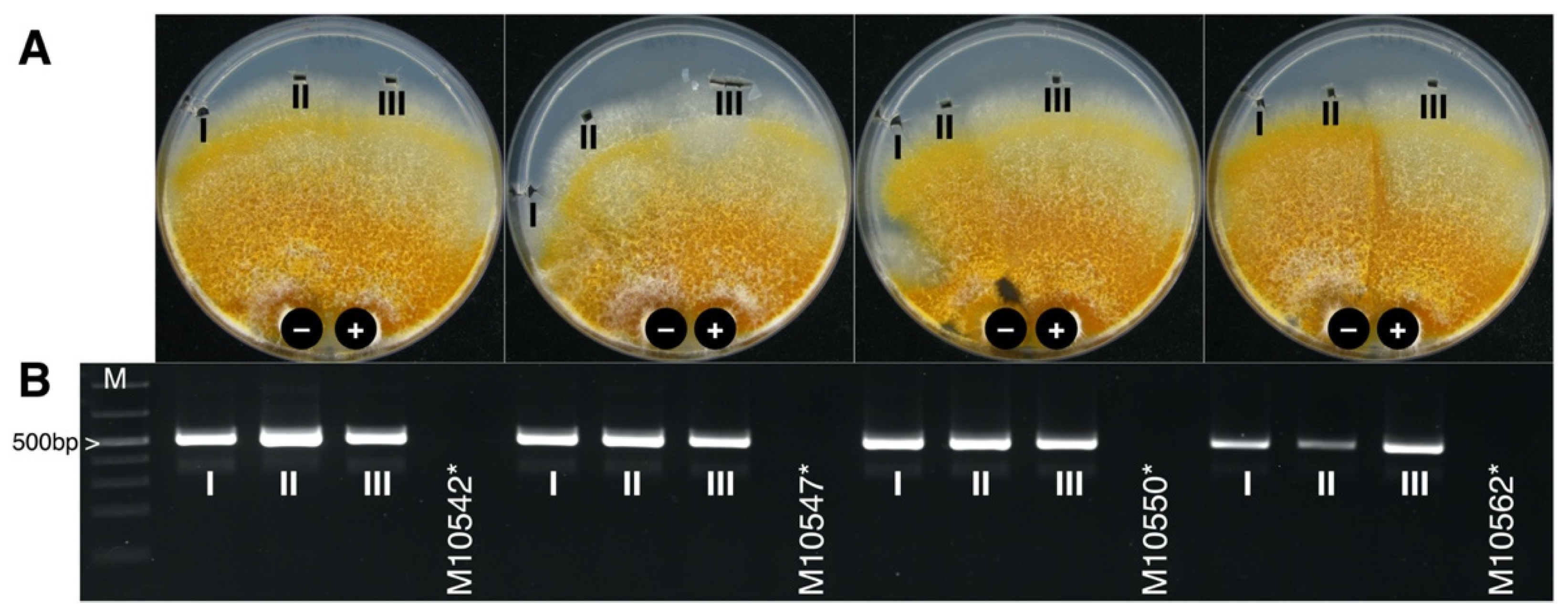



| Species/Host | Collection ID 1 | Country | Year | CnFGV1 Detection 2 | Reference |
|---|---|---|---|---|---|
| C. naterciae | |||||
| Quercus suber | M10535 | Portugal | 1960 | Positive | [21] |
| Q. suber | M10536 | Portugal | 2010 | Positive | This study |
| Q. suber | M10537 | Portugal | 2001 | Negative | [21] |
| Q. suber | M10538 | Portugal | 2001 | Negative | [21] |
| Q. suber | M10539 | Portugal | 2001 | Negative | [21] |
| Q. suber | M10540 | Portugal | 2000 | Negative | [21] |
| Q. suber | M10541 | Portugal | 2000 | Positive | [21] |
| Q. suber | M10542 | Portugal | 2001 | Negative | [21] |
| Q. suber | M10543 | Portugal | 2001 | Negative | [21] |
| Q. suber | M10544 | Portugal | 2005 | Positive | [21] |
| Q. suber | M10545 | Portugal | 2005 | Positive | [21] |
| Q. suber | M10557 | Portugal | 2014 | Negative | This study |
| Q. suber | M10558 | Portugal | 2014 | Negative | This study |
| Q. suber | M10559 | Portugal | 2014 | Positive | This study |
| Q. suber | M10560 | Portugal | 2014 | Positive | This study |
| Q. suber | M10561 | Portugal | 2014 | Positive | This study |
| Q. suber | M10562 | Portugal | 2015 | Negative | This study |
| Q. suber | M10563 | Portugal | 2011 | Negative | This study |
| Q. suber | M10564 | Portugal | 2014 | Positive | This study |
| Q. suber | M10565 | Portugal | 2015 | Positive | This study |
| Castanea sativa | M10546 | Portugal | 2001 | Negative | [21] |
| C. sativa | M10547 | Portugal | 2001 | Negative | [21] |
| C. sativa | M10548 | Portugal | 2001 | Positive | [21] |
| C. sativa | M10549 | Portugal | 2001 | Positive | [21] |
| C. sativa | M10550 | Portugal | 2001 | Negative | [21] |
| C. sativa | M10551 | Portugal | 2001 | Positive | [21] |
| C. sativa | M10552 | Portugal | 2001 | Negative | [21] |
| C. sativa | M10553 | Portugal | 2001 | Negative | [21] |
| C. sativa | M10554 | Portugal | 2001 | Negative | [21] |
| C. sativa | M10555 | Portugal | 2001 | Negative | [21] |
| C. sativa | M10556 | Portugal | 2001 | Negative | [21] |
| C. carpinicola | |||||
| Carpinus sp. | M9290 | Austria | 2009 | Negative | [20] |
| Carpinus betulus | M9615 | Switzerland | 2019 | Negative | [20] |
| C. parasitica | |||||
| C. sativa | M2372 | Switzerland | 1992 | Negative | [40] |
| C. sativa | M2671 | Switzerland | 1992 | Negative | [40] |
| C. sativa | M4023 | Switzerland | 2000 | Negative | [41] |
| C. radicalis | |||||
| C. sativa | M2270 | Switzerland | 1996 | Negative | [42] |
| C. sativa | M4733 | Switzerland | 2001 | Negative | This study |
| Donor | |||||
|---|---|---|---|---|---|
| Cryphonectria naterciae | |||||
| Recipient | Isolate-ID | M10535 | M10544 | M10545 | |
| Within-species tests | C. naterciae | M10542 | 2/3 | 3/3 | 3/3 |
| M10547 | 2/3 | 3/3 | 3/3 | ||
| M10550 | 0/3 | 2/3 | 3/3 | ||
| M10562 | 1/3 | 3/3 | 3/3 | ||
| Cross-species tests | C. carpinicola | M9290 | 0/10 | 0/10 | 2/10 |
| M9615 | 0/10 | 0/10 | 0/10 | ||
| C. radicalis | M2270 | 0/10 | 1/10 | 1/10 | |
| M4733 | 0/10 | 0/10 | 1/10 | ||
| C. parasitica | M2372 | 0/10 | 0/10 | 0/10 | |
| M2671 | 0/10 | 0/10 | 0/10 | ||
| M4023 | 0/10 | 0/10 | 0/10 | ||
| Strain Label (Rate of Vertical Transmission) | |||
|---|---|---|---|
| Species | Experiment I | Experiment II | |
| Donor strain | C. naterciae | M10544 (50/50) | M10545 (50/50) |
| Recipient, isogenic strain | C. radicalis | M2270-14II (1/50) | M2270-EI (44/48) 1 |
| No transmission 2 | M4733-CII (44/50) | ||
| C. carpinicola | No transmission 2 | M9290-CII (24/50) 3 M9290-HII (49/50) 4 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cornejo, C.; Hisano, S.; Bragança, H.; Suzuki, N.; Rigling, D. A New Double-Stranded RNA Mycovirus in Cryphonectria naterciae Is Able to Cross the Species Barrier and Is Deleterious to a New Host. J. Fungi 2021, 7, 861. https://doi.org/10.3390/jof7100861
Cornejo C, Hisano S, Bragança H, Suzuki N, Rigling D. A New Double-Stranded RNA Mycovirus in Cryphonectria naterciae Is Able to Cross the Species Barrier and Is Deleterious to a New Host. Journal of Fungi. 2021; 7(10):861. https://doi.org/10.3390/jof7100861
Chicago/Turabian StyleCornejo, Carolina, Sakae Hisano, Helena Bragança, Nobuhiro Suzuki, and Daniel Rigling. 2021. "A New Double-Stranded RNA Mycovirus in Cryphonectria naterciae Is Able to Cross the Species Barrier and Is Deleterious to a New Host" Journal of Fungi 7, no. 10: 861. https://doi.org/10.3390/jof7100861
APA StyleCornejo, C., Hisano, S., Bragança, H., Suzuki, N., & Rigling, D. (2021). A New Double-Stranded RNA Mycovirus in Cryphonectria naterciae Is Able to Cross the Species Barrier and Is Deleterious to a New Host. Journal of Fungi, 7(10), 861. https://doi.org/10.3390/jof7100861








