Yeast Plasma Membrane Fungal Oligopeptide Transporters Display Distinct Substrate Preferences despite Their High Sequence Identity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Fermentation Conditions
2.2. Strain and Plasmid Construction
2.3. Phenotype Microarray Assays for Di- and Tripeptide Consumption
2.4. Gene Expression Analysis
2.5. Epifluorescence and Confocal Microscopy
2.6. Analysis of Promoter Regions
2.7. Data Treatment and Statistical Analysis
3. Results
3.1. Evaluation of Substrate Specificity in Fot Family Members by Phenotype Microarrays
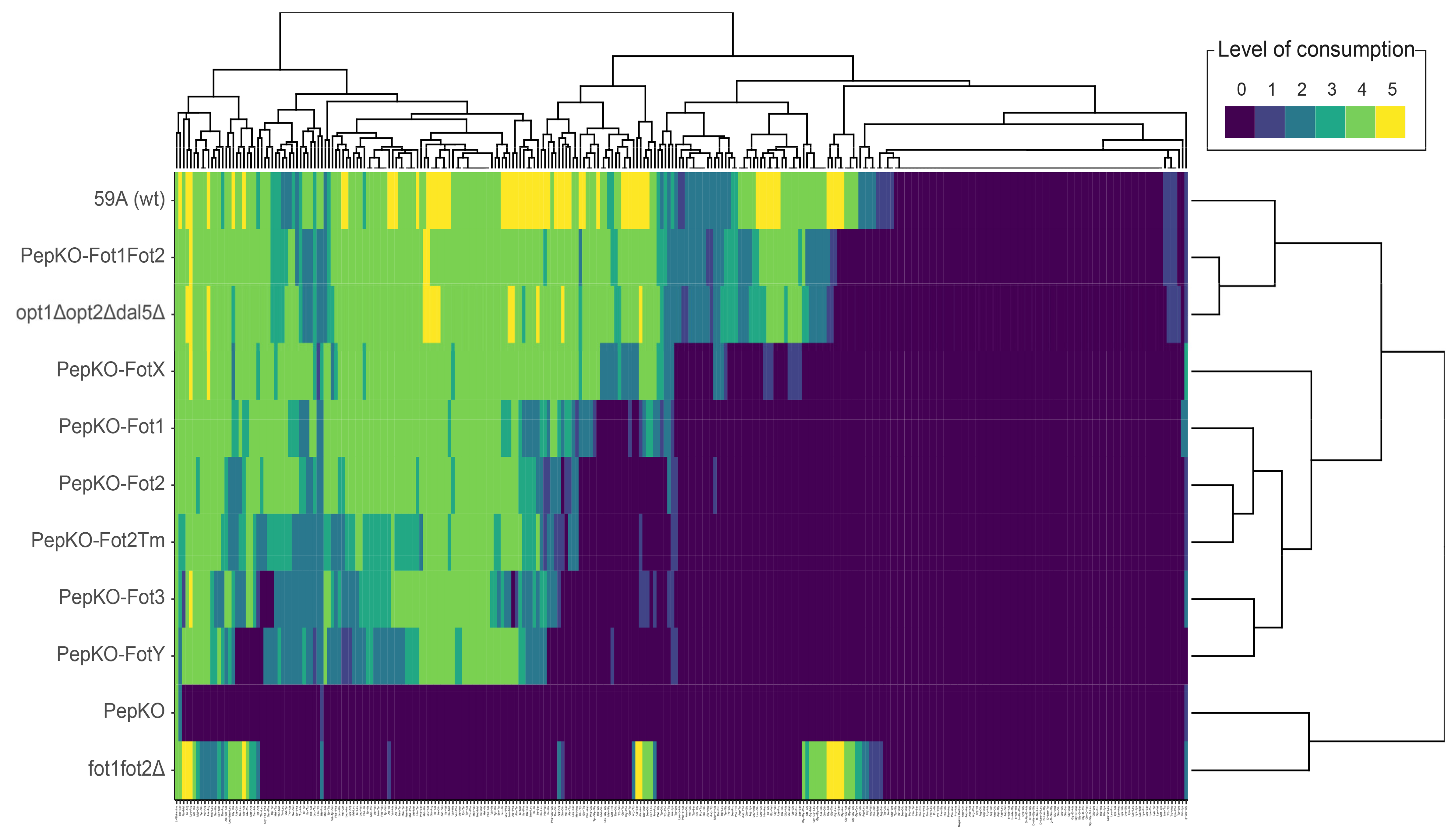
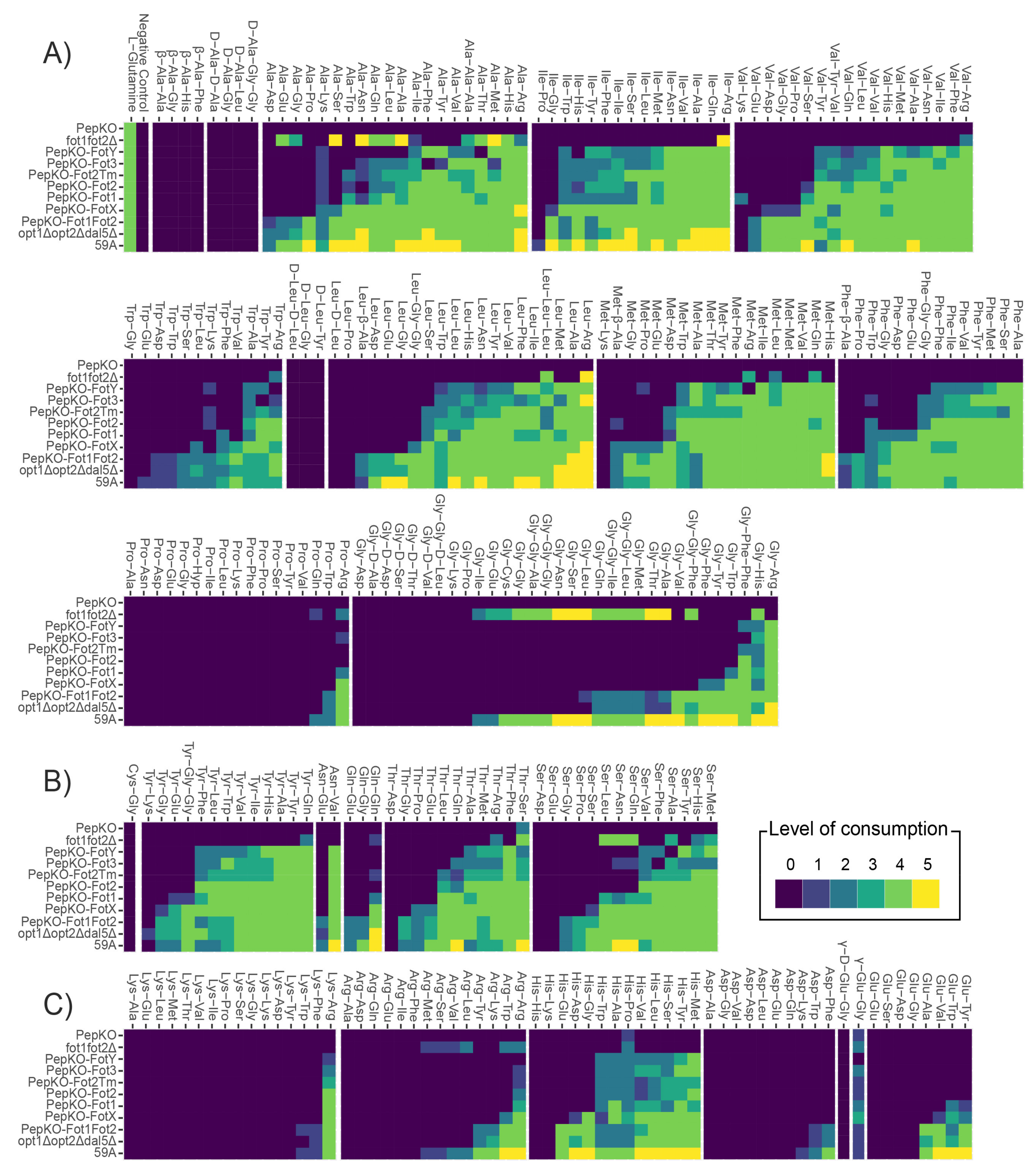
3.2. Specificity of Fot Members Depends on the Type of Amino Acid Located in the Oligopeptide N-Terminus
3.3. FOT Genes Expression Depends on the S. cerevisiae Strain Background, Type of Nitrogen Source and Stage of Enological Fermentation
3.4. Fot1 Is Located in S. cerevisiae Plasma Membrane
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perry, J.R.; Basrai, A.M.; Steiner, H.Y.; Naider, F.; Becker, J.M. Isolation and Characterization of a Saccharomyces cerevisiae Peptide Transport Gene. Mol. Cell. Biol. 1994, 14, 104–115. [Google Scholar] [CrossRef]
- Nelissen, B.; De Wachter, R.; Goffeau, A. Classification of all Putative Permeases and Other Membrane Plurispanners of the Major Facilitator Superfamily Encoded by the Complete Genome of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 1997, 21, 113–134. [Google Scholar] [CrossRef]
- Hauser, M.; Donhardt, A.M.; Barnes, D.; Naider, F.; Becker, J.M. Enkephalins are Transported by a Novel Eukaryotic Peptide Uptake System. J. Biol. Chem. 2000, 275, 3037–3041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourbouloux, A.; Shahi, P.; Chakladar, A.; Delrot, S.; Bachhawat, A.K. Hgt1p, a High Affinity Glutathione Transporter from the Yeast Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 13259–13265. [Google Scholar] [CrossRef] [Green Version]
- Damon, C.; Vallon, L.; Zimmermann, S.; Haider, M.Z.; Galeote, V.; Dequin, S.; Luis, P.; Fraissinet-Tachet, L.; Marmeisse, R. A Novel Fungal Family of Oligopeptide Transporters Identified by Functional Metatranscriptomics of Soil Eukaryotes. ISME J. 2011, 5, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Novo, M.; Bigey, F.; Beyne, E.; Galeote, V.; Gavory, F.; Mallet, S.; Cambon, B.; Legras, J.-L.; Wincker, P.; Casaregola, S.; et al. Eukaryote-to-Eukaryote Gene Transfer Events Revealed by the Genome Sequence of the Wine Yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. USA 2009, 106, 16333–16338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsit, S.; Mena, A.; Bigey, F.; Sauvage, F.-X.; Couloux, A.; Guy, J.; Legras, J.-L.; Barrio, E.; Dequin, S.; Galeote, V. Evolutionary Advantage Conferred by an Eukaryote-to-Eukaryote Gene Transfer Event in Wine Yeasts. Mol. Biol. Evol. 2015, 32, 1695–1707. [Google Scholar] [CrossRef] [Green Version]
- Marsit, S.; Sanchez, I.; Galeote, V.; Dequin, S. Horizontally Acquired Oligopeptide Transporters Favour Adaptation of Saccharomyces cerevisiae Wine Yeast to Oenological Environment. Environ. Microbiol. 2016, 18, 1148–1161. [Google Scholar] [CrossRef] [PubMed]
- Duc, C.; Maçna, F.; Sanchez, I.; Galeote, V.; Delpech, S.; Silvano, A.; Mouret, J.-R. Large-Scale Screening of Thiol and Fermentative Aroma Production during Wine Alcoholic Fermentation: Exploring the Effects of Assimilable Nitrogen and Peptides. Fermentation 2020, 6, 98. [Google Scholar] [CrossRef]
- Becerra-Rodríguez, C.; Marsit, S.; Galeote, V. Diversity of Oligopeptide Transport in Yeast and Its Impact on Adaptation to Winemaking Conditions. Front. Genet. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Ambroset, C.; Petit, M.; Brion, C.; Sanchez, I.; Delobel, P.; Guérin, C.; Chiapello, H.; Nicolas, P.; Bigey, F.; Dequin, S.; et al. Deciphering the Molecular Basis of Wine Yeast Fermentation Traits Using a Combined Genetic and Genomic Approach. G3 Genes Genomes Gen. 2011, 1, 263–281. [Google Scholar] [CrossRef] [Green Version]
- Coi, A.L.; Legras, J.-L.; Zara, G.; Dequin, S.; Budroni, M. A Set of Haploid Strains Available for Genetic Studies of Saccharomyces cerevisiaeflor Yeasts. FEMS Yeast Res. 2016, 16, fow066. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Shirogane, T.; Liu, D.; Harper, J.; Elledge, S.J. Exit from Exit: Resetting the Cell Cycle through Amn1 Inhibition of G Protein Signaling. Cell 2003, 112, 697–709. [Google Scholar] [CrossRef] [Green Version]
- Yvert, G.; Brem, R.B.; Whittle, J.; Akey, J.M.; Foss, E.; Smith, E.N.; Mackelprang, R.; Kruglyak, L. Trans-Acting Regulatory Variation in Saccharomyces cerevisiae and the Role of Transcription Factors. Nat. Genet. 2003, 35, 57–64. [Google Scholar] [CrossRef]
- Bely, M.; Sablayrolles, J.-M.; Barre, P. Automatic Detection of Assimilable Nitrogen Deficiencies during Alcoholic Fermentation in Oenological Conditions. J. Ferment. Bioeng. 1990, 70, 246–252. [Google Scholar] [CrossRef]
- Stovicek, V.; Borodina, I.; Forster, J. CRISPR–Cas System Enables Fast and Simple Genome Editing of Industrial Saccharomyces cerevisiae Strains. Metab. Eng. Commun. 2015, 2, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Mans, R.; Van Rossum, H.M.; Wijsman, M.; Backx, A.; Kuijpers, N.G.; Broek, M.V.D.; Daran-Lapujade, P.; Pronk, J.; van Maris, A.; Daran, J.-M. CRISPR/Cas9: A Molecular Swiss Army Knife for Simultaneous Introduction of Multiple Genetic Modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gietz, R.D.; Schiestl, R.H. High-Efficiency Yeast Transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef]
- Homann, O.R.; Cai, H.; Becker, J.M.; Lindquist, S.L. Harnessing Natural Diversity to Probe Metabolic Pathways. PLoS Genet. 2005, 1, e80. [Google Scholar] [CrossRef]
- Vida, A.T.; Emr, S.D. A New Vital Stain for Visualizing Vacuolar Membrane Dynamics and Endocytosis in Yeast. J. Cell Biol. 1995, 128, 779–792. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Boer, C.G.; Hughes, T.R. YeTFaSCo: A Database of Evaluated Yeast Transcription Factor Sequence Specificities. Nucleic Acids Res. 2012, 40, D169–D179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Galili, T.; O’Callaghan, A.; Sidi, J.; Sievert, C. Heatmaply: An R Package for Creating Interactive Cluster Heatmaps for Online Publishing. Bioinformatics 2018, 34, 1600–1602. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Texeira, M.; Van Zeebroeck, G.; Thevelein, J. Peptides Induce Persistent Signaling from Endosomes by a Nutrient Transceptor. Nat. Chem. Biol. 2012, 8, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Hauser, M.; Narita, V.; Donhardt, A.M.; Naider, F.; Becker, J.M. Multiplicity and Regulation of Genes Encoding Peptide Transporters in Saccharomyces cerevisiae. Mol. Membr. Biol. 2001, 18, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Wiles, A.M.; Cai, H.; Naider, F.; Becker, J.M. Nutrient Regulation of Oligopeptide Transport in Saccharomyces cerevisiae. Microbiology 2006, 152, 3133–3145. [Google Scholar] [CrossRef] [Green Version]
- Ito, K.; Hikida, A.; Kawai, S.; Lan, V.T.T.; Motoyama, T.; Kitagawa, S.; Yoshikawa, Y.; Kato, R.; Kawarasaki, Y. Analysing the Substrate Multispecificity of a Proton-Coupled Oligopeptide Transporter using a Dipeptide Library. Nat. Commun. 2013, 4, 2502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, H.; Hauser, M.; Naider, F.; Becker, J.M. Differential Regulation and Substrate Preferences in Two Peptide Transporters of Saccharomyces cerevisiae. Eukaryot. Cell 2007, 6, 1805–1813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinders, A.; Schulze, W.; Kühn, C.; Barker, L.; Schulz, A.; Ward, J.; Frommer, W.B. Protein–Protein Interactions between Sucrose Transporters of Different Affinities Colocalized in the Same Enucleate Sieve Element. Plant Cell 2002, 14, 1567–1577. [Google Scholar] [CrossRef] [Green Version]
- Alguel, Y.; Cameron, A.D.; Diallinas, G.; Byrne, B. Transporter Oligomerization: Form and Function. Biochem. Soc. Trans. 2016, 44, 1737–1744. [Google Scholar] [CrossRef] [Green Version]
- Duc, C.; Pradal, M.; Sanchez, I.; Noble, J.; Tesnière, C.; Blondin, B. A Set of Nutrient Limitations Trigger Yeast Cell Death in a Nitrogen-Dependent Manner during Wine Alcoholic Fermentation. PLoS ONE 2017, 12, e0184838. [Google Scholar] [CrossRef] [PubMed]
- Devia, J.; Bastías, C.; Kessi, E.; Villarroel, C.A.; De Chiara, M.; Cubillos, F.; Liti, G.; Martínez, C.; Salinas, F. Transcriptional Activity and Protein Levels of Horizontally Acquired Genes in Yeast Reveal Hallmarks of Adaptation to Fermentative Environments. Front. Genet. 2020, 11. [Google Scholar] [CrossRef]
- Bon, E.; Carvajal, E.; Stanbrough, M.; Rowen, D.; Magasanik, B. Asparaginase II of Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 1997, 63–65, 203–212. [Google Scholar] [CrossRef]
- Lubkowitz, M.A.; Barnes, D.; Breslav, M.; Burchfield, A.; Naider, F.; Becker, J.M. Schizosaccharomyces pombe isp4 encodes a Transporter Representing a Novel Family of Oligopeptide Transporters. Mol. Microbiol. 1998, 28, 729–741. [Google Scholar] [CrossRef] [Green Version]
- Magasanik, B.; Kaiser, A.C. Nitrogen Regulation in Saccharomyces cerevisiae. Gene 2002, 290, 1–18. [Google Scholar] [CrossRef]
- Island, M.D.; Naider, F.; Becker, J.M. Regulation of Dipeptide Transport in Saccharomyces cerevisiae by Micromolar Amino Acid Concentrations. J. Bacteriol. 1987, 169, 2132–2136. [Google Scholar] [CrossRef] [Green Version]
- Rai, R.; Genbauffe, F.; Lea, H.Z.; Cooper, T.G. Transcriptional Regulation of the DAL5 Gene in Saccharomyces cerevisiae. J. Bacteriol. 1987, 169, 3521–3524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bleve, G.; Zacheo, G.; Cappello, M.S.; Dellaglio, F.; Grieco, F. Subcellular Localization and Functional Expression of the Glycerol Uptake Protein 1 (GUP1) of Saccharomyces cerevisiae tagged with Green Fluorescent Protein. Biochem. J. 2005, 390, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Feilmeier, B.; Iseminger, G.; Schroeder, D.; Webber, H.; Phillips, G.J. Green Fluorescent Protein Functions as a Reporter for Protein Localization in Escherichia coli. J. Bacteriol. 2000, 182, 4068–4076. [Google Scholar] [CrossRef] [Green Version]
- Hyde, R.; Cwiklinski, E.L.; MacAulay, K.; Taylor, P.M.; Hundal, H.S. Distinct Sensor Pathways in the Hierarchical Control of SNAT2, a Putative Amino Acid Transceptor, by Amino Acid Availability. J. Biol. Chem. 2007, 282, 19788–19798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Albers, T.; Fiumera, H.L.; Gameiro, A.; Grewer, C. A Conserved Na+ Binding Site of the Sodium-coupled Neutral Amino Acid Transporter 2 (SNAT2). J. Biol. Chem. 2009, 284, 25314–25323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gueldener, U. A Second Set of loxP Marker Cassettes for Cre-Mediated Multiple Gene Knockouts in Budding Yeast. Nucleic Acids Res. 2002, 30, 23e. [Google Scholar] [CrossRef] [Green Version]
- Breslow, D.; Cameron, D.M.; Collins, S.; Schuldiner, M.; Stewart-Ornstein, J.; Newman, H.W.; Braun, S.; Madhani, H.; Krogan, N.J.; Weissman, J.S. A Comprehensive Strategy Enabling High-Resolution Functional Analysis of the Yeast Genome. Nat. Methods 2008, 5, 711–718. [Google Scholar] [CrossRef] [Green Version]


| Strain | Genotype | Source/Reference |
|---|---|---|
| 59A | MATa ho amn1Δ::LOXP | [7] |
| fot1fot2Δ | MATa ho amn1Δ::LOXP fot1fot2Δ::KANMX4 | This study |
| opt1Δopt2Δdal5Δ | MATa ho amn1Δ::LOXP opt1Δ opt2Δ dal5Δ | This study |
| PepKO | MATa ho amn1Δ::LOXP fot1fot2Δ::KANMX4 opt1Δ opt2Δ dal5Δ | This study |
| PepKO-Fot1 | MATa ho amn1Δ::LOXP opt1Δ opt2Δ dal5Δ fot1fot2Δ::FOT1 | This study |
| PepKO-Fot2 | MATa ho amn1Δ::LOXP opt1Δ opt2Δ dal5Δ fot1fot2Δ::FOT2 | This study |
| PepKO-Fot3 | MATa ho amn1Δ::LOXP opt1Δ opt2Δ dal5Δ fot1fot2Δ::FOT3 | This study |
| PepKO-FotX | MATa ho amn1Δ::LOXP opt1Δ opt2Δ dal5Δ fot1fot2Δ::FOTX | This study |
| PepKO-FotY | MATa ho amn1Δ::LOXP opt1Δ opt2Δ dal5Δ fot1fot2Δ::FOTY | This study |
| PepKO-Fot2Tm | MATa ho amn1Δ::LOXP opt1Δ opt2Δ dal5Δ fot1fot2Δ::FOT2Tm | This study |
| PepKO-Fot1Fot2 | MATa ho amn1Δ::LOXP opt1Δ opt2Δ dal5Δ fot1fot2Δ::FOT1–FOT2 | This study |
| 59A-GFP | MATa ho amn1Δ::TEFp-GFP-ADH1-NATMX4 | [7] |
| MTF2533 | MATa ho::LOXP | [12] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Becerra-Rodríguez, C.; Taghouti, G.; Portier, P.; Dequin, S.; Casal, M.; Paiva, S.; Galeote, V. Yeast Plasma Membrane Fungal Oligopeptide Transporters Display Distinct Substrate Preferences despite Their High Sequence Identity. J. Fungi 2021, 7, 963. https://doi.org/10.3390/jof7110963
Becerra-Rodríguez C, Taghouti G, Portier P, Dequin S, Casal M, Paiva S, Galeote V. Yeast Plasma Membrane Fungal Oligopeptide Transporters Display Distinct Substrate Preferences despite Their High Sequence Identity. Journal of Fungi. 2021; 7(11):963. https://doi.org/10.3390/jof7110963
Chicago/Turabian StyleBecerra-Rodríguez, Carmen, Géraldine Taghouti, Perrine Portier, Sylvie Dequin, Margarida Casal, Sandra Paiva, and Virginie Galeote. 2021. "Yeast Plasma Membrane Fungal Oligopeptide Transporters Display Distinct Substrate Preferences despite Their High Sequence Identity" Journal of Fungi 7, no. 11: 963. https://doi.org/10.3390/jof7110963
APA StyleBecerra-Rodríguez, C., Taghouti, G., Portier, P., Dequin, S., Casal, M., Paiva, S., & Galeote, V. (2021). Yeast Plasma Membrane Fungal Oligopeptide Transporters Display Distinct Substrate Preferences despite Their High Sequence Identity. Journal of Fungi, 7(11), 963. https://doi.org/10.3390/jof7110963






