Differential Requirement for the Cell Wall Integrity Sensor Wsc1p in Diploids Versus Haploids
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Yeast Techniques, Strain and Plasmid Construction
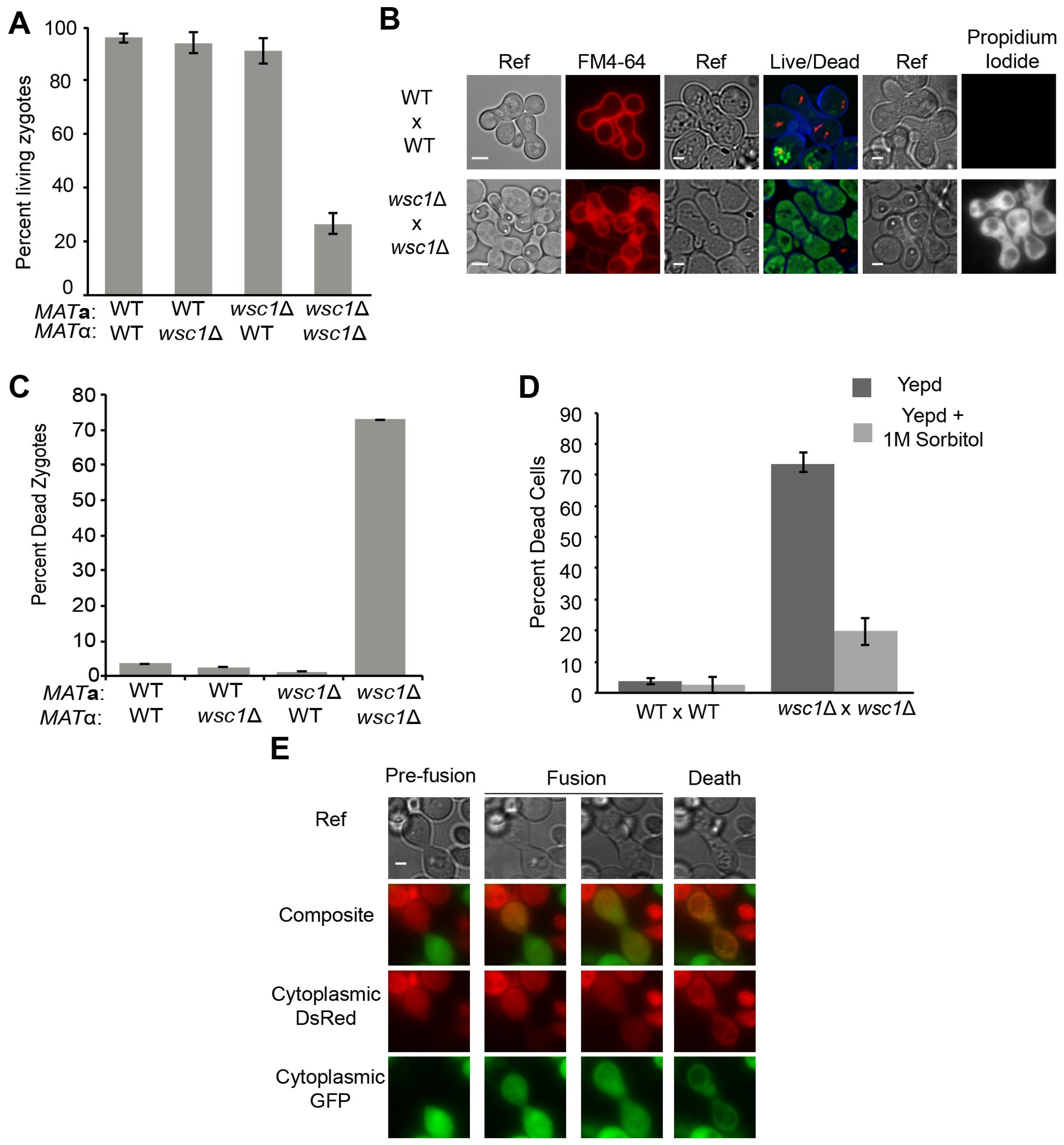
2.2. Yeast Mating Assays
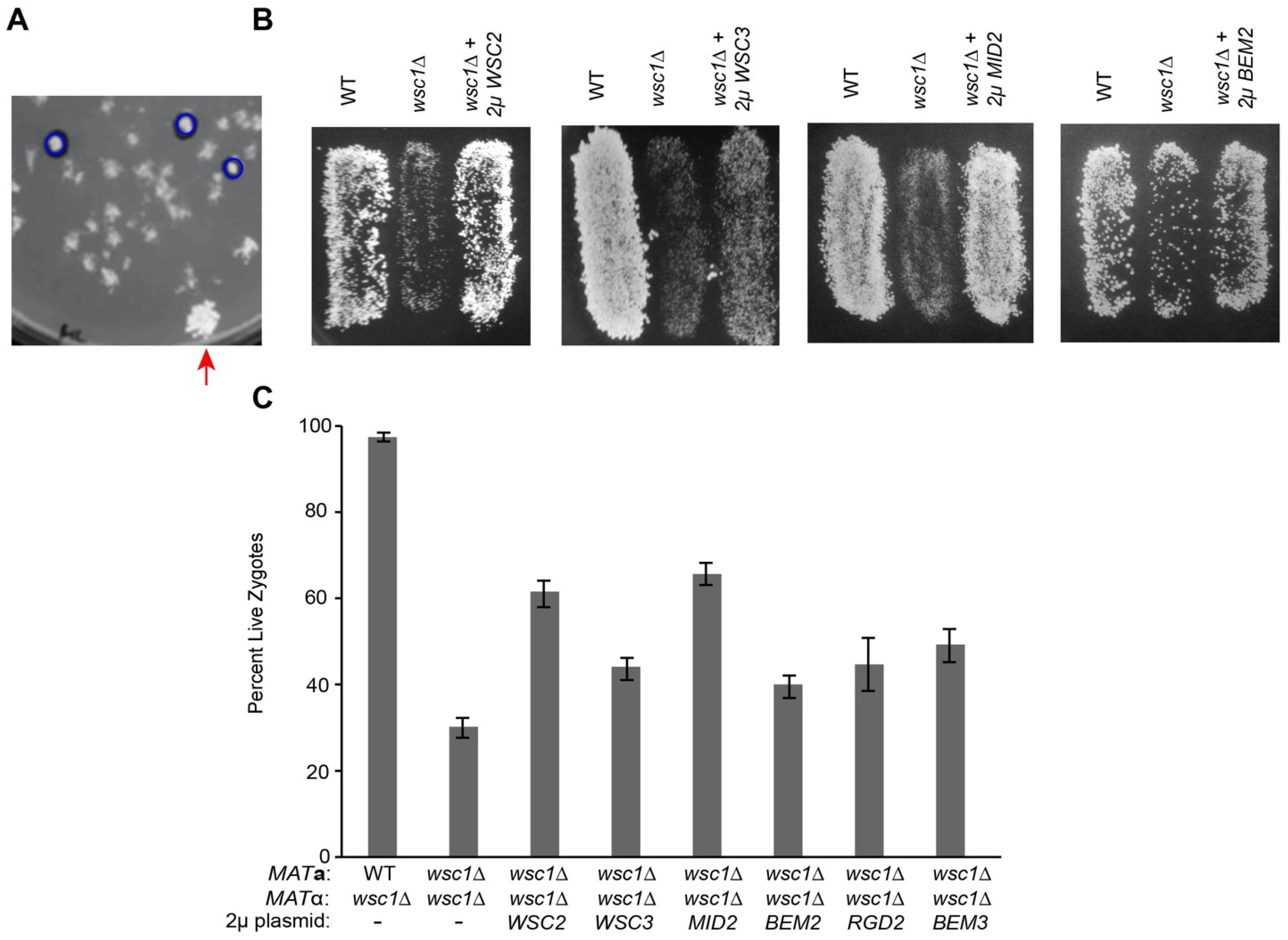
2.3. High Copy Suppression of wsc1∆
2.4. Microscopy
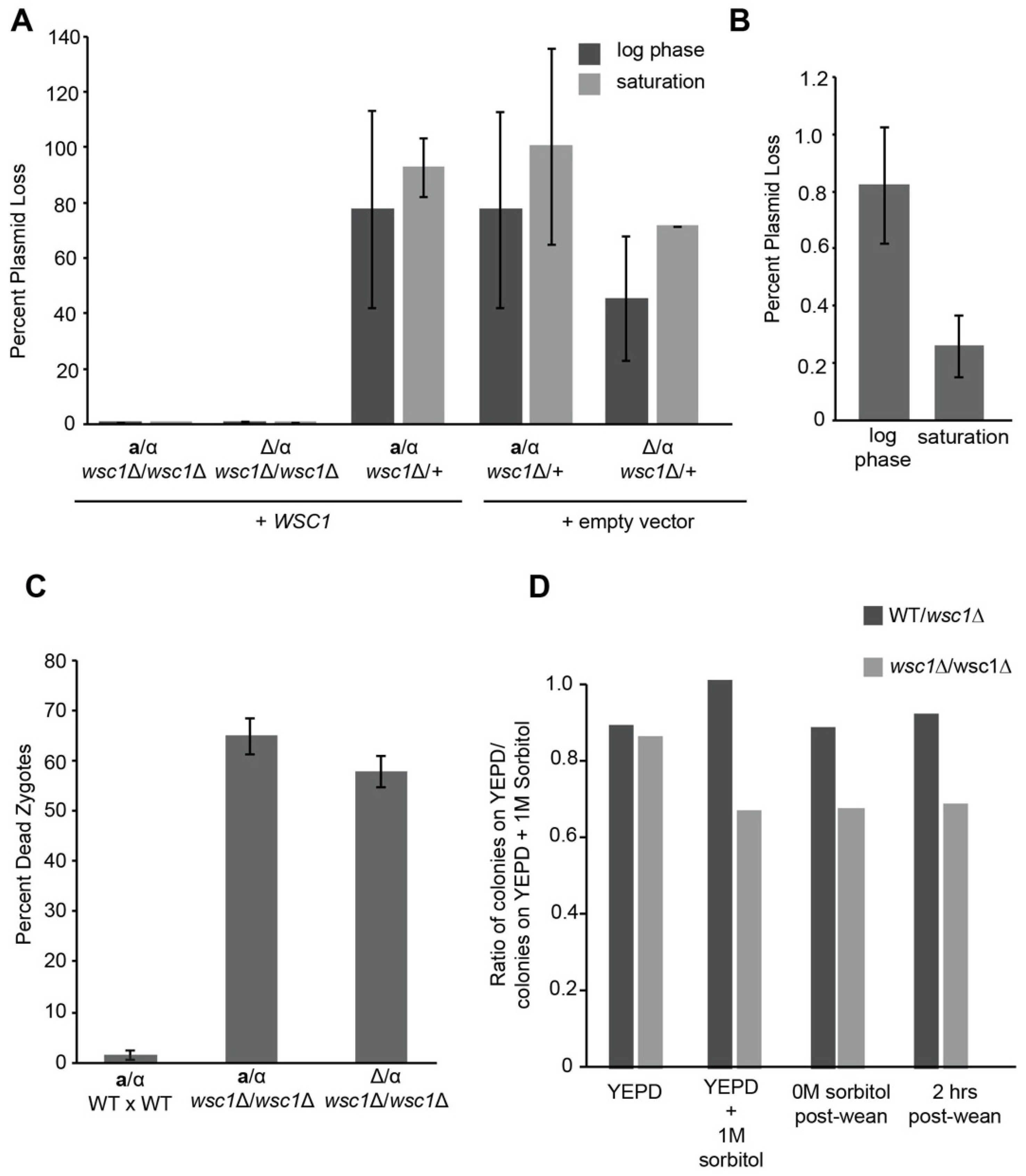
2.5. WSC1 Plasmid Loss and Weaning Experiments
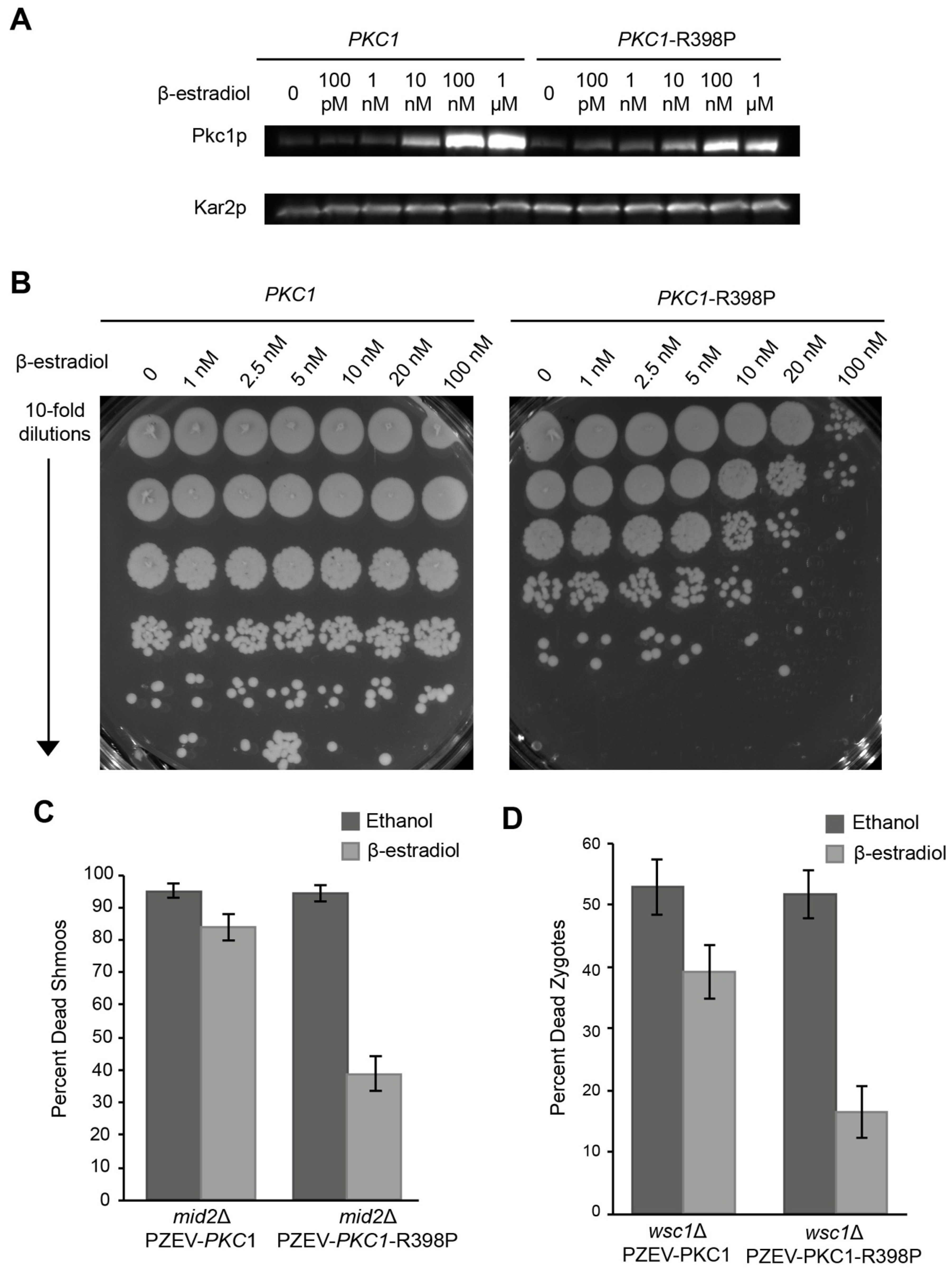
2.6. Western Blotting
3. Results
3.1. WSC1 Is Required for Diploid Survival
3.2. Overexpression of CWI Pathway Components Suppresses wsc1Δ Zygote Death
3.3. Diploid Cells Are Sensitive to Loss of WSC1
3.4. Overexpression of Hyperactivated Pkc1p Causes Growth Defects
4. Discussion
4.1. WSC1 Is the Most Important CWI Sensor in Zygotes
4.2. WSC1 May Be Involved in Maintaining Cell Wall Integrity during Bud Emergence
4.3. Constitutive Activation of the CWI Pathway Is Detrimental to Cells
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cid, V.J.; Cenamor, R.; Sanchez, M.; Nombela, C. A mutation in the Rho1-GAP-encoding gene BEM2 of Saccharomyces cerevisiae affects morphogenesis and cell wall functionality. Microbiology 1998, 144 Pt 1, 25–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klis, F.M.; Boorsma, A.; De Groot, P.W. Cell wall construction in Saccharomyces cerevisiae. Yeast 2006, 23, 185–202. [Google Scholar] [CrossRef] [PubMed]
- Delley, P.A.; Hall, M.N. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 1999, 147, 163–174. [Google Scholar] [CrossRef] [Green Version]
- Levin, D.E. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005, 69, 262–291. [Google Scholar] [CrossRef] [Green Version]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [Green Version]
- Ketela, T.; Green, R.; Bussey, H. Saccharomyces cerevisiae mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J. Bacteriol. 1999, 181, 3330–3340. [Google Scholar] [CrossRef] [Green Version]
- Lodder, A.L.; Lee, T.K.; Ballester, R. Characterization of the Wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics 1999, 152, 1487–1499. [Google Scholar] [CrossRef] [PubMed]
- Philip, B.; Levin, D.E. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol 2001, 21, 271–280. [Google Scholar] [CrossRef] [Green Version]
- Rajavel, M.; Philip, B.; Buehrer, B.M.; Errede, B.; Levin, D.E. Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 3969–3976. [Google Scholar] [CrossRef] [Green Version]
- Hall, A.E.; Rose, M.D. Cell fusion in yeast is negatively regulated by components of the cell wall integrity pathway. Mol. Biol. Cell 2019, 30, 441–452. [Google Scholar] [CrossRef]
- Douglas, C.M.; Foor, F.; Marrinan, J.A.; Morin, N.; Nielsen, J.B.; Dahl, A.M.; Mazur, P.; Baginsky, W.; Li, W.; El-Sherbeini, M. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-D-glucan synthase. Proc. Natl. Acad. Sci. USA 1994, 91, 12907–12911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drgonova, J.; Drgon, T.; Tanaka, K.; Kollár, R.; Chen, G.-C.; Ford, R.A.; Chan, C.S.M.; Takai, Y.; Cabib, E. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science 1996, 272, 277–279. [Google Scholar] [CrossRef]
- Inoue, S.B.; Takewakt, N.; Takasuka, T.; Mio, T.; Adachi, M.; Fujii, Y.; Miyamoto, C.; Arisawa, M.; Furuichi, Y.; Watanabe, T. Characterization and gene cloning of 1,3-beta-D-glucan synthase from Saccharomyces cerevisiae. Eur. J. Biochem. 1995, 231, 845–854. [Google Scholar] [CrossRef]
- Kamada, Y.; Qadota, H.; Python, C.P.; Anraku, Y.; Ohya, Y.; Levin, D. Activation of yeast protein kinase C by Rho1 GTPase. J. Biol. Chem. 1996, 271, 9193–9196. [Google Scholar] [CrossRef] [Green Version]
- Mazur, P.; Baginsky, W. In vitro activity of 1,3-beta-D-glucan synthase requires the GTP-binding protein Rho1. J. Biol. Chem. 1996, 271, 14604–14609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nonaka, H.; Tanaka, K.; Hirano, H.; Fujiwara, T.; Kohno, H.; Umikawa, M.; Mino, A.; Takai, Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995, 14, 5931–5938. [Google Scholar] [CrossRef]
- Ozaki, K.; Tanaka, K.; Imamura, H.; Hihara, T.; Kameyama, T.; Nonaka, H.; Hirano, H.; Matsuura, Y.; Takai, Y. Rom1p and Rom2p are GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996, 15, 2196–2207. [Google Scholar] [CrossRef]
- Qadota, H.; Python, C.P.; Inoue, S.B.; Arisawa, M.; Anraku, Y.; Zheng, Y.; Watanabe, T.; Levin, D.E.; Ohya, Y. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science 1996, 272, 279–281. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.; Zheng, Y.; Bender, L.; Myers, A.; Cerione, R.; Bender, A. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J. Cell Biol. 1994, 127, 1395–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roumanie, O.; Weinachter, C.; Larrieu, I.; Crouzet, M.; Doignon, F. Functional characterization of the Bag7, Lrg1 and Rgd2 RhoGAP proteins from Saccharomyces cerevisiae. FEBS Lett. 2001, 506, 149–156. [Google Scholar] [CrossRef]
- Schmidt, A.; Bickle, M.; Beck, T.; Hall, M.N. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 1997, 88, 531–542. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, D.; Abe, M.; Ohya, Y. Yeast Lrg1p acts as a specialized RhoGAP regulating 1,3-beta-glucan synthesis. Yeast 2001, 18, 943–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, A.; Schmelzle, T.; Hall, M.N. The RHO1-GAPs SAC7, BEM2 and BAG7 control distinct RHO1 functions in Saccharomyces cerevisiae. Mol. Microbiol. 2002, 45, 1433–1441. [Google Scholar]
- Martin, H.; Rodriguez-Pachon, J.M.; Ruiz, C.; Nombela, C.; Molina, M. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 1511–1519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, D.E.; Bowers, B.; Chen, C.Y.; Kamada, Y.; Watanabe, M. Dissecting the protein kinase C/MAP kinase signalling pathway of Saccharomyces cerevisiae. Cell. Mol. Biol. Res. 1994, 40, 229–239. [Google Scholar] [PubMed]
- Paravicini, G.; Friedli, L. Protein-protein interactions in the yeast PKC1 pathway: Pkc1p interacts with a component of the MAP kinase cascade. Mol. Gen. Genet. 1996, 251, 682–691. [Google Scholar]
- Kamada, Y.; Jung, U.S.; Piotrowski, J.; Levin, D.E. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995, 9, 1559–1571. [Google Scholar] [CrossRef] [Green Version]
- Irie, K.; Takase, M.; Lee, K.S.; Levin, D.E.; Araki, H.; Matsumoto, K.; Oshima, Y. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol. Cell. Biol. 1993, 13, 3076–3083. [Google Scholar]
- Lee, K.S.; Irie, K.; Gotoh, Y.; Watanabe, Y.; Araki, H.; Nishida, E.; Matsumoto, K.; Levin, D.E. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol. Cell. Biol. 1993, 13, 3067–3075. [Google Scholar]
- Lee, K.S.; Levin, D.E. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol. Cell. Biol. 1992, 12, 172–182. [Google Scholar]
- Torres, L.; Martín, H.; García-Saez, M.I.; Arroyo, J.; Molina, M.; Sánchez, M.; Nombela, C. A protein kinase gene complements the lytic phenotype of Saccharomyces cerevisiae lyt2 mutants. Mol. Microbiol. 1991, 5, 2845–2854. [Google Scholar] [CrossRef]
- Jacoby, J.J.; Nilius, S.M.; Heinisch, J.J. A screen for upstream components of the yeast protein kinase C signal transduction pathway identifies the product of the SLG1 gene. Mol. Gen. Genet. 1998, 258, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Reinoso-Martin, C.; Schuller, C.; Schuetzer-Muehlbauer, M.; Kuchler, K. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot. Cell 2003, 2, 1200–1210. [Google Scholar] [CrossRef] [Green Version]
- Dupres, V.; Alsteens, D.; Wilk, S.; Hansen, B.; Heinisch, J.J.; Dufrêne, Y.F. The yeast Wsc1 cell surface sensor behaves like a nanospring in vivo. Nat. Chem. Biol. 2009, 5, 857–862. [Google Scholar] [CrossRef]
- Heinisch, J.J.; Dufrene, Y.F. Is there anyone out there?--Single-molecule atomic force microscopy meets yeast genetics to study sensor functions. Integr. Biol. 2010, 2, 408–415. [Google Scholar] [CrossRef]
- Kock, C.; Arlt, H.; Ungermann, C.; Heinisch, J.J. Yeast cell wall integrity sensors form specific plasma membrane microdomains important for signalling. Cell. Microbiol. 2016, 18, 1251–1267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huh, W.-K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef]
- Straede, A.; Heinisch, J.J. Functional analyses of the extra- and intracellular domains of the yeast cell wall integrity sensors Mid2 and Wsc1. FEBS Lett. 2007, 581, 4495–4500. [Google Scholar] [CrossRef] [Green Version]
- Gray, J.V.; Ogas, J.P.; Kamada, Y.; Stone, M.; Levin, D.; Herskowitz, I. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 1997, 16, 4924–4937. [Google Scholar] [CrossRef] [PubMed]
- Verna, J.; Lodder, A.; Lee, K.; Vagts, A.; Ballester, R. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1997, 94, 13804–13809. [Google Scholar] [CrossRef] [Green Version]
- Philips, J.; Herskowitz, I. Osmotic balance regulates cell fusion during mating in Saccharomyces cerevisiae. J. Cell Biol. 1997, 138, 961–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gammie, A.E.; Brizzio, V.; Rose, M.D. Distinct morphological phenotypes of cell fusion mutants. Mol. Biol. Cell 1998, 9, 1395–1410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merlini, L.; Dudin, O.; Martin, S.G. Mate and fuse: How yeast cells do it. Open Biol. 2013, 3, 130008. [Google Scholar] [CrossRef]
- Ydenberg, C.A.; Rose, M.D. Yeast mating: A model system for studying cell and nuclear fusion. Methods Mol. Biol. 2008, 475, 3–20. [Google Scholar] [CrossRef]
- Chen, E.H.; Grote, E.; Mohler, W.; Vignery, A. Cell-cell fusion. FEBS Lett. 2007, 581, 2181–2193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Bortz, E.; Zhong, H.; Leeuw, T.; Leberer, E.; Vershon, A.K.; Hirsch, J.P. Localization and signaling of G(beta) subunit Ste4p are controlled by a-factor receptor and the a-specific protein Asg7p. Mol. Cell. Biol. 2000, 20, 8826–8835. [Google Scholar] [CrossRef] [Green Version]
- Amberg, D.C.; Burke, D.; Strathern, J.N.; Burke, D. Cold Spring Harbor Laboratory. In Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual; Cold Spring Harbor Laboratory Press: ColdSpring Harbor, NY, USA, 2005. [Google Scholar]
- Gammie, A.E.; Rose, M.D. Assays of cell and nuclear fusion. Methods Enzymol. 2002, 351, 477–498. [Google Scholar]
- Broach, J.R.; Strathern, J.N.; Hicks, J.B. Transformation in yeast: Development of a hybrid cloning vector and isolation of the CAN1 gene. Gene 1979, 8, 121–133. [Google Scholar] [CrossRef]
- Grote, E. Cell fusion assays for yeast mating pairs. Methods Mol. Biol. 2008, 475, 165–196. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.D.; Misra, L.M.; Vogel, J.P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 1989, 57, 1211–1221. [Google Scholar] [CrossRef]
- Fischer-Parton, S.; Parton, R.M.; Hickey, P.C.; Dijksterhuis, J.; Atkinson, H.A.; Read, N.D. Confocal microscopy of FM4-64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. J. Microsc. 2000, 198, 246–259. [Google Scholar] [CrossRef] [Green Version]
- Yadav, V.R.; Prasad, S.; Kannappan, R.; Ravindran, J.; Chaturvedi, M.M.; Vaahtera, L.; Parkkinen, J.; Aggarwal, B.B. Cyclodextrin-complexed curcumin exhibits anti-inflammatory and antiproliferative activities superior to those of curcumin through higher cellular uptake. Biochem. Pharmacol. 2010, 80, 1021–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millard, P.J.; Roth, B.L.; Thi, H.P.; Yue, S.T.; Haugland, R.P. Development of the FUN-1 family of fluorescent probes for vacuole labeling and viability testing of yeasts. Appl. Environ. Microbiol. 1997, 63, 2897–2905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deere, D.; Shen, J.; Vesey, G.; Bell, P.; Bissinger, P.; Veal, D. Flow cytometry and cell sorting for yeast viability assessment and cell selection. Yeast 1998, 14, 147–160. [Google Scholar] [CrossRef]
- Bender, A.; Pringle, J.R. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991, 11, 1295–1305. [Google Scholar] [PubMed] [Green Version]
- Kim, Y.J.; Francisco, L.; Chen, G.C.; Marcotte, E.; Chan, C.S. Control of cellular morphogenesis by the Ip12/Bem2 GTPase-activating protein: Possible role of protein phosphorylation. J. Cell Biol. 1994, 127, 1381–1394. [Google Scholar] [CrossRef]
- Zheng, Y.; Cerione, R.; Bender, A. Control of the yeast bud-site assembly GTPase Cdc42. Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J. Biol. Chem. 1994, 269, 2369–2372. [Google Scholar] [CrossRef]
- Zheng, Y.; Hart, M.J.; Shinjo, K.; Evans, T.; Bender, A.; Cerione, R.A. Biochemical comparisons of the Saccharomyces cerevisiae Bem2 and Bem3 proteins. Delineation of a limit Cdc42 GTPase-activating protein domain. J. Biol. Chem. 1993, 268, 24629–24634. [Google Scholar] [CrossRef]
- Schmitz, H.P.; Huppert, S.; Lorberg, A.; Heinisch, J.J. Rho5p downregulates the yeast cell integrity pathway. J. Cell Sci. 2002, 115, 3139–3148. [Google Scholar] [CrossRef]
- McIsaac, R.S.; Oakes, B.L.; Wang, X.; Dummit, K.A.; Botstein, D.; Noyes, M.B. Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast. Nucleic Acids Res. 2013, 41, e57. [Google Scholar] [CrossRef] [Green Version]
- De Godoy, L.M.; Olsen, J.V.; Cox, J.; Nielsen, M.L.; Hubner, N.C.; Fröhlich, F.; Walther, T.C.; Mann, M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 2008, 455, 1251–1254. [Google Scholar] [CrossRef]
- Mukherjee, D.; Sen, A.; Boettner, D.R.; Fairn, G.D.; Schlam, D.; Valentin, F.J.B.; McCaffery, J.M.; Hazbun, T.; Staiger, C.J.; Grinstein, S.; et al. Bem3, a Cdc42 GTPase-activating protein, traffics to an intracellular compartment and recruits the secretory Rab GTPase Sec4 to endomembranes. J. Cell Sci. 2013, 126, 4560–4571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bos, J.L.; Rehmann, H.; Wittinghofer, A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell 2007, 129, 865–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sprang, S.R. G proteins, effectors and GAPs: Structure and mechanism. Curr. Opin. Struct. Biol. 1997, 7, 849–856. [Google Scholar] [CrossRef]
- Kahn, R.A. GAPs: Terminator versus effector functions and the role(s) of ArfGAP1 in vesicle biogenesis. Cell. Logist. 2011, 1, 49–51. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, A.; Gibson, J.; Gregor, I.; Schatz, G. Import of proteins into mitochondria. The precursor of cytochrome c1 is processed in two steps, one of them heme-dependent. J. Biol. Chem. 1982, 257, 13042–13047. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hall, A.E.; Lisci, M.; Rose, M.D. Differential Requirement for the Cell Wall Integrity Sensor Wsc1p in Diploids Versus Haploids. J. Fungi 2021, 7, 1049. https://doi.org/10.3390/jof7121049
Hall AE, Lisci M, Rose MD. Differential Requirement for the Cell Wall Integrity Sensor Wsc1p in Diploids Versus Haploids. Journal of Fungi. 2021; 7(12):1049. https://doi.org/10.3390/jof7121049
Chicago/Turabian StyleHall, Allison E., Miriam Lisci, and Mark D. Rose. 2021. "Differential Requirement for the Cell Wall Integrity Sensor Wsc1p in Diploids Versus Haploids" Journal of Fungi 7, no. 12: 1049. https://doi.org/10.3390/jof7121049
APA StyleHall, A. E., Lisci, M., & Rose, M. D. (2021). Differential Requirement for the Cell Wall Integrity Sensor Wsc1p in Diploids Versus Haploids. Journal of Fungi, 7(12), 1049. https://doi.org/10.3390/jof7121049






