Tools for Assessing Translation in Cryptococcus neoformans
Abstract
:1. Introduction
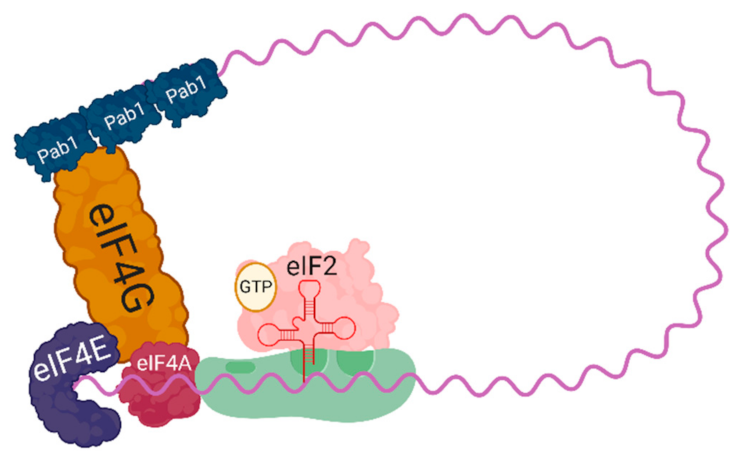
1.1. Translation Initiation
1.2. Translation Elongation
1.3. Regulation of Initiation and Elongation
2. Methods Used in Investigating Translation
2.1. Translation Inhibitors
2.1.1. Cycloheximide
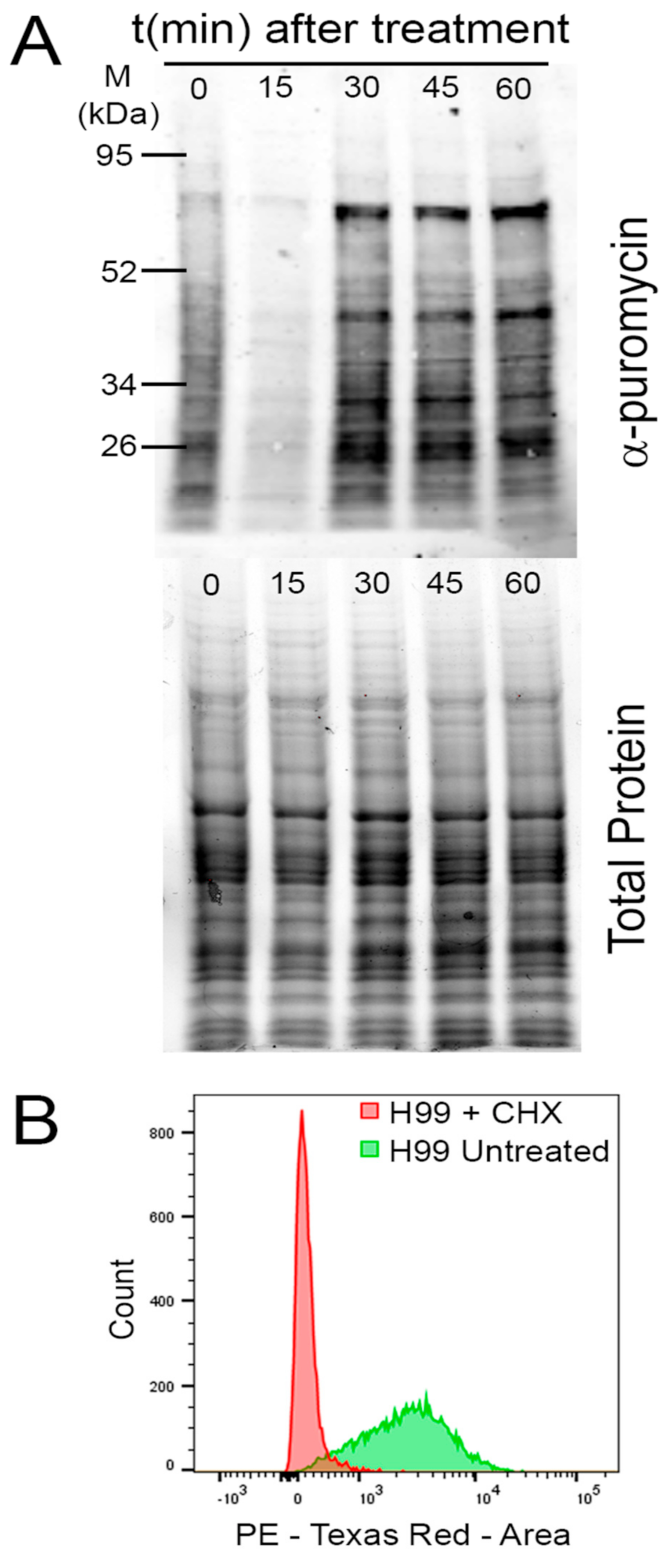
2.1.2. Puromycin
2.1.3. Rocaglates
2.2. Polysome Profiling
2.2.1. Gradient Preparation
- (1)
- Prepare 50 mL each of 10% and 50% sucrose solutions in the following buffer:
- Tris-HCl, pH 8: 20 mM
- KCl: 140 mM
- MgCl2: 5 mM
- DTT: 0.5 mM
- Cycloheximide: 0.1 mg/mL
- Heparin: 0.5 mg/mL
- Sucrose: 5 g and 25 g, respectively
- (2)
- Use the Amersham gradient maker to make 10 mL gradients from 5 mL of 10% sucrose solution and 50% sucrose solution per manufacturer’s instructions. (Alternatively, in the absence of a gradient maker, layer 2 mL each of 10%, 20%, 30%, 40%, and 50% sucrose solutions and allow the gradient to equilibrate at 4°C overnight.)
2.2.2. Culture Preparation
- (1)
- Start cultures in 250 mL baffled flasks at OD600 = 0.15–0.20 in the desired media. A minimum volume of 50 mL is recommended. Incubate cultures, shaking, until midlogarithmic growth phase is reached, an OD600 = 0.55–0.70.
- (2)
- If translation is to be assessed for response to stress or specific compounds, treat cultures appropriately ensuring a no stress/no drug control is also analyzed
- (3)
- Pellet cells by centrifuging for 2 min at 4000 RPM and flash freeze cultures in liquid nitrogen to preserve ribosome position.
2.2.3. Polysomes Extraction and Ultracentrifugation
- (1)
- Prepare polysome lysis buffer and chill on ice:
- Tris-HCl, pH 8: 20 mM
- KCl: 140 mM
- MgCl2: 5 mM
- DTT: 0.5 mM
- Cycloheximide: 0.1 mg/mL
- Heparin: 0.5 mg/mL
- (2)
- Thaw pellets on ice and resuspend in 5 mL lysis buffer. Transfer to a 14 mL snap-cap tube.
- (3)
- Centrifuge at 4000 RPM for 5 min to pellet.
- (4)
- Resuspend pellet in 1 mL lysis buffer and transfer to microfuge tube.
- (5)
- Centrifuge at 4000 RPM for 5 min to pellet.
- (6)
- Aspirate supernatant using a pipette.
- (7)
- Resuspend pellet in 50 μL of lysis buffer.
- (8)
- In an Eppendorf Safe-Lock tube, layer 0.5 mL of 0.5 mm glass disruption beads (RPI). Add resuspended pellet to the top of the beads, and layer with another 0.5 mL of beads.
- (9)
- Lyse in Bullet Blender Tissue Homogenizer, chilled with dry ice, for 5 min on speed 12. (Alternatively, cells can be lysed by vortexing for 30 s, followed by 30 s of incubation on ice for a total of 5 times.)
- (10)
- Add an additional 150 μL of lysis buffer to lysate and beads, and vortex to mix.
- (11)
- Remove lysate from beads and transfer to a new microfuge tube, and centrifuge at 4 °C for 10 min at 15,000 RCF to clear lysate
- (12)
- Transfer supernatant to new microfuge tube
- (13)
- Quantify RNA using a Nanodrop spectrophotometer (or other suitable method).
- (14)
- Layer an equivalent amount of lysate (based on RNA quantification) in equal volumes carefully on top of each gradient.
- 100–250 μg if only a profile is needed.
- Up to 350 μg if fractions will be collected for analysis of nucleic acids or protein.
- (15)
- Centrifuge in an SW41 Rotor at 39,000 RPM for 2 h at 4 °C.
2.2.4. Polysome Profile and Fraction Collection
- (1)
- Prepare Isco UA-6 UV/VIS detector with 254 nm filter along with Isco Retriever 500 fraction collector (if fractions are to be collected) as per manufacturer’s instructions.
- (2)
- Construct a Teledyne tube piercer with tubing connected to a peristaltic pump.
- (3)
- Position the gradient in the tube piercer, pierce the tube, and pump the gradient (0.75 mL/min.) through the flow cell while reading absorbance at 254 nm.
- (4)
- Simultaneously collect 500 μL fractions and denote the beginning and end of each fraction on the polysome profile trace.
- If using the Isco UA-6 detector, the output can be digitally converted using a DATAQ DI-1110 data acquisition device and recorded using the WinDaq recording software.
2.2.5. Protein and Nucleic Acid Extraction
- (1)
- Precipitate RNA and protein complexes by adding three volumes of cold 95–100% ethanol to each fraction and precipitate at −80 °C overnight. (Alternatively, if only protein precipitation is required, 25% w/v TCA can be used in place of ethanol followed by three acetone washes.)
- (2)
- Centrifuge the fractions at 15,000 RCF for 20 min at 4 °C
- (3)
- Aspirate supernatant with a pipette.
- (4)
- Quickly resuspend pellets in 250 μL RNase free water and immediately add 750 μL of TRIzol LS (ThermoFisher, New York, NY, USA). Proceed with manufacturer’s instructions to isolate RNA, protein, or both.
2.3. Measuring Translational Output
2.3.1. Puromycin Incorporation Assay
- (1)
- Start 50 mL cultures in baffled flasks at OD600 = 0.15–0.20 in the desired media. Incubate cultures, shaking, until mid-logarithmic growth phase is reached, an OD600 = 0.55–0.70.
- (2)
- If translation is to be assessed for response to stress or specific compounds, treat cultures appropriately ensuring a no-stress/no-drug control is also analyzed.
- (3)
- 10 min prior to your desired time point, pellet cells for 2 min at 4000 RPM, resuspend in a volume of 5 mL of media (smaller volume used to limit the quantity of puromycin used) with a final concentration of 150 μg/mL.
- (4)
- Allow puromycin to incorporate for 10 min.
- (5)
- Pellet cells by centrifuging for 2 min at 4000 RPM and flash freeze cultures in liquid nitrogen.
- (1)
- Thaw pellets on ice and resuspend pellet in 30 μL lysis buffer.
- HEPES pH 7.4: 15 mM
- KCl: 10 mM
- MgCl2: 5 mM
- Halt Protease Inhibitor (ThermoFisher): 10 μL/mL
- (2)
- In an Eppendorf Safe-Lock tube, layer 0.5 mL of 0.5 mm glass disruption beads (RPI). Add resuspended pellet to the top of the beads, and layer with another 0.5 mL of beads.
- (3)
- Lyse in Bullet Blender Tissue Homogenizer, chilled with dry ice, for 5 min on speed 12.
- (4)
- Add an additional 50 μL of lysis buffer to lysate and beads, and vortex to mix.
- (5)
- Remove lysate from beads and transfer to a new microfuge tube, and centrifuge at 4 °C for 10 min at 15,000 RCF to clear lysate.
- (6)
- Transfer supernatant to new microfuge tube.
- (7)
- Quantify protein using a Qubit fluorometer protein assay (Invitrogen) or other suitable quantification method.
- (1)
- To 25 μg of protein, add an equal amount of 2× Laemmli buffer and boil samples at 95 °C for 5 min.
- (2)
- Load samples onto a Bio-Rad Mini-PROTEAN TGX Stain-free gel, 4–15%.
- (3)
- Run the gel for 5 min at 50 V.
- (4)
- Increase the voltage to 120 V and run until the dye front reaches the bottom of the gel.
- (5)
- Remove gel and image total protein using a Bio-Rad GelDoc.
- (6)
- Transfer using Bio-Rad Trans-Blot Turbo as per manufacturer’s instructions.
- (7)
- Block for 5 min in Bio-Rad EveryBlot blocking buffer.
- (8)
- After 5 min of blocking, add α-puromycin antibody (catalog no. MABE343; Millipore) at a 1:1000 dilution.
- (9)
- Incubate overnight at 4 °C.
- (10)
- Wash the blot with Tris-buffered saline containing 0.5% Tween 20 (TBST) for 5 min (repeat ×3).
- (11)
- Incubate with HRP-conjugated α-mouse secondary antibody (catalog no. 7074S; Cell Signaling Technologies) at a 1:10,000 dilution.
- (12)
- Wash the blot with TBST for 5 min (repeat ×3).
- (13)
- Apply chemiluminescent substrate to the blot and image using Bio-Rad ChemiDoc.
- (14)
- Quantify total signal for each lane and normalize to total protein for each respective lane.
2.3.2. Measuring Translational Output using Click Chemistry
- (1)
- Start 50 mL cultures in baffled flasks at OD600 = 0.15–0.20 in YNB-2% Dextrose (yeast nitrogen based without amino acids)*. Incubate cultures shaking until mid-logarithmic growth phase is reached, an OD600 = 0.55–0.70. *Media used for HPG incorporation must be methionine-free.
- (2)
- If translation is to be assessed for response to stress or specific compounds, treat cultures appropriately ensuring a no-stress/no-drug control is also analyzed.
- (3)
- 10 min prior to your desired time point, aliquot 1 mL of treated cells into a microfuge tube and add HPG to a final concentration of 50 mM.
- (4)
- Allow HPG to incorporate for 20 min.
- (5)
- Pellet cells by centrifuging for 2 min at 4000 RPM and quickly resuspend in 1 mL of ice cold 70% ethanol.
- (6)
- Fix overnight at 4 °C.
- (7)
- Carry out the click chemistry labeling reaction using Invitrogen’s Click-iT Protein Reaction Buffer Kit (catalog number: C10276) as per manufacturer’s instructions
- (8)
- Quantify fluorescent signal by fluorescence microscopy, normalizing to the nuclear mask stain, or by flow cytometry.
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hood, M.I.; Skaar, E.P. Nutritional immunity: Transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 2012, 10, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.H.; Shishkova, E.; Hose, J.; Coon, J.J.; Gasch, A.P. Decoupling Yeast Cell Division and Stress Defense Implicates mRNA Repression in Translational Reallocation during Stress. Curr. Biol. 2018, 28, 2673–2680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bloom, A.L.M.; Solomons, J.T.G.; Havel, V.E.; Panepinto, J.C. Uncoupling of mRNA synthesis and degradation impairs adaptation to host temperature in Cryptococcus neoformans. Mol. Microbiol. 2013, 89, 65–83. [Google Scholar] [CrossRef] [Green Version]
- Jackson, R.J.; Hellen, C.U.T.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef] [Green Version]
- Dever, T.E.; Green, R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol. 2012, 4, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Nierhaus, K.H. The Allosteric Three-Site Model for the Ribosomal Elongation Cycle: Features and Future. Biochemistry 1990, 29, 4997–5008. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.B.F.; Becker, T.; Blau, M.; Anand, M.; Halic, M.; Balar, B.; Mielke, T.; Boesen, T.; Pedersen, J.S.; Spahn, C.M.T.; et al. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature 2006, 443, 663–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harding, H.P.; Zhang, Y.; Ron, D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 1999, 397, 271–274. [Google Scholar] [CrossRef]
- Leipheimer, J.; Bloom, A.L.M.; Campomizzi, C.S.; Salei, Y.; Panepinto, J.C. Translational Regulation Promotes Oxidative Stress Resistance in the Human Fungal Pathogen Cryptococcus neoformans. MBio 2019, 10, 8–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inglis, A.J.; Masson, G.R.; Shao, S.; Perisic, O.; McLaughlin, S.H.; Hegde, R.S.; Williams, R.L. Activation of GCN2 by the ribosomal P-stalk. Proc. Natl. Acad. Sci. USA 2019, 116, 4946–4954. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.C.; Peterson, A.; Zinshteyn, B.; Regot, S.; Green, R. Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate. Cell 2020, 182, 404–416. [Google Scholar] [CrossRef]
- Ishimura, R.; Nagy, G.; Dotu, I.; Chuang, J.H.; Ackerman, S.L. Activation of GCN2 kinase by ribosome stalling links translation elongation with translation initiation. eLife 2016, 5, 1–22. [Google Scholar] [CrossRef]
- Carlberg, U.; Nilsson, A.; Nygard, O. Functional properties of phosphorylated elongation factor 2. Eur. J. Biochem. 1990, 191, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Panepinto, J.C.; Komperda, K.W.; Hacham, M.; Shin, S.; Liu, X.; Williamson, P.R. Binding of Serum Mannan Binding Lectin to a Cell Integrity-Defective Cryptococcus neoformans ccr4Δ Mutant. Infect. Immun. 2007, 75, 4769–4779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwanhäusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [Green Version]
- Iyer, K.R.; Whitesell, L.; Porco, J.A.; Henkel, T.; Brown, L.E.; Robbins, N.; Cowen, L.E. Translation inhibition by rocaglates activates a species-specific cell death program in the emerging fungal pathogen Candida auris. MBio 2020, 11, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider-Poetsch, T.; Ju, J.; Eyler, D.E.; Dang, Y.; Bhat, S.; Merrick, W.C.; Green, R.; Shen, B.; Liu, J.O. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 2010, 6, 209–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obrig, T.G.; Culp, W.J.; McKeehan, W.L.; Hardesty, B. The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J. Biol. Chem. 1971, 246, 174–181. [Google Scholar] [CrossRef]
- D’Orazio, K.N.; Wu, C.C.C.; Sinha, N.; Loll-Krippleber, R.; Brown, G.W.; Green, R. The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during no go decay. eLife 2019, 8, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Nathans, D. Puromycin Inhibition of Protein Synthesis: Incorporation of Puromycin into Peptide Chains. Proc. Natl. Acad. Sci. USA 1964, 51, 585–592. [Google Scholar] [CrossRef] [Green Version]
- Blobel, G.; Sabatini, D. Dissociation of Mammalian Polyribosomes into Subunits by Puromycin. Proc. Natl. Acad. Sci. USA 1971, 68, 390–394. [Google Scholar] [CrossRef] [Green Version]
- Azzam, M.E.; Algranati, I.D. Mechanism of puromycin action: Fate of ribosomes after release of nascent protein chains from polysomes. Proc. Natl. Acad. Sci. USA 1973, 70, 3866–3869. [Google Scholar] [CrossRef] [Green Version]
- Chu, J.; Zhang, W.; Cencic, R.; O’Connor, P.B.F.; Robert, F.; Devine, W.G.; Selznick, A.; Henkel, T.; Merrick, W.C.; Brown, L.E.; et al. Rocaglates Induce Gain-of-Function Alterations to eIF4A and eIF4F. Cell Rep. 2020, 30, 2481–2488.e5. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, S.; Floor, S.N.; Ingolia, N.T. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature 2016, 534, 558–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfe, A.L.; Singh, K.; Zhong, Y.; Drewe, P.; Rajasekhar, V.K.; Sanghvi, V.R.; Mavrakis, K.J.; Jiang, M.; Roderick, J.E.; Van der Meulen, J.; et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 2014, 513, 65–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, L.; Woodard, J.L.; Lucas, D.M.; Fuchs, J.R.; Kinghorn, A.D. Rocaglamide, silvestrol and structurally related bioactive compounds from Aglaia species. Nat. Prod. Rep. 2014, 31, 924–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Firczuk, H.; Kannambath, S.; Pahle, J.; Claydon, A.; Beynon, R.; Duncan, J.; Westerhoff, H.; Mendes, P.; McCarthy, J.E. An in vivo control map for the eukaryotic mRNA translation machinery. Mol. Syst. Biol. 2013, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bloom, A.L.M.; Jin, R.M.; Leipheimer, J.; Bard, J.E.; Wohlfert, E.A.; Panepinto, J.C. Thermotolerance in the pathogen Cryptococcus neoformans is linked to antigen masking via mRNA. Nat. Commun. 2019, 10, 4950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, L.E.; James, P.; Craig, E.A.; Hensold, J.O. The Yeast hsp70 Homologue Ssa Is Required for Translation and Interacts with Sis1 and Pab1 on Translating Ribosomes. J. Biol. Chem. 2001, 276, 14426–14433. [Google Scholar] [CrossRef] [Green Version]
- Meydan, S.; Guydosh, N.R. Disome and Trisome Profiling Reveal Genome-wide Targets of Ribosome Quality Control. Mol. Cell 2020, 79, 588–602. [Google Scholar] [CrossRef]
- Sugiyama, T.; Li, S.; Kato, M.; Ikeuchi, K.; Ichimura, A.; Matsuo, Y.; Inada, T. Sequential Ubiquitination of Ribosomal Protein uS3 Triggers the Degradation of Non-functional 18S rRNA. Cell Rep. 2019, 26, 3400–3415. [Google Scholar] [CrossRef] [PubMed]
- Winz, M.L.; Peil, L.; Turowski, T.W.; Rappsilber, J.; Tollervey, D. Molecular interactions between Hel2 and RNA supporting ribosome-associated quality control. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.S.; Daniels, C.M.; Francis, S.A.; Shih, S.C.; Salerno, W.J.; Hicke, L.; Radhakrishnan, I. Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell 2003, 113, 621–630. [Google Scholar] [CrossRef] [Green Version]
- Wallace, E.W.J.; Maufrais, C.; Sales-Lee, J.; Tuck, L.R.; de Oliveira, L.; Feuerbach, F.; Moyrand, F.; Natarajan, P.; Madhani, H.D.; Janbon, G. Quantitative global studies reveal differential translational control by start codon context across the fungal kingdom. Nucleic Acids Res. 2020, 48, 2312–2331. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.S.; Weissman, J.S. Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science 2009, 324, 218–324. [Google Scholar] [CrossRef] [Green Version]
- Guydosh, N.R.; Green, R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell 2014, 156, 950–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guydosh, N.R.; Kimmig, P.; Walter, P.; Green, R. Regulated Ire1-dependent mRNA decay requires no-go mRNA degradation to maintain endoplasmic reticulum homeostasis in S. Pombe. eLife 2017, 6, e29216. [Google Scholar] [CrossRef]
- Klein, D.J.; Moore, P.B.; Steitz, T.A. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA 2004, 10, 1366–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nolan, R.D.; Arnstein, H.R.V. The Dissociation of Rabbit Reticulocyte Ribosomes into Subparticles Active in Protein Synthesis. Eur. J. Biochem. 1969, 10, 96–101. [Google Scholar] [CrossRef]
- Kondratov, K.; Kurapeev, D.; Popov, M.; Sidorova, M.; Minasian, S.; Galagudza, M.; Kostareva, A.; Fedorov, A. Heparinase treatment of heparin-contaminated plasma from coronary artery bypass grafting patients enables reliable quantification of microRNAs. Biomol. Detect. Quantif. 2016, 8, 9–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, X.H.; Fischer, S.; Keshavjee, S.; Liu, M. Heparin interference with reverse transcriptase polymerase chain reaction of RNA extracted from lungs after ischemia-reperfusion. Transpl. Int. 2000, 13, 146–150. [Google Scholar] [CrossRef]
- Duncan, C.D.S.; Mata, J. Effects of cycloheximide on the interpretation of ribosome profiling experiments in Schizosaccharomyces pombe. Sci. Rep. 2017, 7, 10331. [Google Scholar] [CrossRef]
- Hussmann, J.A.; Patchett, S.; Johnson, A.; Sawyer, S.; Press, W.H. Understanding Biases in Ribosome Profiling Experiments Reveals Signatures of Translation Dynamics in Yeast. PLoS Genet. 2015, 11, e1005732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lareau, L.F.; Hite, D.H.; Hogan, G.J.; Brown, P.O. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife 2014, 2014, 1–16. [Google Scholar] [CrossRef]
- Gerashchenko, M.V.; Gladyshev, V.N. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014, 42, e134. [Google Scholar] [CrossRef] [Green Version]
- Simms, C.L.; Thomas, E.N.; Zaher, H.S. Ribosome-based quality control of mRNA and nascent peptides. Wiley Interdiscip. Rev. RNA 2017, 8, e1366. [Google Scholar] [CrossRef] [Green Version]
- Simms, C.L.; Yan, L.L.; Zaher, H.S. Ribosome Collision Is Critical for Quality Control during No-Go Decay. Mol. Cell 2017, 68, 361–373. [Google Scholar] [CrossRef] [Green Version]
- Juszkiewicz, S.; Slodkowicz, G.; Lin, Z.; Freire-Pritchett, P.; Peak-Chew, S.Y.; Hegde, R.S. Ribosome collisions trigger cis-acting feedback inhibition of translation initiation. eLife 2020, 9, 1–29. [Google Scholar] [CrossRef]
- Yan, L.L.; Zaher, H.S. Ribosome quality control antagonizes the activation of the integrated stress response on colliding ribosomes. Mol. Cell 2021, 81, 614–628. [Google Scholar] [CrossRef]
- Schmidt, E.K.; Clavarino, G.; Ceppi, M.; Pierre, P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 2009, 6, 275–277. [Google Scholar] [CrossRef] [PubMed]
- Goodman, C.A.; Hornberger, T.A. Measuring Protein Synthesis With SUnSET. Exerc. Sport Sci. Rev. 2013, 41, 107–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dieterich, D.C.; Lee, J.J.; Link, A.J.; Graumann, J.; Tirrell, D.A.; Schuman, E.M. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat. Protoc. 2007, 2, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Dieterich, D.C.; Hodas, J.J.L.; Gouzer, G.; Shadrin, I.Y.; Ngo, J.T.; Triller, A.; Tirrell, D.A.; Schuman, E.M. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat. Neurosci. 2010, 13, 897–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knowles, C.M.; McIntyre, K.M.; Panepinto, J.C. Tools for Assessing Translation in Cryptococcus neoformans. J. Fungi 2021, 7, 159. https://doi.org/10.3390/jof7030159
Knowles CM, McIntyre KM, Panepinto JC. Tools for Assessing Translation in Cryptococcus neoformans. Journal of Fungi. 2021; 7(3):159. https://doi.org/10.3390/jof7030159
Chicago/Turabian StyleKnowles, Corey M., Kelcy M. McIntyre, and John C. Panepinto. 2021. "Tools for Assessing Translation in Cryptococcus neoformans" Journal of Fungi 7, no. 3: 159. https://doi.org/10.3390/jof7030159
APA StyleKnowles, C. M., McIntyre, K. M., & Panepinto, J. C. (2021). Tools for Assessing Translation in Cryptococcus neoformans. Journal of Fungi, 7(3), 159. https://doi.org/10.3390/jof7030159




