Abstract
Accurate DNA replication and segregation is key to reproduction and cell viability in all organisms. Autonomously replicating sequence-binding factor 1 (Abf1) is a multifunctional protein that has essential roles in replication, transcription, and regional silencing in the model yeast Saccharomyces cerevisiae. In the opportunistic pathogenic fungus Candida glabrata, which is closely related to S. cerevisiae, these processes are important for survival within the host, for example, the regulation of transcription of virulence-related genes like those involved in adherence. Here, we describe that CgABF1 is an essential gene required for cell viability and silencing near the telomeres, where many adhesin-encoding genes reside. CgAbf1 mediated subtelomeric silencing depends on the 43 C-terminal amino acids. We also found that abnormal expression, depletion, or overexpression of Abf1, results in defects in nuclear morphology, nuclear segregation, and transit through the cell cycle. In the absence of ABF1, cells are arrested in G2 but start cycling again after 9 h, coinciding with the loss of cell viability and the appearance of cells with higher DNA content. Overexpression of CgABF1 causes defects in nuclear segregation and cell cycle progression. We suggest that these effects could be due to the deregulation of DNA replication.
1. Introduction
Chromatin structure in eukaryotes plays a crucial role in DNA metabolism, such as DNA replication, activation, and repression of transcription, and regional silencing, among others. Each process requires different proteins and the cis-acting DNA sequences to which they bind, favoring protein-protein interactions that shape different chromatin structures necessary to trigger specific metabolic functions. In Saccharomyces cerevisiae the proteins ScAbf1 and ScRap1 have been shown to be required for all these processes and, because they perform multiple metabolic functions, they are known as General Regulatory Factors, or GRFs [1,2,3]. ScAbf1 is a sequence-specific DNA-binding protein for which there are many recognition sites (~181) throughout the genome, including promoters of genes activated (50) or repressed (36) by ScAbf1, the silent mating-type loci (HML and HMR), the region X of many yeast telomeres and sites known as ARS (Autonomous Replication Sequences), which confer stability and the ability to replicate, to plasmids [4,5,6]. Many ARSs indeed function as origins of replication in the chromosomes and are recognized by ScAbf1. One special case is the sequences that function as ARSs present in the silent mating-type loci, HMR and HML. These loci are each flanked by cis-acting elements called silencers (E and I) that are essential to mediate regional silencing of the mating information at each locus. In particular, the silencers HMR-E and HML-I both contain different combinations of three specific DNA sequences recognized by the Origin of Recognition Complex (ORC), ScRap1, and ScAbf1, respectively. Indeed, these cis-elements mediate silencing of the genes present at HML and HMR and confer plasmid stability as they are functional ARSs, suggesting a connection between initiation of DNA replication and silencing [6,7]. More recently, it has been shown that ScAbf1, along with other GRFs like ScRap1, regulates the accuracy of transcription initiation by preventing inappropriate initiation and pervasive transcription [8].
The ABF1 gene encodes ScAbf1, an essential and abundant protein that consists of 731 amino acids. ScAbf1 is composed of two regions involved in DNA-binding (an atypical Zinc-finger domain and a DNA-binding region) and a C-terminal domain that is required for all its known functions: transcriptional repression and activation, DNA replication, and regional silencing at the silent mating-type loci [6,9,10].
Candida glabrata is a haploid yeast phylogenetically related to S. cerevisiae, which has emerged as a frequent and important opportunistic pathogen associated with high mortality rates in intensive care units worldwide [11,12,13]. C. glabrata is frequently found as a commensal, tightly adhered to the mucosa of the gastrointestinal or urinary tracts of healthy individuals; however, it can cause severe systemic infections when the mucosal barrier is breached or there is severe immunosuppression of the human host.
The ability to adhere to epithelial or endothelial mucosal tissue of the host is thought to be an important first step for C. glabrata infections, and this ability is largely mediated by a family of cell wall proteins called the Epa adhesins encoded by the EPA genes, most of which are located at subtelomeric regions [14,15]. The EPA1 gene encodes the major adhesin (Epa1) and is the only member of the family expressed in vitro in the standard laboratory strain BG14 [16,17]. Expression of EPA1 is negatively controlled by several layers of negative regulation, including subtelomeric silencing that spreads from the telomere through up to 20 Kb [18,19,20], promoter-specific repression by a cis-acting negative element (NE) downstream from EPA1 [21], and repression by the formation of a chromatin loop between a subtelomeric, cis-acting protosilencer (Sil2126) and sequences flanking the EPA1 ORF [22,23]. We have shown that the C. glabrata ortholog of ScABF1 (CAGL0J01177g) is required for the formation of the chromatin loop that represses EPA1 along with other silencing proteins that participate in subtelomeric silencing such as CgSir2, CgSir3, and CgSir4 (forming the SIR complex), and CgRap1 [23].
However, up to date, CgABF1 has not been characterized; it is not known whether it also participates in general subtelomeric silencing, DNA replication or if it is an essential gene in C. glabrata. Here we show that the CgABF1 gene is essential for cell viability, that CgAbf1 participates in subtelomeric silencing through the C-terminal domain, and is also required for correct DNA replication and normal transit through the cell cycle. We suggest that these are all processes that are likely important for the ability of C. glabrata to persist in its human host.
2. Materials and Methods
2.1. Strains, Plasmids, and Primers
All strains, plasmids, and oligonucleotides used are listed in Supplementary Tables S1–S3, respectively.
2.2. Media
Yeast was grown in standard yeast media as described previously with 2% agar added for plates [24]. Synthetic complete (SC) medium contains 1.7 g/L yeast nutrient base (YNB) without (NH4)2SO4 or amino acids, 5 g/L (NH4)2SO4 and supplemented with 0.6% casamino acids (CAA) and 2% glucose. When needed, Nourseothricin (Gold Biotechnology®. St. Louis Missouri, USA) was added to liquid YPD media at a concentration of 50 µg/mL. Minimal media is YNB without (NH4)2SO4 or amino acids; 2% glucose and 5 g/L (NH4)2SO4 as nitrogen source. For the selection of Nourseothricin resistance (NatR) in minimal media, ammonium sulfate was substituted for 1 g/L of monosodium glutamate. For the plasmids with the shut-off expression from the MET3 promoter, ON media (expression of MET3 promoter) is minimal media as described above or with 1 g/L monosodium glutamate instead of ammonium sulfate. For the OFF media 0.2 mM of each methionine (met) and cysteine (cys) were added to the ON media. To prepare 5-fluoroorotic acid (5-FOA; Toronto Research Chemicals, Toronto, Ontario, Canada) media, 0.9 g of 5-FOA and 25 mg of uracil were added to 1 L of SC medium. 5-FOA compound is toxic to cells that express the URA3 gene.
Yeast extract-peptone-dextrose (YPD) medium contains 10 g/L yeast extract, 20 g/L peptone, and supplemented with 2% glucose. When required, YPD plates were supplemented with nourseothricin (AG Scientific, San Diego, CA, USA) at 100 µg/mL and at 50 µg/mL for liquid YPD media. Bacteria were grown in LB medium as described previously [25]. LB medium contained 5 g/L yeast extract, 10 g/L tryptone, 5 g/L NaCl. All plasmid constructs were introduced into strain DH10 by electroporation, and 100 µg/mL carbenicillin (AG Scientific, San Diego, CA, USA) was added to select for plasmids. For plates, 1.5% agar was used. Both bacteria and yeast were grown at 30 °C.
2.3. Yeast Transformation
Yeast transformations with digested or supercoiled plasmids were performed as previously described using the LiOAc/salmon sperm carrier DNA/PEG method [26,27].
2.4. Growth Assays in Liquid and Solid Media
Cells were grown to stationary phase for 48 h in YPD, CAA, or YNB. To determine the duplication time, cells of each strain were grown in the appropriate liquid media. Stationary phase cultures were adjusted to an OD600 of 0.01 in the corresponding media and 300 µL dispensed in a 100 well plate. Growth was automatically recorded using a Bioscreen C analyser at 30 °C (Oy, Growth Curves) with constant shaking and OD measurements taken every 15 min during a period of 48 h. The doubling time of each strain was calculated as described elsewhere [28]. Growth curves in liquid media were performed 3 separate times and with technical triplicates each repetition. The data shown represents the mean of the three experiments.
For solid media experiments, stationary phase cultures were adjusted to an OD600 of 1.0 with sterile water, and 10-fold serial dilutions were made in a 96-well plate. Five microliters of each dilution were spotted on to YPD plates. For temperature sensitivity assay, a total of 5 µL of each dilution was spotted onto YPD, then incubated at 30, 37 or 45 °C, and photographed every 24 h during three days. All solid media experiments were done in triplicate, and photographs of representative experiments are presented.
2.5. Construction of Null Mutant and Truncated Allele of the ABF1 Gene
Since ScABF1 is essential for cell viability in S. cerevisiae, we designed a strategy to determine whether CgABF1 is an essential gene in C. glabrata. To do this, we first constructed a replicative plasmid containing the full-length ORF of CgABF1 plus the 5′ and 3′ flanking regions of this gene (1 Kb upstream and 800 bp downstream, primers #1589 and #1881, Supplementary Table S3), and the URA3 selection marker (pCI12). We also generated a disruption plasmid for CgABF1. Briefly, the 5′ (primers #1589 and #1881) and 3′ (primers #1561 and #1562, Table S3) flanking intergenic regions of the gene to be deleted were PCR amplified and cloned into pYC44 integrative plasmid [29] on either side of the nourseothricin expression cassette (conserving the relative orientation of the chromosomal locus of ABF1) (pCI42). The plasmid generated was used to replace the wild-type CgABF1 gene with the nourseothricin cassette by homologous recombination in a one-step gene replacement procedure to generate abf1∆::NatR allele. For this, we first transformed the parental strain BG14 (Supplementary Table S1) with the plasmid containing the wild-type CgABF1 gene and URA3 marker (pCI12). This strain was then transformed with the linearized fragment from the knock-out plasmid (pCI42). The plasmid was previously digested with enzymes that cut within both fragments of the cloned 5′ and 3′ flanking regions, generating homologous ends to the ABF1 gene in the C. glabrata genome. Transformants were selected on plates supplemented with nourseothricin (AG Scientific, San Diego, CA, USA) at 100 µg/mL. Homologous recombination and allele replacement of the gene was verified by PCR analysis using primers annealing within the nourseothricin cassette and outside of the cloned 5′ and 3′ flanking regions. We also verified the absence of the gene deleted by the inability to amplify an internal fragment from ABF1 by PCR.
To construct the C-terminal truncated allele of ABF1 (that lacks the last 43 amino acids that correspond to part of the CS2 domain important for silencing in S. cerevisiae [10], we designed primers to amplify the truncated ORF of ABF1 ending at amino acid 436 using primers #1559 and #1880 (Supplementary Table S3). The fragment obtained was cloned into the integrative vector pYC44, generating plasmid pCI30. A 714 bp fragment containing the 3′ UTR region of ABF1 was cloned into pCI30 flanking the nourseothricin cassette (pCI32) [23]. Plasmid pCI32 was digested with BsgI within the region of C. glabrata homology and transformed into the parental BG14 selecting for NatR.
2.6. Plasmid Loss Assay to Determine If ABF1 Is Essential in C. glabrata
We used the null mutant described above, which contains a URA3 plasmid carrying ABF1 (pCI12) and a deletion of the chromosomal copy of ABF1 (abf1∆). This strain, as well as the BG14 parental strain transformed with pCI12, and BG14 with empty vector, were grown at 30 °C for 48 h in YPD and diluted into fresh media and grown for another 24 h, as described previously [28]. This was repeated three times. Ten-fold serial dilutions were plated on YPD plates for viable counts on SC + 5-FOA plates to select for the loss of the URA3 plasmid. Ura+ cells die on SC + 5-FOA plates; therefore, only cells that have lost the URA3 plasmid can grow on SC + 5-FOA. The percentage of cells without plasmid was calculated by counting the number of colonies on SC + 5-FOA divided by the number of colonies on YPD (viable count). The results shown are the mean of three biological replicates ± SD. The statistical analysis used was One-Way ANOVA using GraphPad.
2.7. Plasmid for Tagging Abf1 with Flag or c-Myc under the Repressible MET3 and Inducible MT-1 Promoters
We generated a replicative vector to tag CgAbf1 at the N-terminal end with Flag or c-Myc epitopes followed by a linker (5 repetitions of GA) and under the control of the promoters of either the MET3 gene (METhionine requiring), which is repressed by methionine and cysteine or MT-1 gene (MetalloThionenin 1) which is inducible with CuSO4.
For the construction of the plasmid with the Flag tag under the MET3 promoter, we digested the plasmid pGH5 that contains the full length of ABF1 flanked by ClaI restriction sites; the released fragment was cloned into the plasmid pRM153 [30], which contains the PMET3 promoter followed by the Flag tag and the GA linker to generate the N-terminal translational fusion (pVS1: pPMET3::Flag-ABF1-THIS3.NatR).
We amplified the full-length ABF1 gene with primers #2353 and #2354 (Supplementary Table S3), both of which contain ClaI restriction sites, to clone it into the plasmid pGH3 [23], which contains the PMT1 promoter fusion with the c-Myc tag and the GA linker to generate translational fusions at the N-terminal end of the protein of interest. The final plasmid (pGH8: pPMT-1::c-Myc-ABF1-THIS3.NatR) [23] was transformed into E. coli, and after verifying that the gene fragment was cloned in the correct orientation, it was transformed into C. glabrata (see yeast transformation section). All final plasmids were sequenced to confirm that constructs were correct and had no mutations.
2.8. Growth Assays with the Repressible Promoter MET3 in C. glabrata
We generated a collection of mutants that contain the plasmid pVS1 (pPMET3::Flag-ABF1-THIS3. NatR), the MET3 promoter is repressible by the addition of cysteine and methionine [31]. We conducted a series of growth assays in order to determine the minimum concentration needed to achieve the shutdown of the promoter (Supplementary Table S4). Then we conducted the experiments with two different media: ON medium (MET3 promoter is active), which is YNB (no methionine or cysteine), where the ammonium sulfate ((NH4)2SO4) was replaced with sodium glutamate (C5H8NO4Na) (1 g/L) when Nat had to be added for selection of the plasmids; OFF medium (no expression of the MET3 promoter) is the same as ON media, but adding 0.2 mM of L-cysteine (cys) and L-methionine (met). In SC (which contains casaminoacids) or YPD media, the MET3 promoter was also fully repressed.
2.9. Viability Assays
Stationary phase cultures (48 h in ON media) of strain abf1Δ/pVS1 (pPMET3::Flag-ABF1.Nat) were diluted to OD600 0.1 in either ON (YNB) or OFF (YNB plus 0.2 mM each met and cys) and incubated at 30 °C with shaking. Samples were taken every hour for 6 h in one experiment and from 7 to 12 h in a second experiment, and 24 h samples were also taken from both experiments. Cell suspensions from the samples were serially diluted in sterile water, and the appropriate dilutions were plated on ON (YNB) media. Colonies were counted, and CFUs were calculated per mL of cell suspension. All experiments were performed in triplicate, and the data shown corresponds to the mean of the three independent experiments ± SD. Statistical analyses used were Two-Way ANOVA and Tukey’s test using GraphPad, PRISM.
2.10. Reporter URA3 Gene Expression Assays (5-FOA Sensitivity Assays)
We generated a collection of mutants that contain the URA3 reporter gene inserted near different telomeres. These strains allow us to measure silencing at different positions in the selected subtelomeric regions, as shown in Figure 7 and in Supplementary Figure S4. A plate growth assay was used to determine the level of silencing as described previously [18,19]. Strains were grown in YPD for 48 h to stationary phase and then were adjusted to an OD600 of 1.0 in sterile water, and 10-fold serial dilutions were made in 96-well plates. Subsequently, ~5 µL of each dilution was spotted onto YPD, SC (no uracil), and SC + uracil + 5-FOA plates (with or without CuSO4 50 µM for the induction of PMT-1 containing plasmids), with the replica-plating tool (Frogger) (NUNC™). The plates were incubated for 48 h at 30 °C and then photographed.
2.11. Cell Cycle Progression Experiments by FACS
Strains to be analyzed were grown to stationary phase in the appropriate media and then inoculated in either ON or OFF media at an OD600 0.5 and incubated with shaking at 30 °C. Samples were taken every 3 h and stained with Sytox Green as follows. Cells were centrifuged and 1 × 107 cells were resuspended in 450 µL of sterile water. One-thousand and fifty microliters of 100% ethanol were slowly added and gently mixed, after which cells were incubated from 1–12 h at 4 °C. Cells were collected by centrifugation and washed with 1 mL 50 mM Tris pH 8.0. The cell pellet was resuspended in 500 µL of RNAse (2 mg/mL) for 2 h at 37 °C after which cells were centrifuged and resuspended in 200 µL of proteinase K (5 mg/mL) for 20 to 45 min at 37 °C. Cells were centrifuged and washed with 1 mL 50 mM Tris pH 8.0. Cells were resuspended in 50 µL of 50 mM Tris pH 8.0 and stained with SYTOXTM Green (5 mM, Thermo Fisher Sientific, Watham, MA, USA: 1 µL SYTOXTM Green in 1 mL 50 mM Tris pH 8.0). Cells were analyzed by flow cytometry (FACS) using BD FACSCalibur Flow Cytometer with Cell Quest Pro software, and the data were analyzed with FlowJoTM.
2.12. Fluorescence Microscopy
Strains to be analyzed were grown to stationary phase in the appropriate media and then inoculated in either ON, OFF, with 25 μM CuSO4 or without CuSO4 media at an OD600 0.5 and incubated with shaking at 30 °C. Samples were taken every 3 h until 9 h. 1 mL samples of each strain were centrifuged and washed with PBS, and then fixed with ethanol 70% for 1 h. Cells were washed with PBS and stained with DAPI for nuclei visualization. Cells were analyzed with Axio Imager.m2 Microscope (Carl Zeiss). Images were analyzed with the software AxioVision v. 4.8.2.0 image browser. At least 100 cells were counted for each strain and condition except in the presence of copper, where 65 and 49 cells at 6 h and 9 h were counted, respectively. Representative images are shown.
3. Results
3.1. Candida glabrata Encodes the ABF1 Gene
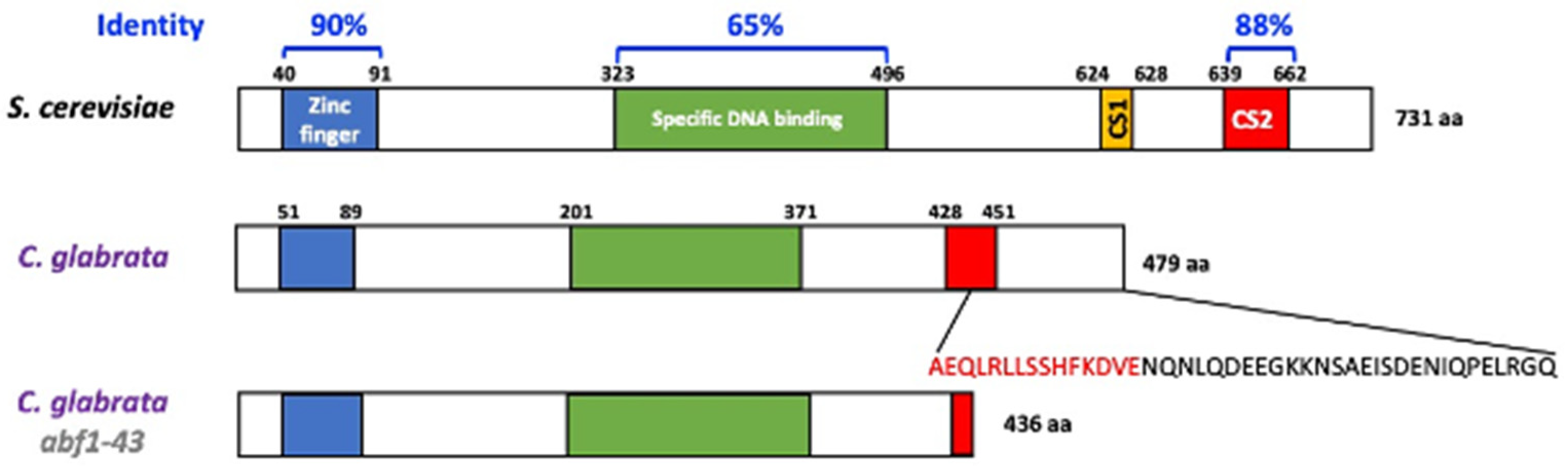
The Abf1protein of S. cerevisiae is involved in several important processes, including, among others, DNA replication, transcriptional regulation (activation and repression), and silencing of the silent mating loci HMR and HML [5,6,7,10]. The C. glabrata genome harbors the orthologous CgABF1 (CAGL0J01177g) gene, which encodes a predicted protein of 479 amino acids with 63.87% overall identity with ScABF1 (Figure 1). Similar to ScAbf1, CgAbf1 is predicted to contain a bipartite DNA-binding domain: Zn finger domain (residues 51–89) and a specific DNA-binding domain (residues 201–371) sharing 90 and 65% identity with the corresponding domains of ScAbf1 (Figure 1). CgAbf1 also contains a conserved C-terminal activation domain called CS2 (residues 428–451) that is 88% identical to the ScAbf1 CS2 domain, which in S. cerevisiae is involved in chromatin remodeling and transcriptional regulation [5,10] (Figure 1 and Supplementary Figure S1). When deletions are made in the C-terminal end of the S. cerevisiae protein, silencing and other functions are impaired. We generated a C-terminal deletion allele (abf1-43) that lacks the last 43 amino acids, which have been shown to be necessary and sufficient for silencing at the HMR-E locus in S. cerevisiae [10]. This deletion removes the last 15 amino acids from the CS2 domain to the end of the protein (28 additional amino acids for a total of 43 removed) (Figure 1).
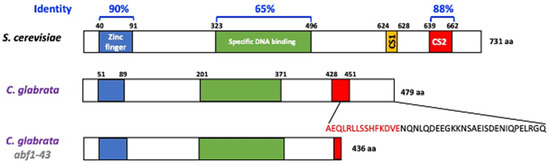
Figure 1.
C. glabrata encodes the CgAbf1 protein with conserved DNA-binding and regulatory domains. Schematic representation of Abf1 from S. cerevisiae and C. glabrata, CgAbf1 has 479 aa and ScAbf1 has 731 aa. The overall identity between ScAbf1 and CgAbf1 across the entire length of the proteins is 63.87%. The blue and green rectangles correspond to the DNA binding domains, the yellow rectangle corresponds to the CS1 domain in ScAbf1 and the red rectangles to the CS2 domain responsible for chromatin remodeling and transcriptional regulation. Alignments were performed using ClustalW with the program McVector. We generated a CgABF1 truncated allele, Cgabf1-43, which lacks the last 43 amino acids of CgAbf1, deleting 15 highly conserved residues at the CS2 domain (in red letters) that mediate transcriptional silencing in S. cerevisiae (See Materials and Methods). The structure of the Abf1-43 truncation mutant protein is shown at the bottom (total of 436 amino acids).
3.2. Abf1 Is Essential for Cell Growth in C. glabrata
To determine whether CgAbf1 is essential for cell viability in C. glabrata, we designed two different experiments: (1) a plasmid loss assay and (2) a conditional expression system (“shut off plasmid”) using the repressible promoter from the MET3 gene (PMET3) (see Materials and Methods).
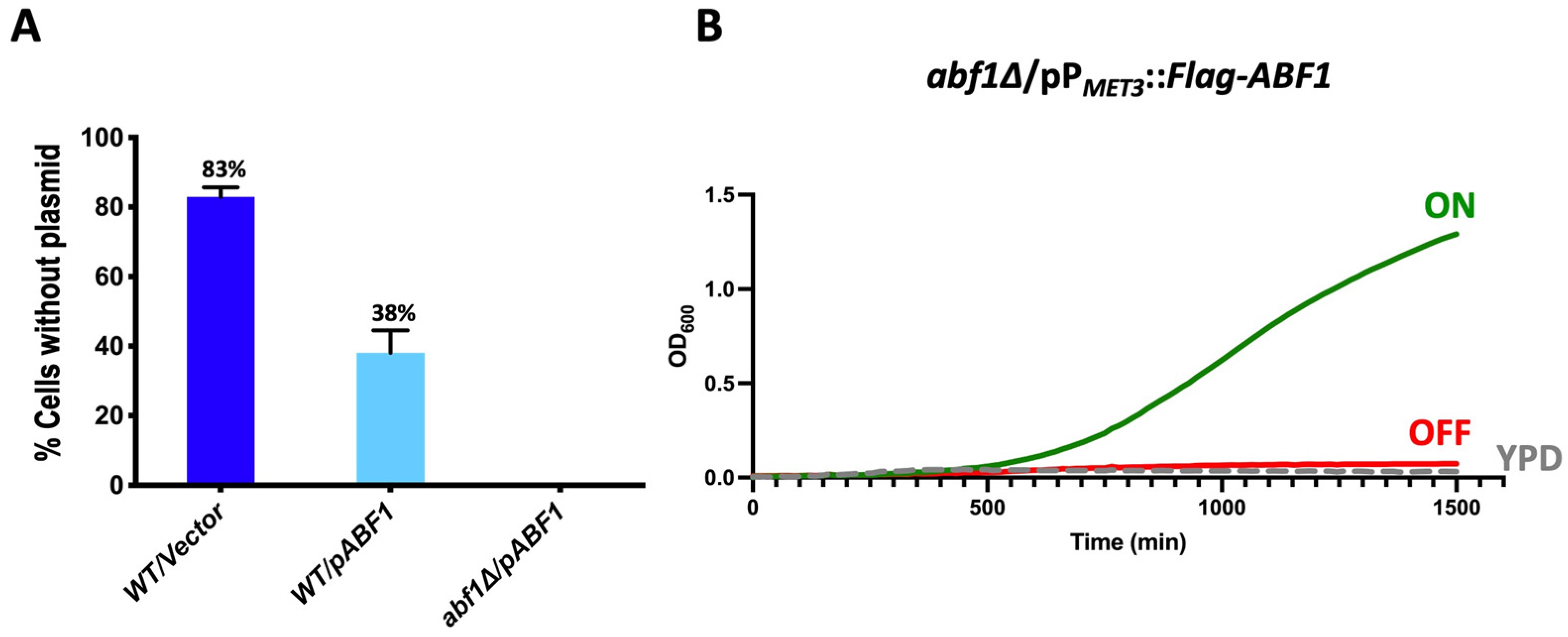
For the plasmid loss assay, we first introduced a replicative plasmid in the parental strain (BG14) with the ABF1 gene under the control of its own promoter and the counter selectable marker URA3 (pCI12). In this strain, we deleted the native ABF1 gene by homologous recombination using a knock-out plasmid (pCI45), containing the NatR cassette flanked by the intergenic 5′ and 3′ regions of ABF1 (Supplementary Figure S2A). In a second step, we eliminated the NatR cassette generating strain CGM3584 (Supplementary Table S1). If ABF1 is essential for viability in C. glabrata, we expected that the strain abf1∆/pABF1.URA3 would not be able to lose the plasmid with the ABF1 gene. Conversely, if ABF1 is not essential, this strain would lose the plasmid after repeated cycles of growth in the absence of selective pressure (rich media). To determine the frequency of plasmid loss, we used the plate growth assay on SC + 5-FOA plates since cells expressing URA3 die in the presence of 5-FOA and only cells that have lost the URA3 plasmid (Ura− cells) can grow on these plates. Figure 2A shows that the strain abf1∆/pABF1.URA3, cannot lose the complementing plasmid as measured by the absence of 5-FOAR colonies. This result is in agreement with the fact that ScABF1 is also essential for viability in S. cerevisiae [6,10].
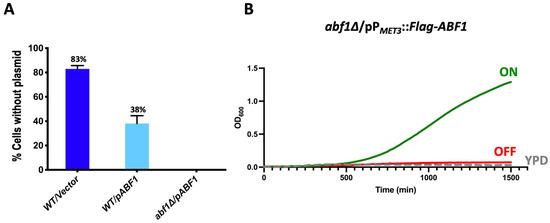
Figure 2.
CgAbf1 is an essential protein for growth in C. glabrata. (A) The indicated strains were grown in YPD rich media (without selection) for 72 h, diluting in fresh media every 12 h after which cells were diluted and plated on YPD and incubated 48 h at 30 °C. Plates were replica-printed onto SC + 5-FOA, SC—uracil and YPD plates to determine the number of Ura− colonies. The graph shows the percentage of cells that lost the plasmid in each culture, and the numbers represent the mean of three separate experiments ± SD. (B) Cells from strain abf1∆/pPMET3::Flag-ABF1 (shut off plasmid) were grown in YNB minimal media (ON) for 48 h. Cells were adjusted to an OD600 of 0.01 and inoculated into three media: ON (in green) (YNB minimal media); OFF (in red) (YNB + 0.2 mM each cys and met), and YPD rich media (in broken gray lines). The OD600 was recorded every 15 min in Bioscreen C equipment. The graph represents the mean of three biological replicates with three technical replicates each (see Section 2.4 and Section 2.6).
It should be noted that in the parental strain, more than half of the cells (62%) retained an extra copy of ABF1 gene driven by its own promoter (the endogenous and the episomal copies), suggesting that a higher level of transcription of ABF1 may be advantageous under these conditions.
For the “shut off plasmid” in which we cloned the full-length ABF1 gene fused with the Flag epitope at the N-terminal end and driven by the repressible promoter from the MET3 gene (pVS1: pPMET3::Flag-ABF1). This plasmid was transformed into the abf1∆/pABF1.URA3 strain followed by plasmid shuffling (Materials and Methods and Supplementary Figure S2A). The resulting strain (abf1∆/pPMET3::Flag-ABF1.NatR) allows us to use a conditional expression system where ABF1 is expressed in minimal media without amino acids and completely repress the expression of ABF1 when methionine and cysteine are added to the media. Figure 2B shows the growth of the null ABF1 mutant strain with the repressible ABF1 plasmid (abf1∆/pPMET3::Flag-ABF1.NatR) in three different media: YPD (rich media), ON (minimal media without amino acids), and OFF (minimal media supplemented with met and cys). If ABF1 is essential for growth in C. glabrata as suggested by the inability to lose the ABF1 plasmid in the abf1∆ strain, we expected that repressing the PMET3 promoter would prevent the growth of this strain. Figure 2B shows that this strain only grew in ON medium but not in the OFF or YPD media, both of which contain amino acids, indicating that ABF1 is essential for cell growth in C. glabrata.
3.3. Repression of ABF1 Results in Loss of Cell Viability
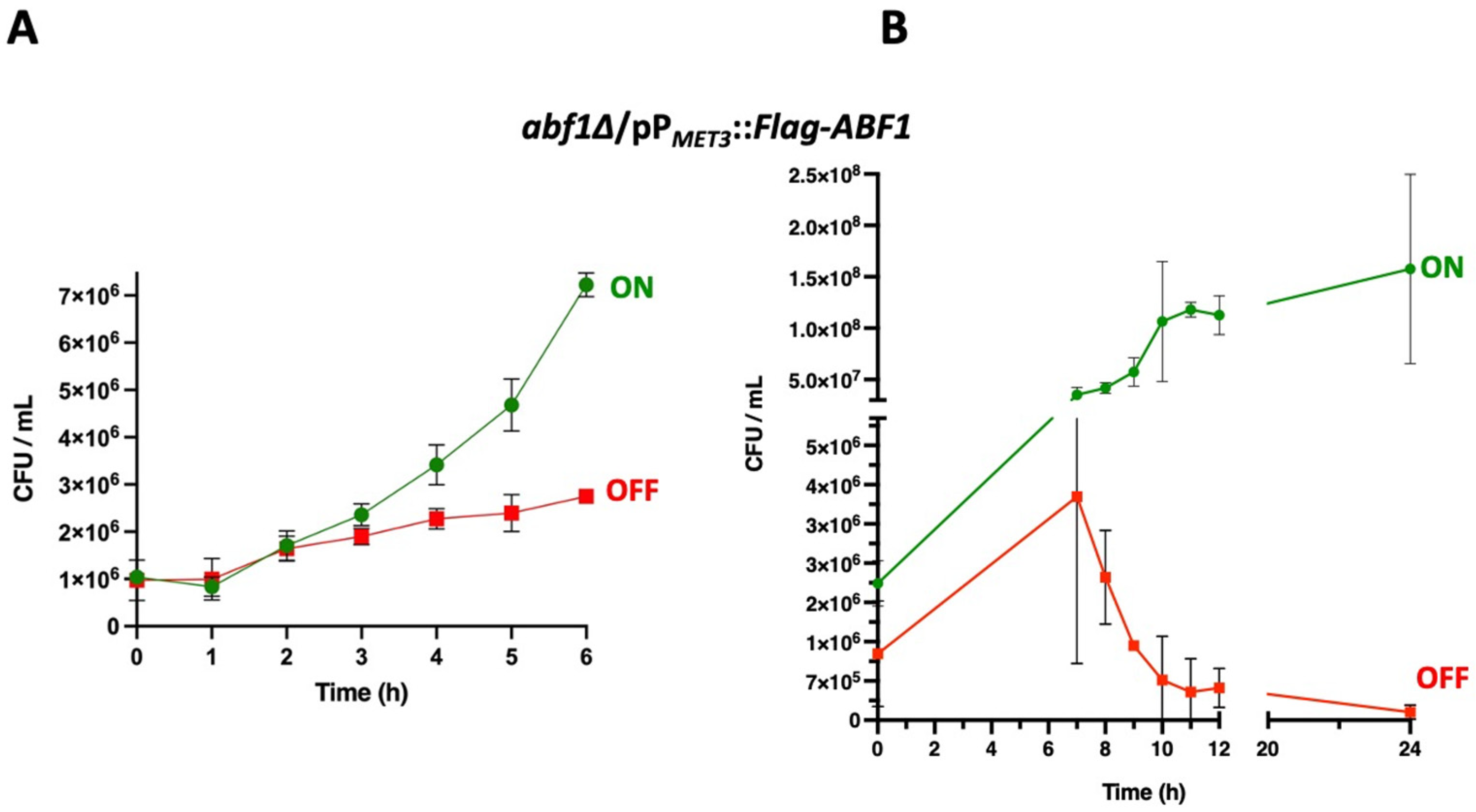
We used the ABF1 shut-off plasmid to determine whether the absence of Abf1 causes cell death. To do this, we grew the abf1∆ strain containing the plasmid with Flag-ABF1 fusion under the control of the MET3 promoter in minimal media in the absence of amino acids (ON media). We then added methionine and cysteine to repress the promoter and took aliquots every 60 min for 12 h and then at 24 h, to determine cell viability. As shown in Figure 3A, during the first 6 h of repression of ABF1 in the abf1∆/pPMET3::Flag-ABF1 strain, there is only a two-fold increase in the number of colony forming units (CFU), but after 8 h of repression, viability starts decreasing, and after 24 h, there is a ±10-fold drop (Figure 3B). However, the cells incubated in ON media continue replicating (Figure 3A,B and Supplementary Table S5).
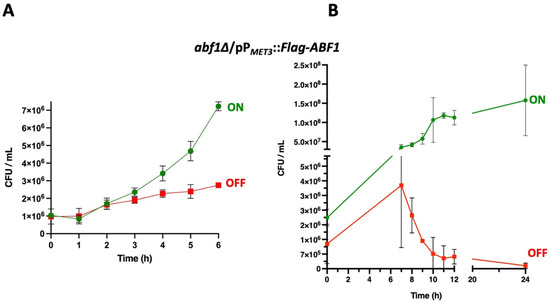
Figure 3.
Repression of ABF1 results in loss of cell viability. (A) Stationary phase cultures of strain abf1∆/pPMET3::Flag-ABF1 were adjusted to an OD600 of 0.1 and diluted in either YNB minimal media (ON in green) or in YNB + 0.2 mM each cys and met (OFF in red) and incubated at 30 °C with shaking. Samples were taken to determine viable counts (expressed as colony-forming units or CFU) every hour for the first six hours. (B) In an independent set of experiments, samples were taken to determine CFUs as in section A, every hour starting at 7 h after inoculation into each medium until 12 h and another sample at 24 h (graph on the right). All the results shown represent the mean of three independent experiments, and bars correspond to the SD. Analysis of the data was done with Two-Way ANOVA and Tukey’s test using GraphPad, PRISM (See Section 2.9).
3.4. Overexpression of ABF1 Is Toxic in C. glabrata
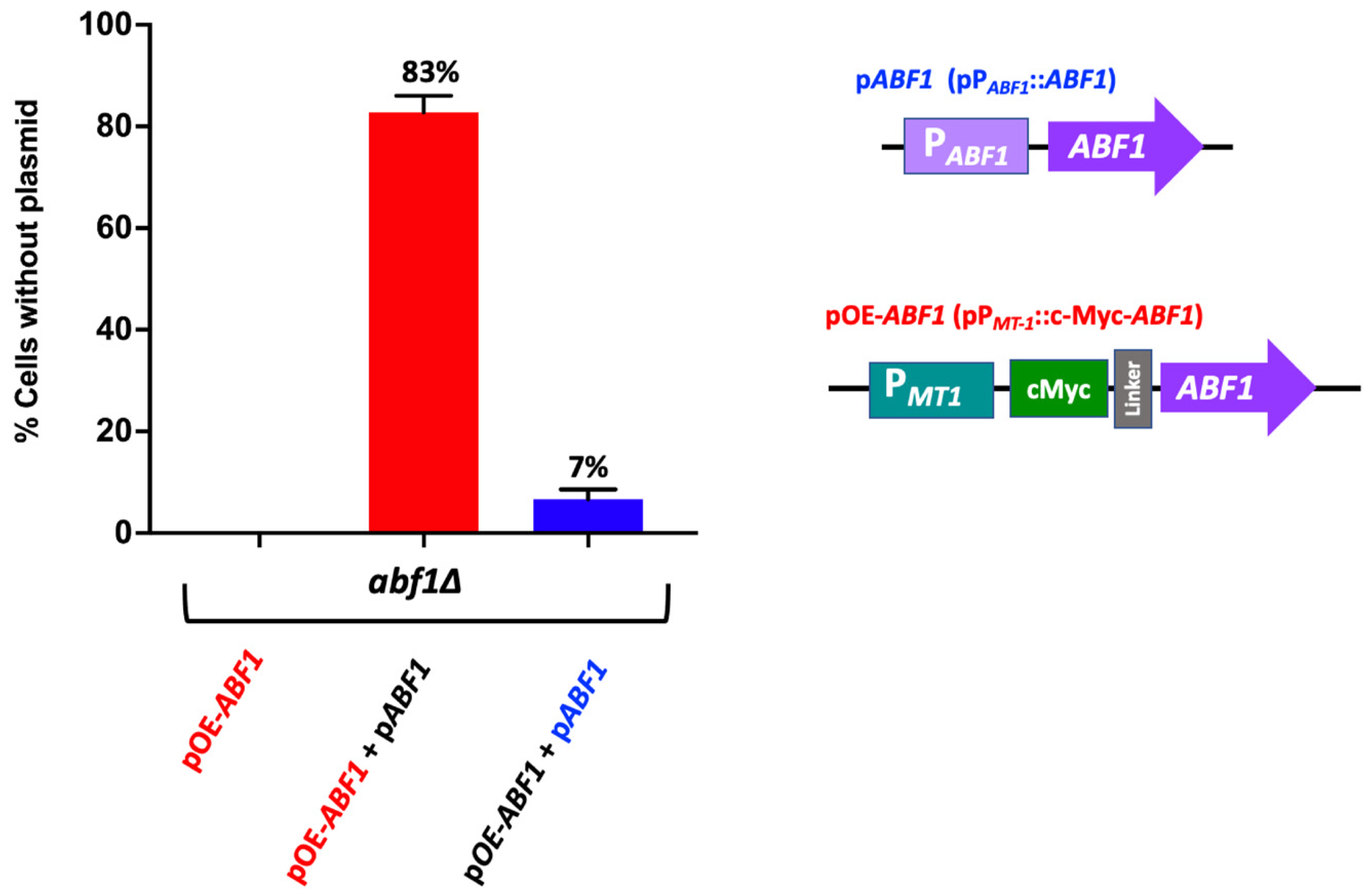
To determine whether overexpression of Abf1 results in toxicity in C. glabrata as has been reported for S. cerevisiae [32,33], we generated an epitope-tagged version of Abf1 by constructing a plasmid that contains a translational fusion of the c-Myc tag at the 5′ end of ABF1, separated by a linker, and under the control of the copper inducible promoter PMT-1 from the MT-1 gene (CAGL0D01265g) (pGH8) (Supplementary Figure S2B). This construct complements the abf1∆ strain as determined by the ability to exchange the pPABF1::ABF1.URA3 plasmid for the plasmid carrying the c-Myc-ABF1 tagged version in the abf1∆ background (Supplementary Figure S2). We also showed that the c-Myc tagged Abf1 (pGH8) is functional for silencing activity mediated by the protosilencer Sil2126 when the Sil-reporter system is placed 32 kb away from telomere E-R (See López-Fuentes et al. [23]. The abf1∆ strain carrying both plasmids lost the plasmid with the inducible promoter at a much higher frequency (83% of cells lost the pPMT1::c-Myc-ABF1 plasmid), than the plasmid where ABF1 is driven by its own promoter (pPABF1::ABF1) under non-inducing conditions (Figure 4).
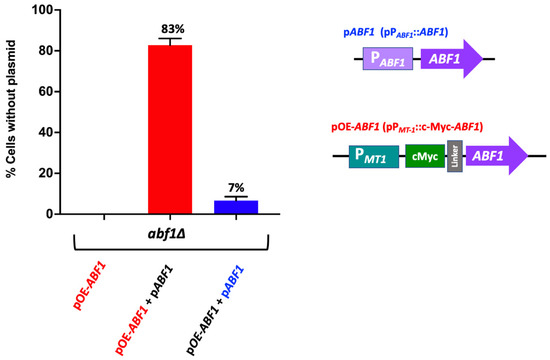
Figure 4.
Over expression of ABF1 is toxic in C. glabrata. Schematic representation of the plasmids transformed in the abf1∆ strain. The first plasmid contains the full-length ABF1 gene driven by its own promoter (pABF1+) and a second plasmid with the amino-terminal tagged version (c-Myc) of ABF1 driven by the overexpressing promoter from MT-1 gene (pOE-ABF1). These plasmids were transformed simultaneously, and the loss of each plasmid was determined after passages every 12 h for 72 h in rich media YPD (without selection). The graph shows the percentage of cells that lost the corresponding plasmid (the mean of three independent experiments ± SD, see in Section 2.6). The red bar indicates loss of the over-expressing plasmid pc-Myc-ABF1 (OE-ABF1); the blue bar indicates loss of the plasmid pABF1.
We also determined the growth rate of the parental (ABF1) and the abf1∆ strains in the presence of the overexpressing plasmid PMT-1::c-Myc-ABF1 under inducing (with 50 µM CuSO4) or non-inducing conditions (without CuSO4) in different media SC, YNB or YPD. Table 1 shows that overexpression of ABF1 by induction with copper in the medium, decreases the growth rate both, in the parental strain (duplication time from 66 min to 89 under inducing conditions), as well as in the strain carrying the C-terminal truncation of the last 43 amino acids (abf1-43) where the duplication time increased from 70 to 106 min under the induction of the MT-1 promoter.

Table 1.
Duplication time of strains overexpressing c-Myc-ABF1 (pPMT1::c-Myc-ABF1).
3.5. Misexpression of Abf1 Results in Cells with Aberrant Nucleus Morphology and Segregation and Delayed Passage through the Cell Cycle in C. glabrata
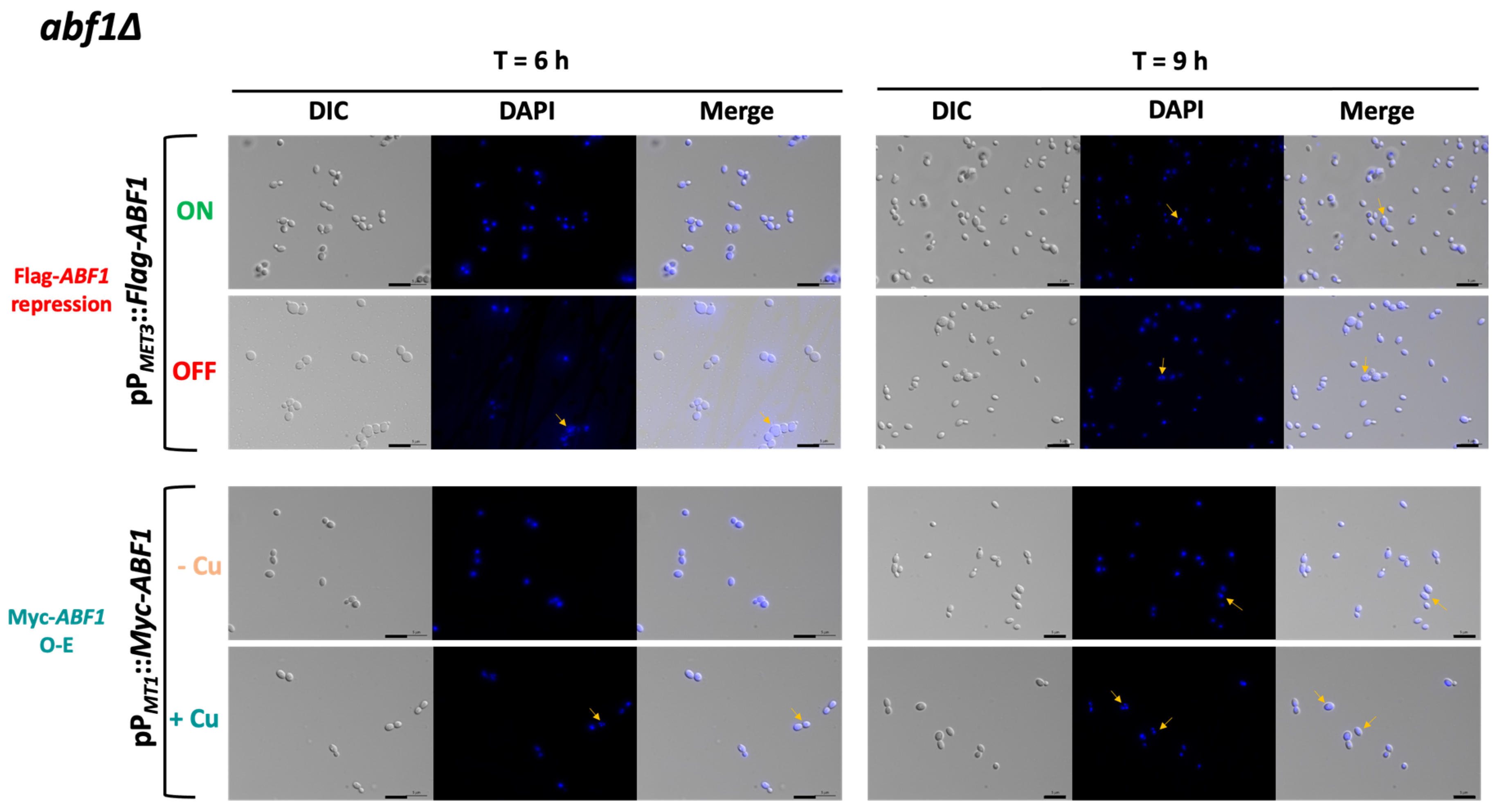
Since ScABF1 is required for DNA synthesis, we asked whether deprivation or overexpression of ABF1 results in changes in nuclear morphology or cell cycle defects. To do this, we first used the abf1∆ strain with the ABF1 repression plasmid (pVS1: pPMET3::Flag-ABF1) to determine the nuclear morphology at different time points after the repression of ABF1. Figure 5 (top panels) shows that after 6 h of Flag-ABF1 repression, some cells appear to have a less well-defined nucleus with fuzzier DAPI staining (~16%), and after 9 h of the absence of Flag-ABF1, there are some cells with more than one nucleus (2.5%). At both time points, the cells appear larger in the absence of Flag-Abf1 than in ON media, and there were more budding cells (small budded and large budded) compared to the condition where Flag-ABF1 is expressed (57.5% in OFF vs. 18.5% in ON at 9 h) (Supplementary Table S6), which suggests that cells in OFF media spend more time in the S or S/G2 phases of the cell cycle than cells in ON media. It should be noted that even under conditions where the MET3 promoter is active, around 1.3% of the cells at 9 h also appear to have more than one nucleus, suggesting that this promoter is not as strong as the ABF1 native promoter. Furthermore, cells from the wild-type parental strain did not show abnormal nuclear morphology or more than one nucleus.
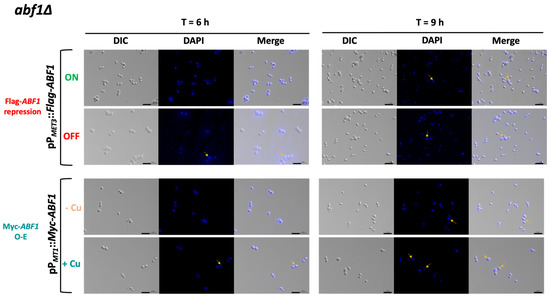
Figure 5.
CgAbf1 is required for nuclear integrity. Cells were grown to stationary phase in ON media; then, the OD600 was adjusted to 0.5 and incubated at 30 °C in the following media depending on the strain: ON (in green, YNB minimal media) or OFF (in red, YNB plus 0.2 mM each cys and met) or in—Cu (in orange, YNB minimal media) or +Cu (cyan, YNB + 25 µM CuSO4) as indicated. Samples were taken at 3, 6, and 9 h of incubation, stained with DAPI, and visualized and photographed under an Axio Imager.m2 fluorescence microscope. Images of the 6 and 9 h are shown. Yellow arrows point to cells with nuclei defects. From left to right, DIC (differential interference contrast), DAPI (blue), Merge (DIC + DAPI). Scale bar corresponds to 5 μm.
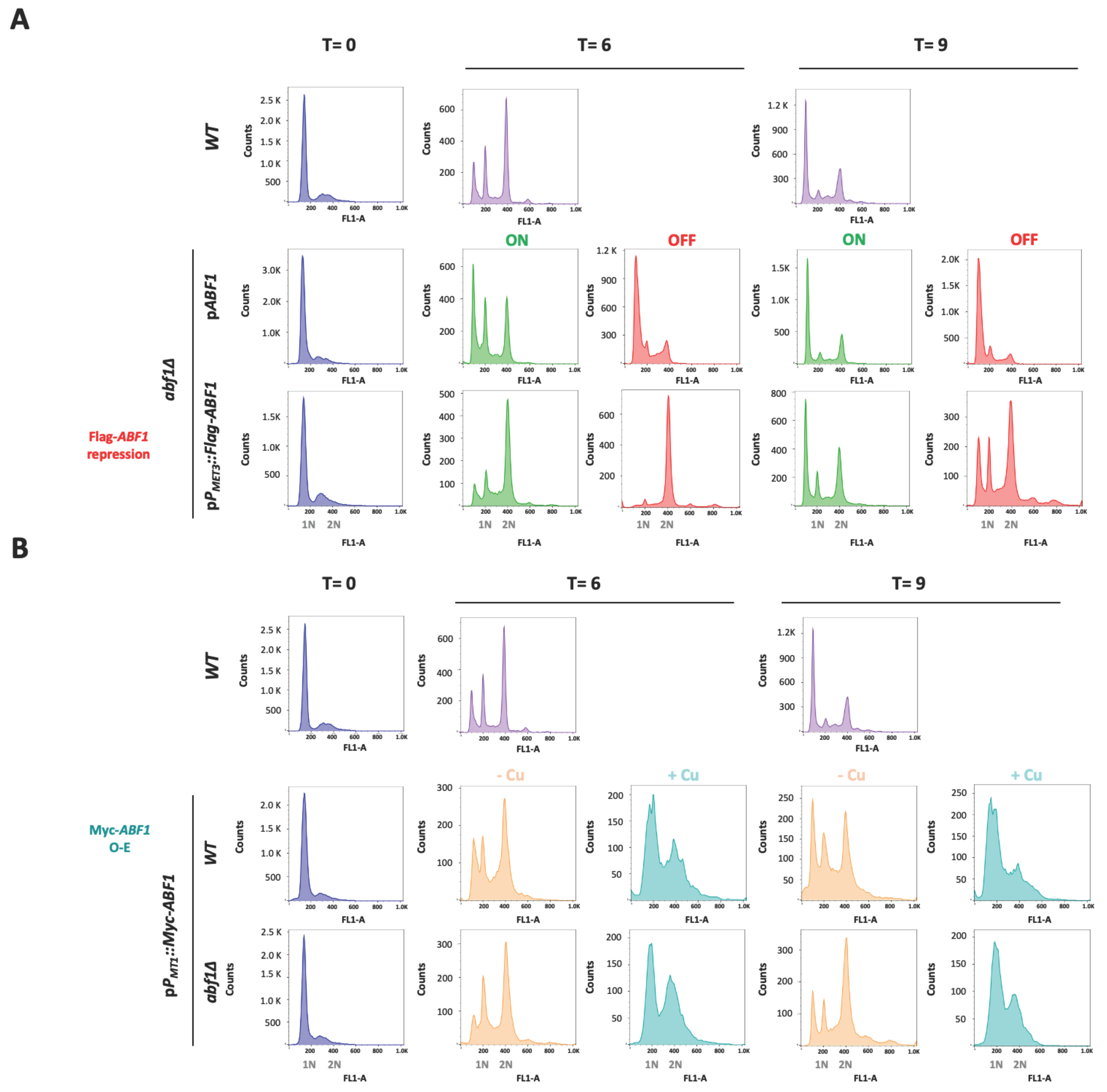
We also determined the cell cycle progression of the cells in the absence of Flag-ABF1 compared to the abf1∆ complemented with the plasmid expressing ABF1 under its own promoter (pCI12), as well as with the parental strain (ABF1). We used stationary phase cells of each strain (T = 0 h) and then diluted each culture into fresh ON or OFF media and took samples after 6 h or 9 h to determine DNA content by flow cytometry. As shown in Figure 6A, 6 h after Flag-ABF1 repression, cells accumulate in the G2 DNA content phase while the cells in the ON media have started to divide, showing a small proportion of cells with G1 DNA content. After 9 h in OFF media, the cells have apparently bypassed the arrest in G2 and started cycling so that a relatively large proportion of cells with G1 DNA content are detected. In addition, a few cells appear to have higher DNA content equivalent to 3 C and 4 C (visible as small peaks at 600 and 800 fluorescence intensity units in the FL-1A channel, Figure 6A bottom panels), consistent with some observed cells containing two or three nuclei at this time point in the DAPI staining (Figure 5, top panels). In contrast, the cells with the plasmid driving ABF1 with its own promoter continue cycling albeit at a slower rate since the OFF media causes slow growth as it contains met and cys (Supplementary Table S4).
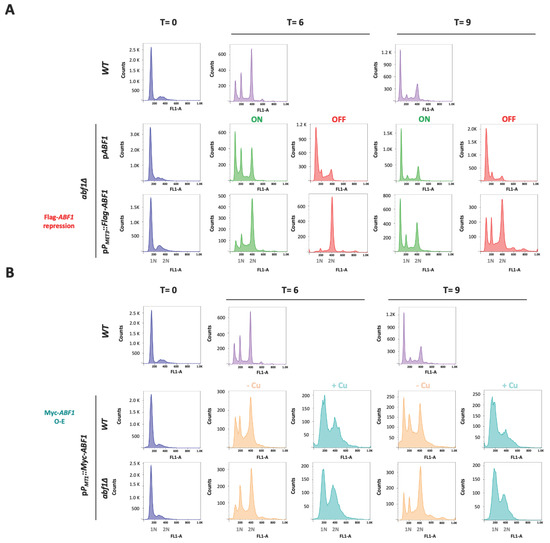
Figure 6.
Absence or overexpression of CgAbf1 causes defects in the cell cycle progression. Stationary phase (SP) cells (T = 0) grown in YNB minimal medium were adjusted to an OD600 0.5 in ON (in green, YNB minimal media); OFF (in red, YNB + 0.2 mM each cys and met),—Cu (Orange, YNB minimal media) or + Cu (teal blue, YNB + 25 μM CuSO4) media, depending on the strain, and then incubated 9 h at 30 °C. Samples were taken every three hours and stained with SYTOX™ Green for DNA content analysis by Flow Cytometry (FACS). Representative histograms for DNA content quantification with SYTOX Green are shown; “Y” axis shows the cells count, “X” axis corresponds to the intensity of fluorescence signal from SITYOX Green. 1N and 2N marks in the “X” axis correspond to G1 and G2 DNA content. (A) From left to right, T = 0 h (in blue) corresponds to histograms from the indicated strains in SP. T = 6 h histograms show the DNA content after 6 h incubation of: WT strain (light purple), abf1Δ strain with the native ABF1 promoter pPABF1::ABF1, or shut-off plasmid (pPMET3::Flag-ABF1), in ON (green) or OFF (red) media. T = 9 h are the same as T = 6 h but after 9 h of incubation in each media. (B) Same as in A but using the overexpression plasmid as follows. From left to right, T = 0 (blue) histograms show the DNA content of strains from stationary phase cultures; T = 6 correspond to samples after 6 h of WT strain (light purple), strains with the overexpression plasmid (pPMT1::Myc-ABF1) in WT and abf1Δ backgrounds in—Cu (orange, non-inducing conditions) and + Cu (teal blue, inducing conditions) media. T = 9 same as T = 6 but after 9 h of incubation. To facilitate the comparison with the WT strain, these strain’s histograms were duplicated for panels A and B, as all the strains were run together in this experiment and the same WT culture served as a control for the rest of the strains and conditions. Experiments were performed three times and the histograms from a representative experiment are shown. Data were analyzed with the software FlowJo.
To test whether over-expression also causes aberrant nuclear morphology and defects in the transit through the cell cycle, we transformed both the parental (ABF1) and the abf1∆ strains with the over-expression plasmid pGH8, where Myc-ABF1 is induced by adding copper to the medium. Cells from each culture were grown to stationary phase and then diluted into non-inducing (−Cu) and inducing media (+Cu). Figure 5 shows (bottom panels) that overexpression of Myc-ABF1 in both strains causes the appearance of multiple cells (~16%) with more than one nucleus after 9 h of c-Myc-Abf1 induction, compared to the medium in the absence of copper (~1.25%). In addition, passage through the cell cycle is abnormal, since stationary phase cells (T = 0 h) start cycling 6 h after dilution into media containing 25 µM of CuSO4 (Figure 6B); however, there is a large population of cells that have an intermediate DNA content between the characteristic G1 and G2 peaks, which correspond to cells in S-phase, compared to the cells without copper or the wild-type, untransformed strain. After 9 h of overexpression, a population with higher DNA content starts appearing, particularly in the wild-type strain (WT) with the pPMT-1::Myc-ABF1 plasmid in which, in addition to the native ABF1 promoter driving normal levels of expression of ABF1, there is additional over-expression of c-Myc-ABF1 from the induced MT-1 promoter. This is consistent with the large percentage of budded cells under inducing conditions vs. non-inducing conditions at 6 h (65.1% vs. 35.1%). These effects are most likely due to overexpression of ABF1 and not an effect caused by CuSO4 since the WT strain in the presence of this relatively low concentration of copper shows normal duplication time (Table 1) and a very similar pattern of DAPI staining in the absence or presence of copper.
3.6. Subtelomeric Silencing at Telomeres E-R and B-L Require the C-Terminal 43 Amino Acids of Abf1
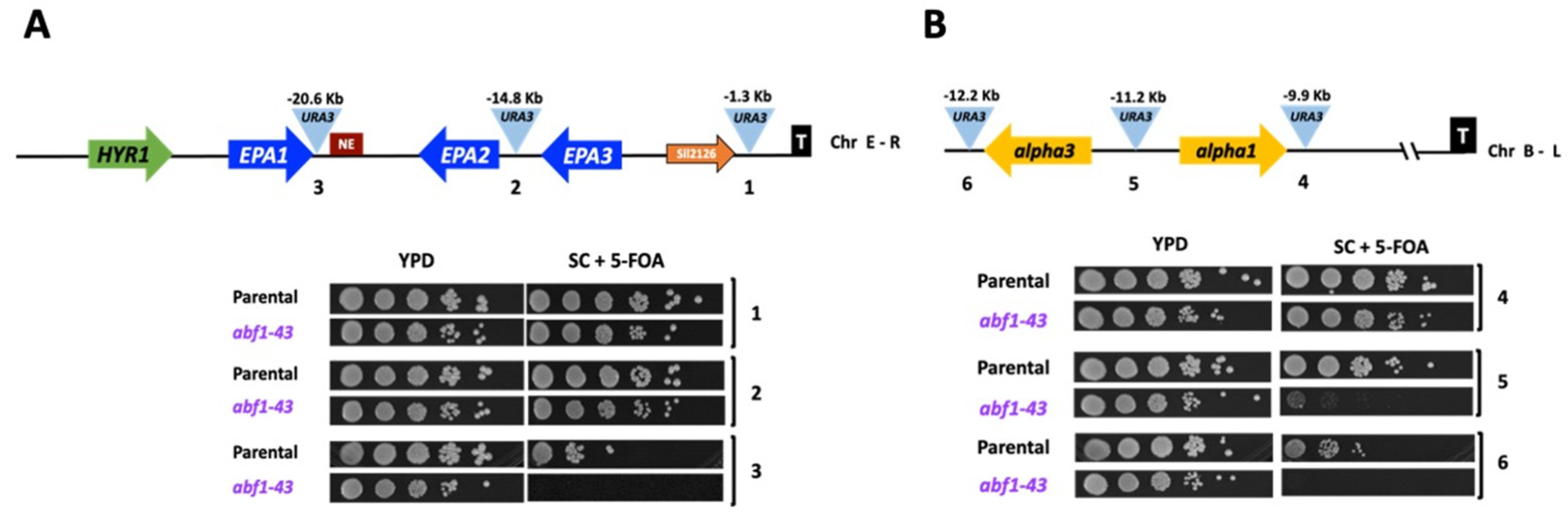
ScAbf1 has been shown to play a role in silencing the silent mating-type cassettes (HML and HMR) in S. cerevisiae, and the C-terminal end of ScAbf1 is known to be required for silencing [6,10]. We constructed a truncation mutant in ABF1, called abf1-43, lacking the last 43 amino acids that eliminate almost all the CS2 conserved region (Figure 1). This mutant is viable but has a longer duplication time compared to the parental strain, and it is also temperature-sensitive (Table 1 and Supplementary Figure S3). To determine whether CgAbf1 is required for silencing, we constructed different strains containing the URA3 reporter gene integrated at various distances from different telomeres in the wild-type background and in the abf1-43 mutant. Silencing activity by abf1-43 was assayed by the ability of the strains carrying each reporter insertion to grow on plates lacking uracil (SC-ura plates) where only cells that express the URA3 reporter can grow and on plates containing 5-FOA where cells expressing URA3 convert 5-FOA to a toxic compound and die. As shown in Figure 7A, insertions 1 and 2, where the reporter gene URA3 is closer to the telomere (1.3 and 14.8 Kb, respectively), were efficiently silenced in the abf1-43 mutant. However, when the reporter was inserted 20.6 Kb away from the telomere (insertion three), the level of silencing was clearly decreased in the abf1-43 mutant compared to the parental (ABF1 strain). In addition, the abf1-43 strain is also defective in silencing at telomere B-L (where the silent MTL3 locus is localized) when the URA3 reporter is inserted at a distance of 11 Kb or more from the telomere (Figure 7B, insertions 5 and 6), although the insertion closest to the telomere is not affected by the absence of the last 43 amino acids of CgAbf1. Instead, at the Chr I-R where EPA4 and EPA5 form a 15 Kb inverted repeat, the strong silencing of the reporter URA3 at different positions in this telomere was as efficient in the parental strain as in the abf1-43 mutant (Supplementary Figure S4).
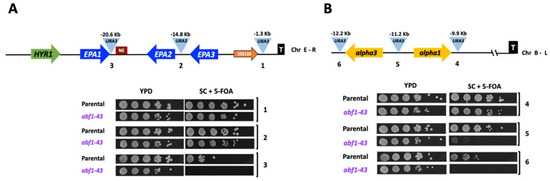
Figure 7.
Abf1 plays a role in silencing at different subtelomeric regions in C. glabrata. (A) Schematic representation of the subtelomeric region of the Chr E-R in C. glabrata. The light blue triangles represent the position of three independent insertions of the URA3 reporter, and the distance from the telomere is indicated above each insertion. The numbers beneath the triangles correspond to the insertions carried by each strain in the plate growth assay below. 5 µL of ten-fold serial dilutions of cells grown to stationary phase were spotted onto the media indicated, incubated at 30 °C for 48 h, and photographed. (B) Schematic representation of the subtelomeric region of Chr B-L. Yellow arrows represent alpha3 and alpha1 genes at the MTL3 silent locus and their relative orientation at the Chr B-L. The numbers below the insertions correspond to the numbers in the plate growth assay as in Figure 7A. The experiments were performed three independent times, and photographs from a representative experiment are shown (see in Section 2.10).
4. Discussion
The organization of DNA into chromatin and the particular spatial arrangement of the eukaryotic chromatin has a profound impact on the regulation of gene expression and other essential DNA metabolic functions like DNA replication, DNA damage repair, and regional silencing (reviewed in [34]).
In S. cerevisiae, ScAbf1 is an essential protein, classified as a GRF since it participates in a variety of important metabolic functions such as transcriptional activation and repression of many genes involved in diverse metabolic pathways, regional silencing, ribosome biogenesis, the establishment of the pre-replicative complex during the cell cycle and DNA repair, as it is part of the nucleotide excision repair subcomplex [5,35,36,37]. ScAbf1 has also been shown to locally alter chromatin structure by generating regions of altered nucleosome occupancy in the vicinity of ScAbf1 binding sites throughout the genome near promoters regulated by ScAbf1 [38]. In addition, ScAbf1 and ScRap1 prevent ectopic initiation of transcription at promoters by binding and occluding possible inappropriate pre-initiation complex formation [8]. Involvement in all of these processes could explain why ScAbf1 is essential in S. cerevisiae [10,39].
CgABF1 is an essential gene in C. glabrata.
The C. glabrata ortholog of ScABF1 (CgAbf1) has conserved the three most important domains for the function of ScAbf1: the Zinc finger domain at the N-terminus of the protein, which is 90% identical with the corresponding domain in ScAbf1, the specific DNA binding domain near the center of the protein (65% identical) and the CS2 domain at the C-terminus (88% identical) (Figure 1). The CS2 domain is required for all the known functions of ScAbf1, including transcriptional regulation, DNA replication, regional silencing, and reorganization of the chromatin structure [10].
In this work, we have determined that, as is the case in S. cerevisiae, in C. glabrata, CgABF1 is also required for cell viability since (a) a null abf1∆ mutant is unable to lose the complementing CgABF1 plasmid (Figure 2A) and (b) repression of Flag-CgABF1 from a shut-off plasmid results in the absence of growth and a large drop in viability in the abf1∆ background (Figure 2B and Supplementary Table S5). Therefore, it is possible that in C. glabrata, CgAbf1 also participates in essential functions such as DNA replication, transcriptional regulation, and regional silencing.
Misexpression of CgABF1 causes defects in cell cycle progression.
We found that repression of CgABF1 results in an abnormal cell cycle progression as abf1∆ cells complemented with the plasmid containing Flag-ABF1 driven by the repressible promoter from the MET3 gene, arrest transiently when Flag-ABF1 is repressed, but then continue cycling, which results in a small peak with DNA content equivalent to 3C and 4C (Figure 6A bottom panels). Indeed, repression of Abf1 results in the appearance of cells with more than one nucleus and also many cells with abnormally stained nuclei that appear as fragmented nuclei (Figure 5, top panels).
On the other hand, the overexpression of CgABF1 also leads to an abnormal cell cycle, characterized by the initial accumulation of cells in S and G2 phases, after which cells override the partial arrest and continue cycling, resulting in the accumulation of cells in G1 and S phases (Figure 6B). This correlates with a large proportion of budded cells after 6 h (~65%) and the appearance of cells with two and sometimes three nuclei (Figure 5). In addition, we observed a small number of cells that display higher than G2 DNA content (Figure 6B). We also found that overexpression of CgABF1 significantly increases the duplication time in the parental ABF1 background as well as in the C-terminal truncation mutant, which removes most of the CS2 conserved region (Table 1). These data are in agreement with the reported toxicity of ScABF1 overexpression experiments [32,33].
The abnormal DAPI staining and nuclear segregation of either depletion or overexpression of CgAbf1 might explain the drop in cell viability after CgAbf1 repression and point to an important role of CgAbf1 in DNA replication and passage through the cell cycle in C. glabrata. These data agree well with the described role of ScAbf1 in DNA replication, whereby studying point mutations in ScAbf1 and mutants in the cis-acting ARS binding sites in a plasmid showed that these mutations resulted in plasmid instability [40]. The C-terminal end of the protein is required for DNA replication activity, in particular the CS2 conserved domain [10].
Previous work has shown that the C. glabrata cell cycle progression shares many features with the corresponding process in S. cerevisiae. There are 253 origins of replication throughout the C. glabrata genome and a well-conserved ARS consensus sequence (ACS) [41] as well as high conservation of the functional domains of both Abf1 proteins. This is all consistent with the idea that CgAbf1 participates in DNA replication in C. glabrata.
Recently, Shor et al. [42] described that C. glabrata cells continue dividing in the presence of DNA damage induced by the addition of methyl methane sulfonate (MMS), which produces a substantial loss of viability. In our experiments, we also observed the absence of a prolonged arrest under repression or overexpression of CgABF1, which probably contributes to the large drop in viability after depletion of CgAbf1.
It should be pointed out that the promoter from the MET3 gene is not strong enough to provide normal growth to the abf1∆ null strain, since even under conditions where it is expressed (minimal media without amino acids), the cells grow slowly compared to the same strain with the plasmid with ABF1 driven by its own promoter (127 min vs. 76 min; Supplementary Table S4). Therefore, in C. glabrata, CgAbf1 is probably also an abundant protein whose levels are tightly controlled during the cell cycle. Additionally, the lack of proper regulation of the MET3 promoter could also account for part of the abnormal growth of the abf1∆ null strain containing this plasmid.
The C-terminal domain of CgAbf1 participates in subtelomeric silencing in C. glabrata.
In this work, we found that CgAbf1 participates in subtelomeric silencing in at least two different subtelomeric regions: E-R and B-L, and this function is dependent on the last 43 amino acids of CgAbf1, which include most of the highly conserved CS2 domain (Figure 7). The role CgAbf1 plays in subtelomeric silencing is only detected at distances >11 Kb in B-L telomere and >20 Kb in the case of the subtelomeric E-R region. We have shown previously that in C. glabrata, the propagation of the silent chromatin from the telomeres can extend up to >20 Kb (depending on the telomere) [18,19,20]. It is possible that the effect of CgAbf1 in silencing can only be detected at positions sufficiently distant from the respective telomere so that the silencing propagating from the telomere is diminished. At regions closer to the telomere, there is likely redundant silencing activity of all the silencing proteins acting at subtelomeric regions, and the contribution of CgAbf1 to silencing is masked. Consistent with the contribution of CgAbf1 to subtelomeric silencing at the E-R region, we have determined that CgAbf1 and CgRap1 are recruited throughout this region, where a cluster of the adhesin-encoding genes EPA1, EPA2, and EPA3 reside [23].
On the other hand, at telomere I-R where silencing propagates over 23 Kb from the telomere, the effect in the silencing of the URA3 reporter by CgAbf1-43 is not observed. This is probably because silencing at this telomere is even stronger than at other telomeres since a stem and loop structure may be formed, as EPA4 and EPA5 form an inverted repeat, contributing to a strong silencing at this telomere, which would hide the possible contribution of CgAbf1 silencing at this telomere (Supplementary Figure S4).
In this regard, CgAbf1 also conserves the function in the silencing of ScAbf1, where it contributes to regional silencing shown in experiments using an artificial HMR-E silencer containing an ARS consensus sequence and two consensus binding sites for Abf1 [43].
One of the proposed mechanisms of ScAbf1 action is that its binding to the B-domain of ARS1 of S. cerevisiae prevents nucleosome positioning in the adjacent A domain [44]. In fact, ScAbf1 can compete with histones to generate nucleosome-free regions to favor the access of other proteins and regulate transcription or DNA replication [39].
Therefore, when ScAbf1 binds to the different binding sites throughout the genome, it can change the local chromatin structure [38]. This effect may be driven by regulating nucleosome positioning at the regions where this protein binds. Nucleosome distribution is not random nor determined only by favorable DNA sequences. Instead, the action of different proteins like transcription factors, and general regulatory factors such as Abf1 and Rap1, usually have a strong effect and determine nucleosome positioning [45]. It is possible that CgAbf1 acts similarly to ScAbf1, changing the local chromatin structure by controlling nucleosome positioning in the vicinity of its binding sites near promoters, ARSs, or silencers and protosilencers.
Future experiments will be directed at answering some of these questions.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof7121005/s1, Figure S1: CgAbf1 is similar to its ortholog ScAbf1 and conserves the functional domains. Figure S2: Plasmid shuffling scheme to generate a “shut off ABF1 system” or an “ABF1 overexpression system” (OE-ABF1) in the abf1∆ background. Figure S3: The abf1-43 mutant has a longer duplication time and is temperature-sensitive at 45 °C. Figure S4: CgAbf1 is not required for silencing at telomere I-R where a stem-loop structure may be formed. Table S1: Candida glabrata and Escherichia coli strains used in this study; Table S2: plasmids used in this study; Table S3: oligonucleotides used in this study; Table S4: Duplication times of strains containing ABF1 driven by the MET3 promoter; Table S5: Cell viability of strains expressing ABF1 under the repressible MET3 promoter; Table S6: Percentage of unbudded and budded cells under repression or overexpression of ABF1.
Author Contributions
G.H.-H.: constructed most of the strains, performed the silencing assays, DNA content experiments, the plasmid shuffling, and plasmid loss experiments, constructed the over-expression plasmids and determined the growth and morphology phenotypes of strains containing them, and also contributed to draft preparation, writing and reviewing of the manuscript. L.A.V.-S.: constructed strains and shut off plasmids, performed growth and viability assays, morphological characterization, and DAPI and DNA content experiments, and also contributed to draft preparation and reviewing of the manuscript. L.C.: constructed complementing plasmids and abf1-43 truncation allele and initial characterization of the mutants. E.L.-F.: constructed mutants and performed initial characterization of mutants. G.G.-E.: constructed mutant strains and vectors, performed flow cytometry experiments, and project administration. A.D.L.P.: contributed to conceptualization, writing, reviewing, and editing of the manuscript. I.C.: Conceptualized and designed the experiments, acquired funding, wrote, reviewed, and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by CONACYT, grant number FORDECYT-PRONACES—Ciencia de Frontera 2019 No. 610281” and CB No. 2014—239629 to I.C. “The APC was funded by IPICYT fellowship No. IPI-SAC-FMT-BEAE-009—2470_1_PV to G.H.-H.”. G.H.-H., L.A.V.-S., L.C., and E.L.-F. received fellowships from CONACYT No. 712172, 104640, 448801, and 261740, respectively.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
You might choose to exclude this statement if the study did not report any data.
Acknowledgments
The authors wish to thank the Laboratorio Nacional de Biotecnología Agrícola, Médica y Ambiental (LANBAMA) and Verónica Zárate for excellent technical assistance with sample sequencing. Nicolás Gomez for invaluable technical advice and Olga Araceli Patrón-Soberano for excellent technical assistance with fluorescence microscopy.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Buchman, A.R.; Kimmerly, W.J.; Rine, J.; Kornberg, R.D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol. Cell. Biol. 1988, 8, 210–225. [Google Scholar] [CrossRef] [PubMed]
- Kimmerly, W.; Buchman, A.; Kornberg, R.; Rine, J. Roles of two DNA-binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. Embo J. 1988, 7, 2241–2253. [Google Scholar] [CrossRef] [PubMed]
- Yarragudi, A.; Miyake, T.; Li, R.; Morse, R.H. Comparison of ABF1 and RAP1 in Chromatin Opening and Transactivator Potentiation in the Budding Yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 2004, 24, 9152–9164. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.I.; Rinaldi, N.J.; Robert, F.; Odom, D.T.; Bar-Joseph, Z.; Gerber, G.K.; Hannett, N.M.; Harbison, C.T.; Thompson, C.M.; Simon, I.; et al. Transcriptional Regulatory Networks in Saccharomyces cerevisiae. Science 2002, 298, 799–804. [Google Scholar] [CrossRef]
- Miyake, T.; Reese, J.; Loch, C.M.; Auble, D.T.; Li, R. Genome-wide Analysis of ARS (Autonomously Replicating Sequence) Binding Factor 1 (Abf1p)-mediated Transcriptional Regulation in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 34865–34872. [Google Scholar] [CrossRef] [PubMed]
- Rhode, P.R.; Sweder, K.S.; Oegema, K.F.; Campbell, J.L. The gene encoding ARS-binding factor I is essential for the viability of yeast. Genes Dev. 1989, 3, 1926–1939. [Google Scholar] [CrossRef]
- Loo, S.; Laurenson, P.; Foss, M.; Dillin, A.; Rine, J. Roles of ABF1, NPL3, and YCL54 in silencing in Saccharomyces cerevisiae. Genetics 1995, 141, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Challal, D.; Barucco, M.; Kubik, S.; Feuerbach, F.; Candelli, T.; Geoffroy, H.; Benaksas, C.; Shore, D.; Libri, D. General Regulatory Factors Control the Fidelity of Transcription by Restricting Non-coding and Ectopic Initiation. Mol. Cell 2018, 72, 955–969.e7. [Google Scholar] [CrossRef]
- Cho, G.; Kim, J.; Rho, H.M.; Jung, G. Structure-function analysis of the DNA binding domain of Saccharomyces cerevisiae ABF1. Nucleic Acids Res. 1995, 23, 2980–2987. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miyake, T.; Loch, C.M.; Li, R. Identification of a Multifunctional Domain in Autonomously Replicating Sequence-Binding Factor 1 Required for Transcriptional Activation, DNA Replication, and Gene Silencing. Mol. Cell. Biol. 2002, 22, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Kullberg, B.J.; Arendrup, M.C. Invasive Candidiasis. N. Engl. J. Med. 2015, 373, 1445–1456. [Google Scholar] [CrossRef] [PubMed]
- Pappas, P.G.; Lionakis, M.S.; Arendrup, M.C.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive candidiasis. Nat. Rev. Dis. Prim. 2018, 4, 18026. [Google Scholar] [CrossRef] [PubMed]
- Toda, M.; Williams, S.R.; Berkow, E.L.; Farley, M.M.; Harrison, L.H.; Bonner, L.; Marceaux, K.M.; Hollick, R.; Zhang, A.Y.; Schaffner, W.; et al. Population-Based Active Surveillance for Culture-Confirmed Candidemia—Four Sites, United States, 2012–2016. MMWR. Surveill. Summ. 2019, 68, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Green, B.; Benoit, N.; Schatz, M.; Wheelan, S.; Cormack, B. De novo genome assembly of Candida glabrata reveals cell wall protein complement and structure of dispersed tandem repeat arrays. Mol. Microbiol. 2020, 113, 1209–1224. [Google Scholar] [CrossRef]
- Xu, Z.; Green, B.; Benoit, N.; Sobel, J.D.; Schatz, M.C.; Wheelan, S.; Cormack, B.P. Cell wall protein variation, break-induced replication, and subtelomere dynamics in Candida glabrata. Mol. Microbiol. 2021, 116, 260–276. [Google Scholar] [CrossRef] [PubMed]
- Cormack, B.P.; Falkow, S. Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics 1999, 151, 979–987. [Google Scholar] [CrossRef]
- Cormack, B.P.; Ghori, N.; Falkow, S. An Adhesin of the Yeast Pathogen Candida glabrata Mediating Adherence to Human Epithelial Cells. Science 1999, 285, 578–582. [Google Scholar] [CrossRef]
- Castano, I.; Pan, S.-J.; Zupancic, M.; Hennequin, C.; Dujon, B.; Cormack, B.P. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol. Microbiol. 2004, 55, 1246–1258. [Google Scholar] [CrossRef]
- Peñas, A.D.L.; Pan, S.-J.; Castaño, I.; Alder, J.; Cregg, R.; Cormack, B.P. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 2003, 17, 2245–2258. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Hernández, L.L.; Juárez-Reyes, A.; Arroyo-Helguera, O.-E.; Penas, A.D.L.; Pan, S.-J.; Cormack, B.P.; Castaño, I. yKu70/yKu80 and Rif1 Regulate Silencing Differentially at Telomeres in Candida glabrata. Eukaryot. Cell 2008, 7, 2168–2178. [Google Scholar] [CrossRef] [PubMed]
- Gallegos-García, V.; Pan, S.-J.; Juárez-Cepeda, J.; Ramírez-Zavaleta, C.Y.; Martin-Del-Campo, M.B.; Martínez-Jiménez, V.; Castano, I.; Cormack, B.; Penas, A.D.L. A Novel Downstream Regulatory Element Cooperates with the Silencing Machinery to Repress EPA1 Expression in Candida glabrata. Genetics 2012, 190, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Reyes, A.; Ramírez-Zavaleta, C.Y.; Medina-Sánchez, L.; Peñas, A.D.L.; Castaño, I. A Protosilencer of Subtelomeric Gene Expression in Candida glabrata with Unique Properties. Genetics 2012, 190, 101–111. [Google Scholar] [CrossRef] [PubMed]
- López-Fuentes, E.; Hernández-Hernández, G.; Castanedo, L.; Gutiérrez-Escobedo, G.; Oktaba, K.; Penas, A.D.L.; Castaño, I. Chromatin Loop Formation Induced by a Subtelomeric Protosilencer Represses EPA Genes in Candida glabrata. Genetics 2018, 210, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Sherman, F.; Fink, G.R.; Hicks, J.B. Methods in Yeast Genetics; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1986. [Google Scholar]
- Ausubel, F.R.; Brent, R.E.; Kingston, d.D.; Moore, J.G.; Seidman, J.A.; Struhl, K. Current Protocols in Molecular Biology; Wiley & Sons, Inc.: New York, NY, USA, 2001. [Google Scholar]
- Castaño, I.; Kaur, R.; Pan, S.; Cregg, R.; Peñas, A.D.L.; Guo, N.; Biery, M.C.; Craig, N.L.; Cormack, B.P. Tn7-Based Genome-Wide Random Insertional Mutagenesis of Candida glabrata. Genome Res. 2003, 13, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol. Biol. 2014, 1205, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Escobedo, G.; Orta-Zavalza, E.; Castaño, I.; Peñas, A.D.L. Role of glutathione in the oxidative stress response in the fungal pathogen Candida glabrata. Curr. Genet. 2013, 59, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Carrillo, P.; Orta-Zavalza, E.; Gutiérrez-Escobedo, G.; Patron-Soberano, A.; Peñas, A.D.L.; Castaño, I. Expression vectors for C-terminal fusions with fluorescent proteins and epitope tags in Candida glabrata. Fungal Genet. Biol. 2015, 80, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Robledo-Márquez, K.; Gutiérrez-Escobedo, G.; Yáñez-Carrillo, P.; Vidal-Aguiar, Y.; Orta-Zavalza, E.; Briones-Martín-Del-Campo, M.; Peñas, A.D.L.; Calderone, R.; Castaño, I. Candida glabrata encodes a longer variant of the mating type (MAT) alpha2 gene in the mating type-like MTL3 locus, which can form homodimers. FEMS Yeast Res. 2016, 16, fow082. [Google Scholar] [CrossRef] [PubMed]
- Zordan, R.E.; Ren, Y.; Pan, S.-J.; Rotondo, G.; Penas, A.D.L.; Iluore, J.; Cormack, B.P. Expression Plasmids for Use in Candida glabrata. G3 Genes Genomes Genet. 2013, 3, 1675–1686. [Google Scholar] [CrossRef] [PubMed]
- Sopko, R.; Huang, D.; Preston, N.; Chua, G.; Papp, B.; Kafadar, K.; Snyder, M.; Oliver, S.G.; Cyert, M.; Hughes, T.R.; et al. Mapping Pathways and Phenotypes by Systematic Gene Overexpression. Mol. Cell 2006, 21, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, L.F.; Kennedy, B.; Harlow, E. A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc. Natl. Acad. Sci. USA 2001, 98, 3946–3951. [Google Scholar] [CrossRef] [PubMed]
- Bonev, B.; Cavalli, G. Organization and function of the 3D genome. Nat. Rev. Genet. 2016, 17, 661–678. [Google Scholar] [CrossRef]
- Fermi, B.; Bosio, M.C.; Dieci, G. Promoter architecture and transcriptional regulation of Abf1-dependent ribosomal protein genes inSaccharomyces cerevisiae. Nucleic Acids Res. 2016, 44, 6113–6126. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kawasaki, Y.; Kojima, A.; Seki, T.; Sugino, A.; Kim, H.-D. Reconstitution ofSaccharomyces cerevisiaeprereplicative complex assemblyin vitro. Genes Cells 2006, 11, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Reed, S.H.; Akiyama, M.; Stillman, B.; Friedberg, E.C. Yeast autonomously replicating sequence binding factor is involved in nucleotide excision repair. Genes Dev. 1999, 13, 3052–3058. [Google Scholar] [CrossRef]
- Ganapathi, M.; Palumbo, M.J.; Ansari, S.A.; He, Q.; Tsui, K.; Nislow, C.; Morse, R.H. Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucleic Acids Res. 2010, 39, 2032–2044. [Google Scholar] [CrossRef] [PubMed]
- Yarragudi, A.; Parfrey, L.W.; Morse, R.H. Genome-wide analysis of transcriptional dependence and probable target sites for Abf1 and Rap1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2006, 35, 193–202. [Google Scholar] [CrossRef]
- Rhode, P.R.; Elsasser, S.; Campbell, J.L. Role of multifunctional autonomously replicating sequence binding factor 1 in the initiation of DNA replication and transcriptional control in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992, 12, 1064–1077. [Google Scholar] [CrossRef]
- Descorps-Declère, S.; Saguez, C.; Cournac, A.; Marbouty, M.; Rolland, T.; Ma, L.; Bouchier, C.; Moszer, I.; Dujon, B.; Koszul, R.; et al. Genome-wide replication landscape of Candida glabrata. BMC Biol. 2015, 13, 69. [Google Scholar] [CrossRef] [PubMed]
- Shor, E.; Garcia-Rubio, R.; DeGregorio, L.; Perlin, D.S. A Noncanonical DNA Damage Checkpoint Response in a Major Fungal Pathogen. mBio 2020, 11, e03044-20. [Google Scholar] [CrossRef] [PubMed]
- Rivier, D.H.; Ekena, J.L.; Rine, J. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics 1999, 151, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Venditti, P.; Costanzo, G.; Negri, R.; Camilloni, G. ABFI contributes to the chromatin organization of Saccharomyces cerevisiae ARS1 B-domain. Biochim. et Biophys. Acta (BBA)-Gene Struct. Expr. 1994, 1219, 677–689. [Google Scholar] [CrossRef]
- Lai, W.K.M.; Pugh, W.K.M.L.B.F. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol. 2017, 18, 548–562. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).