Re-Evaluation of the Taxonomy of Talaromyces minioluteus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains
2.2. DNA Isolation, Amplification and Sequence Analysis
2.3. Morphology
3. Results
3.1. Phylogeny
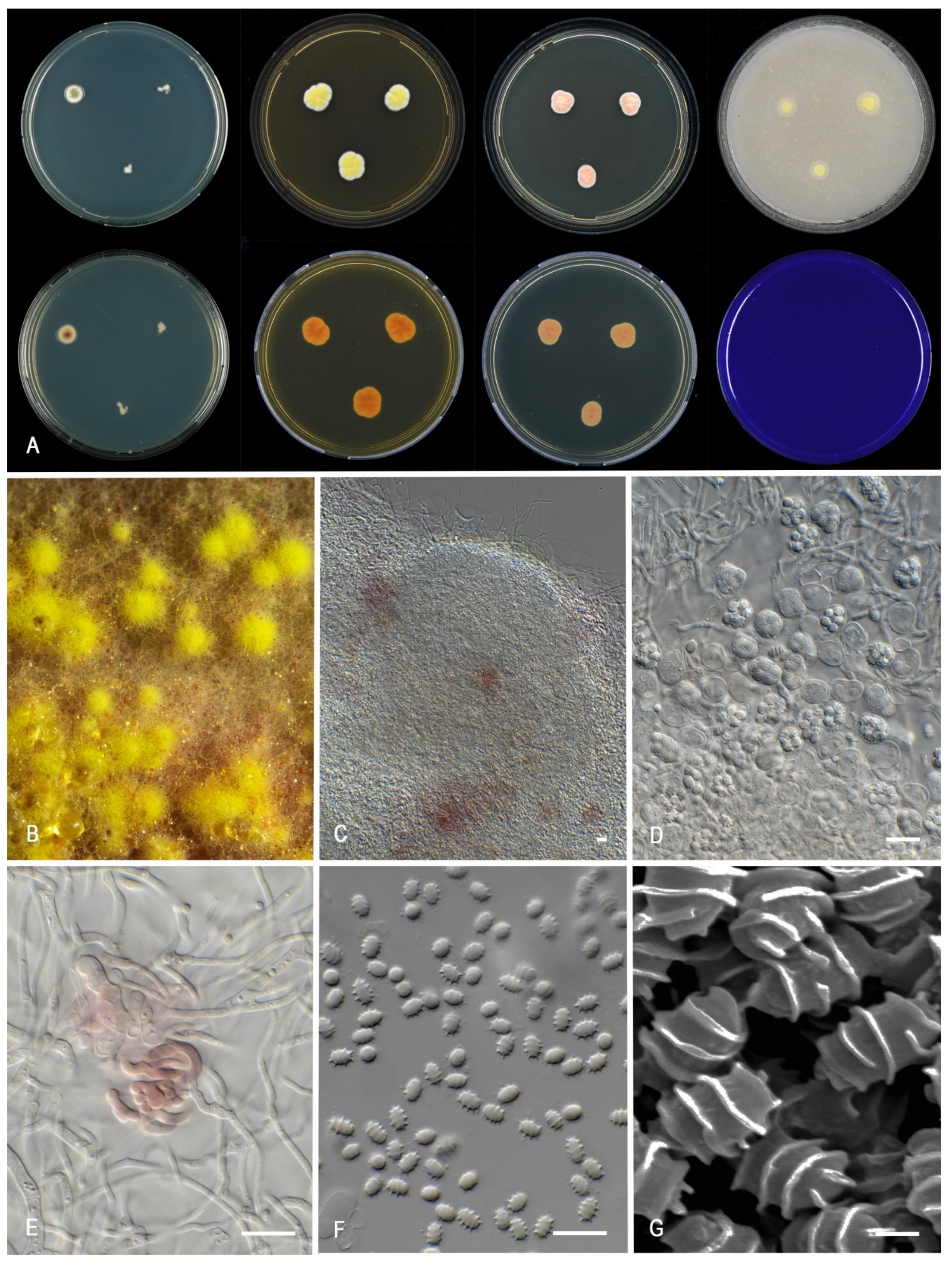
3.2. Morphology
3.3. Taxonomy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stošić, S.; Ristić, D.; Gašić, K.; Starović, M.; Ljaljević Grbić, M.; Vukojević, J.; Živković, S. Talaromyces minioluteus: New postharvest fungal pathogen in Serbia. Plant Dis. 2020, 104, 656–667. [Google Scholar] [CrossRef]
- Birkinshaw, J.H.; Raistrick, H. Studies in the biochemistry of micro-organisms: The metabolic products of Penicillium minioluteum Dierckx. Minioluteic acid. Biochem. J. 1934, 28, 828–836. [Google Scholar] [CrossRef] [Green Version]
- van Reenen-Hoekstra, E.S.; Frisvad, J.C.; Samson, R.A.; Stolk, A.C. The Penicillium funiculosum Complex—Well Defined Species and Problematic Taxa. In Modern Concepts in Penicillium and Aspergillus Classification; Samson, R.A., Pitt, J.I., Eds.; Springer: Boston, MA, USA, 1990; pp. 173–192. [Google Scholar]
- Yilmaz, N.; Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 2014, 78, 175–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitt, J.I. The Genus Penicillium and Its Teleomorphic States Eupenicillium and Talaromyces; Academic Press: London, UK, 1980. [Google Scholar]
- Malloch, D. The Trichocomaceae: Relationships with other Ascomycetes. In Advances in Penicillium and Aspergillus Systematics; Samson, R.A., Pitt, J.I., Eds.; Plenum Press: New York, NY, USA, 1985; pp. 365–382. [Google Scholar]
- Berbee, M.L.; Yoshimura, A.; Sugiyama, J. Is Penicillium monophyletic? An evaluation of phylogeny in the family Trichocomaceae from 18S, 5.8S and ITS ribosomal DNA sequence data. Mycologia 1995, 87, 210–222. [Google Scholar] [CrossRef]
- Houbraken, J.; Samson, R.A. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud. Mycol. 2011, 70, 1–51. [Google Scholar] [CrossRef] [PubMed]
- Houbraken, J.; Kocsubé, S.; Visagie, C.M. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A.; Yilmaz, N.; Houbraken, J.; Spierenburg, H.; Seifert, K.A.; Peterson, S.W.; Varga, J.; Frisvad, J.C. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud. Mycol. 2011, 70, 159–183. [Google Scholar] [CrossRef] [Green Version]
- Dierckx, R.P. Un Essai de revision du genre Penicillium Link. Ann. Soc. Sci. 1901, 25, 83–89. [Google Scholar]
- Raper, K.B.; Thom, C. Manual of the Penicillia; Williams & Wilkins Co.: Baltimore, MD, USA, 1949. [Google Scholar]
- Yilmaz, N.; Houbraken, J.; Hoekstra, E.S.; Frisvad, J.C.; Visagie, C.M.; Samson, R.A. Delimitation and characterisation of Talaromyces purpurogenus and related species. Persoonia 2012, 29, 39–54. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, C.; Martinez, A.T. Four new species of Penicillium isolated from different substrata. Mycopathologia 1981, 74, 163–171. [Google Scholar] [CrossRef]
- Quintanilla, J.A. Three new species of Penicillium belonging to subgenus Biverticillium Dierckx, isolated from different substrates. Mycopathologia 1985, 91, 69–78. [Google Scholar] [CrossRef]
- Guevara-Suarez, M.; Sutton, D.A.; Gené, J.; García, D.; Wiederhold, N.; Guarro, J.; Cano-Lira, J.G. Four new species of Talaromyces from clinical sources. Mycoses 2017, 60, 651–662. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Wang, X.C.; Zhuang, W.Y.; Cheng, X.H.; Zhao, P. New species of Talaromyces (Fungi) isolated from soil in Southwestern China. Biology 2021, 10, 745. [Google Scholar] [CrossRef] [PubMed]
- Visagie, C.M.; Hirooka, Y.; Tanney, J.B.; Whitfield, E.; Mwange, K.; Meijer, M.; Amend, A.S.; Seifert, K.A.; Samson, R.A. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud. Mycol. 2014, 78, 63–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [Green Version]
- Hong, S.-B.; Go, S.-J.; Shin, H.-D.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 2005, 97, 1316–1329. [Google Scholar] [CrossRef]
- Masclaux, F.; Guého, E.; de Hoog, G.S. Phylogenetic relationships of human-pathogenic Cladosporium (Xylohypha) species inferred from partial LS rRNA sequences. J. Med. Vet. Mycol. 1995, 33, 327–338. [Google Scholar] [CrossRef]
- de Hoog, G.S.; Gerrits van den Ende, A.H.G. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycosis 1998, 41, 183–189. [Google Scholar] [CrossRef]
- Houbraken, J.; Spierenburg, H.; Frisvad, J.C. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antonie Van Leeuwenhoek 2012, 101, 403–421. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Conference on Extreme Science and Engineering Discovery Environment: Gateway to Discovery, San Diego, CA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samson, R.A.; Visagie, C.M.; Houbraken, J.; Hong, S.B.; Hubka, V.; Klaassen, C.H.; Perrone, G.; Seifert, K.A.; Susca, A.; Tanney, J.B.; et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014, 78, 141–173. [Google Scholar] [CrossRef] [Green Version]
- Kornerup, A.; Wanscher, J.H. Methuen Handbook of Colour, 2nd ed.; Methuen: London, UK, 1967. [Google Scholar]
- Stolk, A.C.; Samson, R.A. The genus Talaromyces—Studies on Talaromyces and related genera II. Stud. Mycol. 1972, 2, 1–65. [Google Scholar]
- Thom, C. The Penicillium luteum-purpurogenum group. Mycologia 1915, 7, 134–142. [Google Scholar] [CrossRef]
- Peterson, S.W.; Jurjević, Ž. The Talaromyces pinophilus species complex. Fungal Biol. 2019, 123, 745–762. [Google Scholar] [CrossRef] [PubMed]
- Tranchida, M.C.; Centeno, N.D.; Stenglein, S.A.; Cabello, M.N. First record of Talaromyces udagawae in soil related to decomposing human remains in Argentina. Rev. Argent. Microbiol. 2016, 48, 86–90. [Google Scholar] [CrossRef] [Green Version]
- Taylor, J.W.; Jacobson, D.J.; Kroken, S.; Kasuga, T.; Geiser, D.M.; Hibbett, D.S.; Fisher, M.C. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 2000, 31, 21–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hubka, V.; Barrs, V.; Dudová, Z.; Sklenář, F.; Kubátová, A.; Matsuzawa, T.; Yaguchi, T.; Horie, Y.; Nováková, A.; Frisvad, J.C.; et al. Unravelling species boundaries in the Aspergillus viridinutans complex (section Fumigati): Opportunistic human and animal pathogens capable of interspecific hybridization. Persoonia 2018, 41, 142–174. [Google Scholar] [CrossRef]
- Prigione, V.; Trocini, B.; Spina, F.; Poli, A.; Romanisio, D.; Giovando, S.; Varese, G.C. Fungi from industrial tannins: Potential application in biotransformation and bioremediation of tannery wastewaters. Appl. Microbiol. Biotechnol. 2018, 102, 4203–4216. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, J.; Kiyuna, T.; An, K.-D.; Nagatsuka, Y.; Handa, Y.; Tazato, N.; Hata-Tomita, J.; Nishijima, M.; Koide, T.; Yaguchi, Y.; et al. Microbiological survey of the stone chambers of Takamatsuzuka and Kitora tumuli, Nara Prefecture, Japan: A milestone in elucidating the cause of biodeterioration of mural paintings. In Study of Environmental Conditions Surrounding Cultural Properties and Their Protective Measures; Sano, C., Ed.; National Research Institute for Cultural Properties: Tokyo, Japan, 2009; pp. 51–73. [Google Scholar]












| Species Name | Section | Strain Numbers | Location, Substrate | GenBank Accession Number 1 | |||
|---|---|---|---|---|---|---|---|
| BenA | CaM | RPB2 | ITS | ||||
| Talaromyces calidominioluteus | Trachyspermi | DTO 039-I2 = CBS 113167 = IBT 18572 | Unknown location; air in cake factory | OK338785 | OK338816 | OK338836 | OK339611 |
| T. calidominioluteus | Trachyspermi | DTO 052-G3 = CBS 147313 | Imported from Brazil to the Netherlands; melon; type of T. calidominioluteus | OK338786 | OK338817 | OK338837 | OK339612 |
| T. calidominioluteus | Trachyspermi | DTO 265-H8 | Iran, Ajabshir; grapevine | OK338787 | KU711894 | OK338838 | OK339601 |
| T. calidominioluteus | Trachyspermi | DTO 265-I2 = CBS 147341 | Iran, Bonab; grapevine | OK338788 | KU711896 | OK338839 | OK339602 |
| T. calidominioluteus | Trachyspermi | DTO 266-A5 | Iran, Malekan; grapevine | OK338789 | KU711900 | OK338840 | OK339603 |
| T. calidominioluteus | Trachyspermi | DTO 269-H1 | Thailand; house dust | OK338790 | OK338818 | OK338841 | OK339613 |
| T. calidominioluteus | Trachyspermi | DTO 270-A5 = CBS 147342 | Thailand; house dust | KP330045 | OK338815 | OK338835 | OK339600 |
| T. calidominioluteus | Trachyspermi | DTO 390-E9 | Nigeria, Ibadan; cocoa beans | MN787900 | MN787896 | OK338847 | MN788104 |
| T. calidominioluteus | Trachyspermi | DTO 390-F1 | Nigeria, Ibadan; cocoa beans | MN787901 | MN787895 | OK338848 | MN788103 |
| T. calidominioluteus | Trachyspermi | DTO 390-I9 | Nigeria, Ibadan; cocoa beans | MN787911 | MN787885 | OK338849 | MN788115 |
| T. calidominioluteus | Trachyspermi | DTO 391-A5 | Nigeria, Ibadan; cocoa beans | MN787914 | MN787883 | OK338850 | MN788111 |
| T. gaditanus | Trachyspermi | CBS 138.84 = CECT 2773 = IMI 282405 | Spain, Valladolid; apple (Malus sylvestris) damaged by insect | OK338791 | OK338819 | OK338851 | OK339604 |
| T. gaditanus | Trachyspermi | DTO 226-A9 = CBS 104.71 | The Netherlands, Lisse; tulip | OK338792 | OK338820 | OK338852 | OK339614 |
| T. gaditanus | Trachyspermi | DTO 226-B1 = CBS 996.72 | The Netherlands; jute sugar bag | OK338774 | OK338813 | OK338826 | MH860641 |
| T. gaditanus | Trachyspermi | DTO 226-B3 = CBS 442.89 | Denmark, Lyngby, soil | OK338793 | OK338821 | OK338853 | OK339615 |
| T. gaditanus | Trachyspermi | DTO 228-B8 = CBS 169.81 = IJFM 5146 | Spain, Madrid; air; type of P. gaditanum | OK338775 | OK338802 | OK338827 | MH861318 |
| T. gaditanus | Trachyspermi | DTO 333-A5 = CBS 144771 | The Netherlands; sputum of cystic fibroses patient | OK338794 | OK338822 | OK338842 | OK339616 |
| T. gaditanus | Trachyspermi | DTO 050-F7 = CBS 444.89 | Imported from USA to Denmark; cranberry | OK338776 | OK338803 | OK338828 | OK339597 |
| T. minioluteus | Trachyspermi | DTO 304-C4 = CBS 642.68 = CCRC 31698 = IMI 089377 = LSHB P44 = MUCL 28666 = NRRL 1714 | Unknown source and location; neotype of P. minioluteum | MN969409 | KJ885273 | JF417443 | JN899346 |
| T. minnesotensis | Trachyspermi | DTO 423-A7 = CBS 142381 = UTHSC DI16-144 = FMR 14265 | USA, Minnesota; human ear; type of T. minnesotensis | LT559083 | LT795604 | LT795605 | LT558966 |
| T. minnesotensis | Trachyspermi | DTO 055-D3 | Germany; bread | OK338795 | OK338810 | OK338854 | OK339617 |
| T. minnesotensis | Trachyspermi | DTO 055-D2 = CBS 147315 | Germany; wallboard | OK338796 | OK338811 | OK338843 | OK339618 |
| T. minnesotensis | Trachyspermi | DTO 340-G2 = CBS 141838 | China; soil | OK338797 | OK338823 | OK338855 | OK339605 |
| T. samsonii | Trachyspermi | DTO 304-C3 = DTO 169-G6 = CBS 137.84 = CECT 2772 = IMI 282404 = IMI 327872 | Spain, Valladolid; apple (Malus sylvestris) damaged by insect; type of P. samsonii | OK338798 | OK338824 | OK338844 | MH861709 |
| T. samsonii | Trachyspermi | DTO 392-I9 = CBS 147356 | The Netherlands; soil | OK338777 | OK338804 | OK338829 | OK339598 |
| T. samsonii | Trachyspermi | DTO 420-B2 = CBS 147357 = ATHUM 9801 | Greece, air in house | OK338778 | OK338805 | OK338830 | OK339599 |
| Talaromyces sp. | Trachyspermi | DTO 111-B3 = CBS 282.59 | Unknown location; jute treated with copper-naphthenate | OK338779 | OK338806 | OK338831 | OK339607 |
| Talaromyces sp. | Trachyspermi | DTO 162-E5 = CBS 147336 | Germany; lemon solution | OK338780 | OK338814 | OK338846 | OK339608 |
| T. germanicus | Trachyspermi | DTO 055-D1 = CBS 147314 | Germany; indoor environment | OK338799 | OK338812 | OK338845 | OK339619 |
| T. chongqingensis | Trachyspermi | DTO 060-C9 = DTO 013-A5 = CBS 270.35 = CBS 147316 | USA, Virginia, Castle Rock; Zea mays | OK338781 | OK338807 | OK338832 | OK339609 |
| T. chongqingensis | Trachyspermi | CGMCC 3.20482 | China; soil, type of T. chongqingensis | MZ361343 | MZ361350 | MZ361357 | MZ358001 |
| T. africanus | Trachyspermi | DTO 179-C5 = KAS 3859 = CBS 147340 | South Africa; house dust | OK338782 | OK338808 | OK338833 | OK339610 |
| T. trachyspermus | Trachyspermi | DTO 149-H6 = CBS 373.48 = ATCC 10497 = IFO 31757 = IMI 040043 = NRRL 1028 = QM 7682 | USA; unknown substrate; type of T. trachyspermus | OK338800 | KJ885281 | JF417432 | MH856401 |
| T. udagawae | Trachyspermi | DTO 302-A8 = CBS 579.72 = FRR 1727 = IFO 8808 = IMI 197482 = NHL 6089 | Japan, Misugimura; soil; type of T. udagawae | OK338783 | KX961260 | MN969148 | JN899350 |
| T. flavus | Talaromyces | DTO 310-A2 = CBS 310.38 | New Zealand; unknown substrate; type of T. flavus | JX494302 | KF741949 | JF417426 | MH867464 |
| T. sayulitensis | Talaromyces | DTO 304-E4 = ATCC 4713 = ATCC 52244 = FRR 1064 = IBT 4302 = MUCL 29225 = NRRL 1064 = NRRL 1142 = UPSC 3133 | USA, Virginia, Castle Rock; Zea mays; type of P. purpurogenum var. rubrisclerotium | OK338784 | OK338809 | OK338834 | KM066172 |
| T. sayulitensis | Talaromyces | DTO 245-H1 = CBS 138204 | Mexico, Sayulita; house dust; type of T. sayulitensis | OK338801 | OK338825 | MN969146 | OK339606 |
| Species Name | Ascomata | Growth Rate (mm) | CYA | CYA | MEA | Conidia | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| CYA 30 | CYA | MEA | Reverse | Soluble Pigment | Colony Texture | Ornamentation | Shape | Size (μm) | ||
| T. africanus | absent | 18–20 | 21–23 | 26–28 | reddish brown (8E6) | absent | velvety to weakly floccose | finely roughened | subglobose | 2.5–3.5 × 2–2.5 |
| T. calidominioluteus | absent | (20–)25–29 | 20–30 | (22–)25–30 | concentric rings of light brown (6D8), then brownish orange (6C6), dark brown (6F8), edge orange (6B6–7) | absent | velvety to weakly floccose or cottony | smooth | broadly ellipsoidal | 2.5–4 × 2–2.5 |
| T. chongqingensis | absent | 6–8 | 17–18 | 28–29 | violet brown (10D8), edge red (10B8) | red to dark red (10B8–10C8) | densely cottony | smooth | ovoidal | 2.5–3.5 × 1.5–2.5 |
| T. gaditanus | absent | 12–16 | 19–25 | 25–28 | brown (6D8) at the center, orange (5B8–6B8) elsewhere | absent | floccose | smooth | fusiform | 2.5–4 × 2–2.5(–3) |
| T. germanicus | absent | 6–7 | 20–22 | 23–25 | violet brown (10F7) centrally, brownish red (10D7) elsewhere | cherry red (10B8) | velvety | smooth | narrow ellipsoidal to slightly fusiform | 2.5–3.5(–4) × 1.5–2.5 |
| T. minioluteus | absent | 9–14 | 17–19 | 21–26 | light brown (6D8) in center, dark brown (6F8) in a ring under dense sporulation, edge sometimes orange (6B8) | light brow (6D8), brownish orange (7C6–8) | funiculose | smooth | broadly fusiform or ellipsoidal | 2.5–3.5 × 1.5–2.5 |
| T. minnesotensis | absent | 20–24 | 26–27 | 28–30 | dark brown (8F7) at the center, reddish orange (7A7) | absent | granulose | smooth | ellipsoidal | 2.5–3.5(–4.5) × 1.5–2.5 |
| T. samsonii | absent | 11–16 | 15–21 | 22–26 | violet brown to deep red (10E8–F8, 7B–C8) | vivid red to red (10A8–B8) | floccose | smooth | ellipsoidal to fusiform | 2.5–4(–5) × 1.5–3 |
| T. udagawae | present | 5–8 | 5–9 | 15–19 | brownish orange (5C5) | absent | floccose | smooth | subglobose to ellipsoidal | 3–4 × 2–3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pyrri, I.; Visagie, C.M.; Soccio, P.; Houbraken, J. Re-Evaluation of the Taxonomy of Talaromyces minioluteus. J. Fungi 2021, 7, 993. https://doi.org/10.3390/jof7110993
Pyrri I, Visagie CM, Soccio P, Houbraken J. Re-Evaluation of the Taxonomy of Talaromyces minioluteus. Journal of Fungi. 2021; 7(11):993. https://doi.org/10.3390/jof7110993
Chicago/Turabian StylePyrri, Ioanna, Cobus M. Visagie, Piera Soccio, and Jos Houbraken. 2021. "Re-Evaluation of the Taxonomy of Talaromyces minioluteus" Journal of Fungi 7, no. 11: 993. https://doi.org/10.3390/jof7110993
APA StylePyrri, I., Visagie, C. M., Soccio, P., & Houbraken, J. (2021). Re-Evaluation of the Taxonomy of Talaromyces minioluteus. Journal of Fungi, 7(11), 993. https://doi.org/10.3390/jof7110993







