Real-Time PCR Assay for the Detection of Dermatophytes: Comparison between an In-House Method and a Commercial Kit for the Diagnosis of Dermatophytoses in Patients from Dakar, Senegal
Abstract
:1. Introduction
2. Patients and Methods
2.1. Study Design and Sites
2.2. Conventional Diagnosis (CD)
2.3. DNA Extraction
2.4. In-House PCR Assay
2.5. Commercial Kit Assay: EurobioPlex Dermatophytes Real-Time PCR
2.6. ITS Sequencing
2.7. Gold Standard (GS)
2.8. Statistical Analysis
3. Results
3.1. Characteristics of the Samples
3.2. Conventional Methods
3.3. In-House (IH) PCR Assay
3.4. Commercial Kit (CK) EurobioPlex Dermatophytes PCR Assay
3.5. Diagnostic Performance According to Sample Type
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alshahni, M.M.; Yamada, T. Genetic Manipulations in Dermatophytes. Mycopathologia 2017, 182, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Ndiaye, D.; Ndiaye, M.; Badiane, A.; Seck, M.C.; Faye, B.; Ndiaye, J.L.; Tine, R.; Ndir, O. Dermatophytosis diagnosed at the laboratory of parasitology and mycology of Le Dantec Hospital in Dakar between 2007 and 2011. J. Mycol. Med. 2013, 23, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Verrier, J.; Monod, M. Diagnosis of Dermatophytosis Using Molecular Biology. Mycopathologia 2017, 182, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Levitt, J.O.; Levitt, B.H.; Akhavan, A.; Yanofsky, H. The sensitivity and specificity of potassium hydroxide smear and fungal culture relative to clinical assessment in the evaluation of tinea pedis: A pooled analysis. Dermatol. Res. Pract. 2010, 2010. [Google Scholar] [CrossRef] [PubMed]
- Paugam, A.; L’Ollivier, C.; Viguié, C.; Anaya, L.; Mary, C.; De Ponfilly, G.; Ranque, S. Comparison of real-time PCR with conventional methods to detect dermatophytes in samples from patients with suspected dermatophytosis. J. Microbiol. Methods 2013, 95, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Kondori, N.; Tehrani, P.A.; Strömbeck, L.; Faergemann, J. Comparison of Dermatophyte PCR Kit with Conventional Methods for Detection of Dermatophytes in Skin Specimens. Mycopathologia 2013, 176, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Hayette, M.P.; Seidel, L.; Adjetey, C.; Darfouf, R.; Wéry, M.; Boreux, R.; Sacheli, R.; Melin, P.; Arrese, J. Clinical evaluation of the DermaGeniusR Nail real-time PCR assay for the detection of dermatophytes and Candida albicans in nails. Med. Mycol. 2019, 57, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Zaman, M.; Singh, J. Fast and sensitive detection of Trichophyton rubrum DNA from the nail samples of patients with onychomycosis by a double-round polymerase chain reaction-based assay. Br. J. Dermatol. 2007, 157, 698–703. [Google Scholar] [CrossRef] [PubMed]
- Uchida, T.; Makimura, K.; Ishihara, K.; Goto, H.; Tajiri, Y.; Okuma, M.; Fujisaki, R.; Uchida, K.; Abe, S.; Iijima, M. Comparative study of direct polymerase chain reaction, microscopic examination and culture-based morphological methods for detection and identification of dermatophytes in nail and skin samples. J. Dermatol. 2009, 36, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Ross, I.L.; Weldhagen, G.F.; Kidd, S.E. Detection and identification of dermatophyte fungi in clinical samples using a commercial multiplex tandem PCR assay. Pathology 2020, 52, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Diongue, K.; Bréchard, L.; Diallo, M.A.; Seck, M.C.; Ndiaye, M.; Badiane, A.S.; Ranque, S.; Ndiaye, D. A Comparative Study on Phenotypic versus ITS-Based Molecular Identification of Dermatophytes Isolated in Dakar, Senegal. Int. J. Microbiol. 2019, 2019, 6754058. [Google Scholar] [CrossRef] [PubMed]
- Chabasse, D.; Bouchara, J.-P.; de Gentile, L.; Brun, S.; Cimon, B.; Penn, P. Les Dermatophytes. 2004. Available online: https://lesbiologistesmedicaux.fr/images/cahiers/2004-Bioforma-31-Les%20dermatophytes.pdf (accessed on 27 September 2021).
- White, T.J.; Lee, S.B.; Bruns, T.D.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. PCR Protoc. 1990, 18, 315–332. [Google Scholar]
- Brillowska-Dabrowska, A.; Saunte, D.M.; Arendrup, M.C. Five-hour diagnosis of dermatophyte nail infections with specific detection of Trichophyton rubrum. J. Clin. Microbiol. 2007, 45, 1200–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gräser, Y.; Scott, J.; Summerbell, R. The new species concept in dermatophytes—A polyphasic approach. Mycopathologia 2008, 166, 239–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gräser, Y.; Saunte, D.M.L. A hundred years of diagnosing superficial fungal infections: Where do we come from, where are we now and where would we like to go? Acta Derm. Venereol. 2020, 100, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Verrier, J.; Krähenbühl, L.; Bontems, O.; Fratti, M.; Salamin, K.; Monod, M. Dermatophyte identification in skin and hair samples using a simple and reliable nested polymerase chain reaction assay. Br. J. Dermatol. 2013, 168, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Ohst, T.; Kupsch, C.; Gräser, Y. Detection of common dermatophytes in clinical specimens using a simple quantitative real-time TaqMan polymerase chain reaction assay. Br. J. Dermatol. 2016, 174, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Motamedi, M.; Mirhendi, H.; Zomorodian, K.; Khodadadi, H.; Kharazi, M.; Ghasemi, Z.; Reza, M.; Koichi, S. Clinical evaluation of β-tubulin real-time PCR for rapid diagnosis of dermatophytosis, a comparison with mycological methods. Mycoses 2017, 60, 692–696. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Ran, M.; Wang, X.; Wan, Z.; Li, R. Development and Evaluation of a Novel Real-Time PCR for Pan-Dermatophyte Detection in Nail Specimens. Mycopathologia 2016, 181, 51–57. [Google Scholar] [CrossRef] [PubMed]
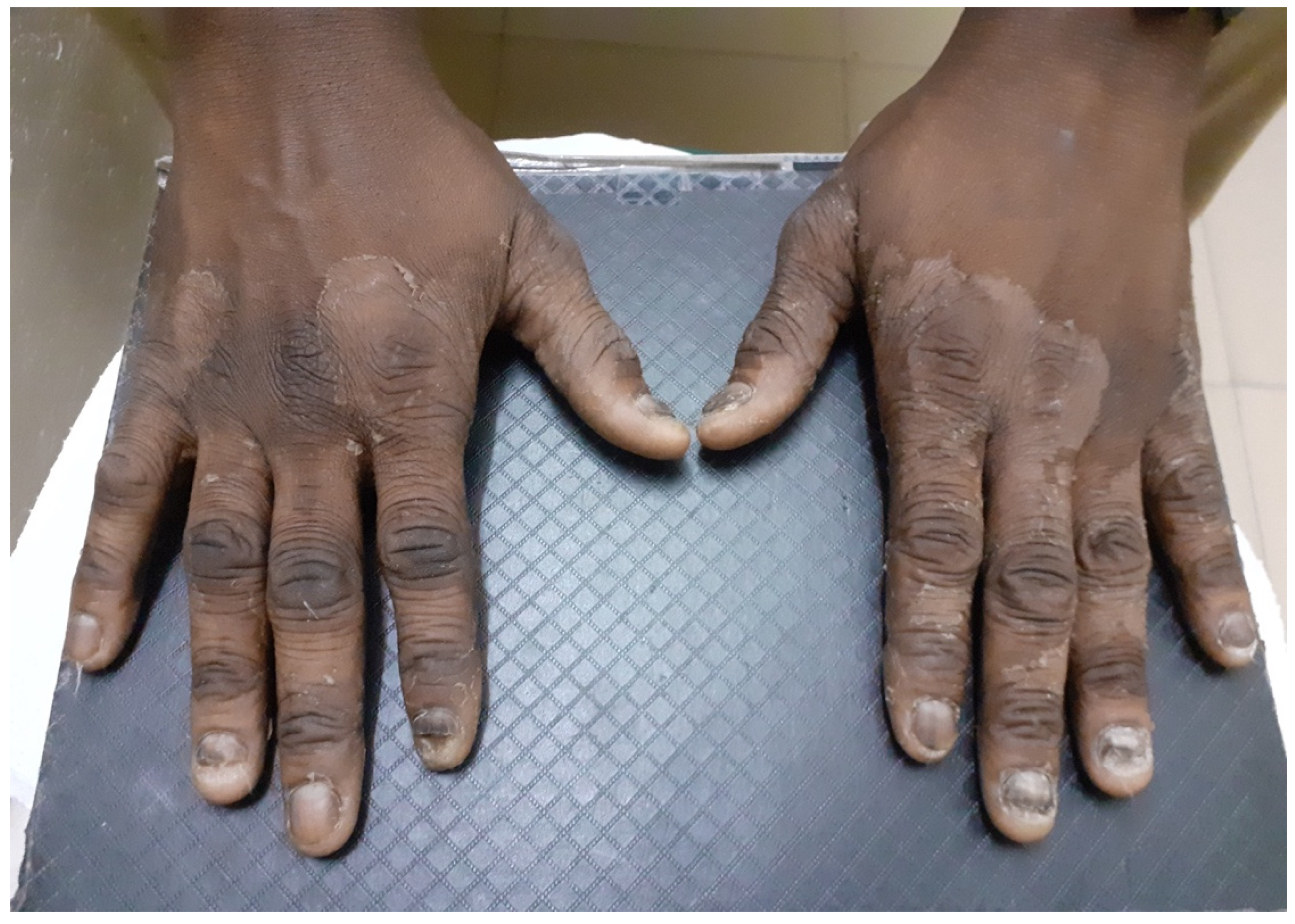
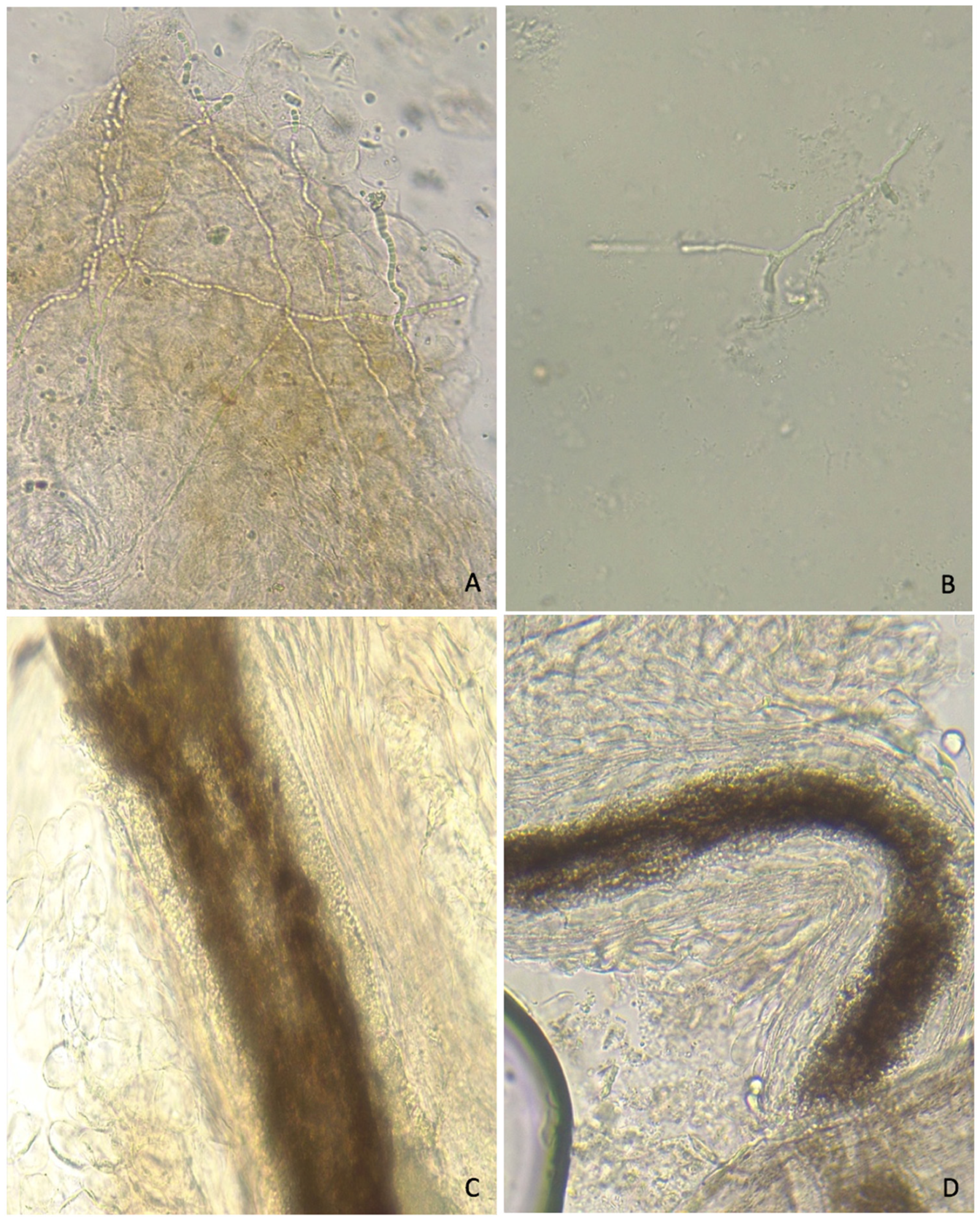
| Direct Microscopic Examination (n = 51) | Hair (n = 62) | Skin (n = 24) | Nail (n = 19) | |
|---|---|---|---|---|
| 21 (33.87) | 19 (79.2) | 11 (57.89) | ||
| Culture (n = 45) | Dermatophytes (n = 26) | |||
| T. soudanense | 11 | 0 | 1 | |
| M. audouinii * | 4 | 0 | 0 | |
| T. mentagrophytes | 3 | 0 | 1 | |
| T. rubrum | 3 | 1 | 1 | |
| T. interdigitale | 0 | 1 | 0 | |
| Yeasts (n = 15) | ||||
| C. albicans | 0 | 3 | 3 | |
| Non-albicans Candida | 0 | 4 | 3 | |
| Trichosporon sp. | 0 | 1 | 1 | |
| Non-dermatophyte filamentous fungi (n = 4) | ||||
| Fusarium sp. | 0 | 2 | 0 | |
| Fusarium sp./C. albicans | 0 | 1 | 1 | |
| IH-PCR | CK-PCR | |||||
|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Total | ||
| Gold Standard | Positive | 23 | 3 | 17 | 9 * | 26 |
| Negative | 15 | 64 | 4 | 75 | 79 | |
| Total | 38 | 67 | 21 | 84 | 105 | |
| Parameters | IH | 95% CI | CK | 95% CI |
|---|---|---|---|---|
| Sensitivity | 0.89 | [0.73–0.97] | 0.65/(0.77 *) | [0.47–0.84] |
| Specificity | 0.82 | [0.77–0.85] | 0.95 | [0.90–0.99] |
| PPV | 0.63 | [0.52–0.69] | 0.85 | [0.70–1.0] |
| NPV | 0.96 | [0.89–0.998] | 0.88 | [0.81–0.95] |
| Youden’s index | 0.71 | [0.49–0.82] | 0.43 | [0.21–0.60] |
| NND | 1.41 | [1.22–2.03] | 2.33 | [1.67–4.68] |
| DOR | 36.57 | [8.67–179.04] | 10.92 | [3.29–37.63] |
| IH | 95% CI | CK | 95% CI | |
|---|---|---|---|---|
| Sensitivity | 0.86 | [0.68–0.96] | 0.57 (0.82 *) | [0.36–0.78] |
| Specificity | 0.85 | [0.76–0.91] | 0.93 | [0.84–0.98] |
| PPV | 0.75 | [0.59–0.84] | 0.75 | [0.46–0.93] |
| NPV | 0.92 | [0.82–0.98] | 0.76 | [0.69–0.80] |
| Youden’s index | 0.71 | [0.44–0.86] | 0.36 | [0.10–0.51] |
| NND | 1.41 | [1.16–2.29] | 2.81 | [1.96–9.62] |
| DOR | 35.0 | [6.63–218.02] | 9.50 | [1.89–53.92] |
| IH | 95% CI | CK | 95% CI | |
|---|---|---|---|---|
| Sensitivity | 1.00 | [0.21–1.00] | 1.00 | [0.210–1.00] |
| Specificity | 0.73 | [0.66–0.73] | 0.86 | [0.79–0.86] |
| PPV | 0.25 | [0.05–0.25] | 0.40 | [0.08–0.40] |
| NPV | 1.00 | [0.90–1.00] | 1.00 | [0.92–1.00] |
| Youden’s index | 0.73 | [−0.14–0.73] | 0.73 | [−0.15–0.73] |
| NND | 1.385 | [1.38–7.23] | 1.16 | [1.16–611.70] |
| DOR | +∞ | [0.50–+∞] | +∞ | [1.01–+∞] |
| IH | 95% CI | CK | 95% CI | |
|---|---|---|---|---|
| Sensitivity | 1.00 | [0.34–1.00] | 1.00 | [0.36–1.00] |
| Specificity | 0.81 | [0.69–0.81] | 0.94 | [0.82–0.94] |
| PPV | 0.50 | [0.17–0.50] | 0.750 | [0.27–0.75] |
| NPV | 1.00 | [0.85–1.00] | 1.00 | [0.87–1.00] |
| Youden’s index | 0.81 | [0.03–0.81] | 0.94 | [0.17–0.94] |
| NND | 1.23 | [1.2–31.49] | 1.07 | [1.07–5.80] |
| DOR | 1.155 | [−∞–+∞] | +∞ | [2.46–+∞] |
| Hair (n = 62) | IH (+) = 24 | CD (+) = 18 | FP = 4 | |
| CD (−) = 6 | Seq (+) = 2 | |||
| Seq (−) = 4 | ||||
| IH (−) = 38 | CD (−) = 35 | FN = 3 | ||
| CD (+) = 3 | ||||
| CK (+) = 14 | CD (+) = 12 | FP = 0 | ||
| CD (−) = 2 | Seq (+) = 2 | |||
| Seq (−) = 0 | ||||
| CK (−) = 48 | CD (−) = 41 | FN = 5 * | ||
| CD (+) = 7 | Seq (−) = 5 | |||
| Seq (+) = 2 | ||||
| Skin (n = 24) | IH (+) = 9 | CD (+) = 2 | FP = 4 | |
| CD (−) = 7 | Seq (+) = 3 | |||
| Seq (−) = 4 | ||||
| IH (−) = 15 | CD (−) = 15 | FN = 0 | ||
| CD (+) = 0 | ||||
| CK (+) = 4 | CD (+) = 2 | FP = 1 | ||
| CD (−) = 2 | Seq (+) = 1 | |||
| Seq (−) = 1 | ||||
| CK (−) = 20 | CD (−) = 16 | FN = 2 | ||
| CD (+) = 4 | Seq (−) = 2 | |||
| Seq (+) = 2 | ||||
| Nails (n = 19) | IH (+) = 6 | CD (+) = 3 | FP = 3 | |
| CD (−) = 3 | Seq (+) = 0 | |||
| Seq (−) = 3 | ||||
| IH (−) = 13 | CD (−) = 13 | FN = 0 | ||
| CD (+) = 0 | ||||
| CK (+) = 3 | CD (+) = 3 | FP = 0 | ||
| CD (−) = 0 | ||||
| CK (−) = 16 | CD (−) = 16 | FN = 0 | ||
| CD (+) = 0 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kabtani, J.; Diongue, K.; Dione, J.-N.; Delmas, A.; L’Ollivier, C.; Amoureux, M.-C.; Ndiaye, D.; Ranque, S. Real-Time PCR Assay for the Detection of Dermatophytes: Comparison between an In-House Method and a Commercial Kit for the Diagnosis of Dermatophytoses in Patients from Dakar, Senegal. J. Fungi 2021, 7, 949. https://doi.org/10.3390/jof7110949
Kabtani J, Diongue K, Dione J-N, Delmas A, L’Ollivier C, Amoureux M-C, Ndiaye D, Ranque S. Real-Time PCR Assay for the Detection of Dermatophytes: Comparison between an In-House Method and a Commercial Kit for the Diagnosis of Dermatophytoses in Patients from Dakar, Senegal. Journal of Fungi. 2021; 7(11):949. https://doi.org/10.3390/jof7110949
Chicago/Turabian StyleKabtani, Jihane, Khadim Diongue, Jean-Noël Dione, Anne Delmas, Coralie L’Ollivier, Marie-Claude Amoureux, Daouda Ndiaye, and Stéphane Ranque. 2021. "Real-Time PCR Assay for the Detection of Dermatophytes: Comparison between an In-House Method and a Commercial Kit for the Diagnosis of Dermatophytoses in Patients from Dakar, Senegal" Journal of Fungi 7, no. 11: 949. https://doi.org/10.3390/jof7110949
APA StyleKabtani, J., Diongue, K., Dione, J.-N., Delmas, A., L’Ollivier, C., Amoureux, M.-C., Ndiaye, D., & Ranque, S. (2021). Real-Time PCR Assay for the Detection of Dermatophytes: Comparison between an In-House Method and a Commercial Kit for the Diagnosis of Dermatophytoses in Patients from Dakar, Senegal. Journal of Fungi, 7(11), 949. https://doi.org/10.3390/jof7110949








