High Frequency of Enterocytozoon bieneusi Genotype WL12 Occurrence among Immunocompromised Patients with Intestinal Microsporidiosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Collection of Fecal Specimens
2.2. DNA Extraction and PCR Amplification
2.3. Genotyping of E. bieneusi Isolates by ITS Sequencing
2.4. Phylogenetic Analyses
2.5. Statistical Analyses
3. Results
3.1. E. bieneusi is the Major Species Infecting Intestinal Microsporidia-Positive Immunocompromised Patients
3.2. ITS Sequencing Reveals High-Frequency Occurrence of E. Bieneusi Genotype WL12
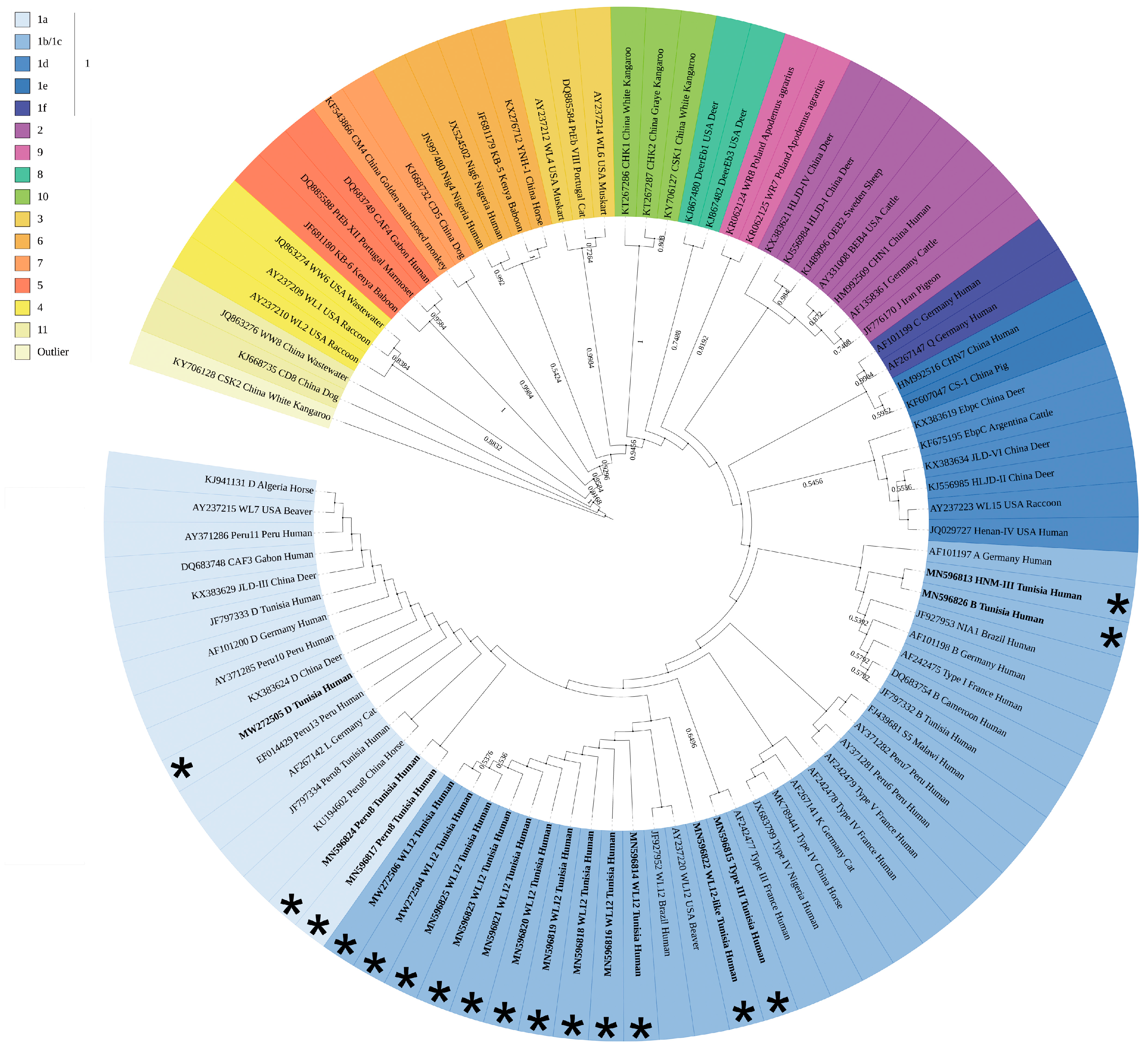
3.3. Phylogenetic Analyses Assign all E. bieneusi Strains to Zoonotic Group 1
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weiss, L.M.; Becnel, J.J. Microsporidia: Pathogens of Opportunity; Wiley: Hoboken, NJ, USA, 2014. [Google Scholar]
- Graczyk, T.K.; Sunderland, D.; Awantang, G.N.; Mashinski, Y.; Lucy, F.E.; Graczyk, Z.; Chomicz, L.; Breysse, P.N. Relationships among bather density, levels of human waterborne pathogens, and fecal coliform counts in marine recreational beach water. Parasitol Res. 2010, 106, 1103–1108. [Google Scholar] [CrossRef]
- Izquierdo, F.; Castro Hermida, J.A.; Fenoy, S.; Mezo, M.; Gonzalez-Warleta, M.; del Aguila, C. Detection of microsporidia in drinking water, wastewater and recreational rivers. Water Res. 2011, 45, 4837–4843. [Google Scholar] [CrossRef] [PubMed]
- Javanmard, E.; Mirjalali, H.; Niyyati, M.; Jalilzadeh, E.; Seyed Tabaei, S.J.; Asadzadeh Aghdaei, H.; Nazemalhosseini-Mojarad, E.; Zali, M.R. Molecular and phylogenetic evidences of dispersion of human-infecting microsporidia to vegetable farms via irrigation with treated wastewater: One-year follow up. Int. J. Hyg. Environ. Health 2018, 221, 642–651. [Google Scholar] [CrossRef] [PubMed]
- Karanis, P.; Kourenti, C.; Smith, H. Waterborne transmission of protozoan parasites: A worldwide review of outbreaks and lessons learnt. J. Water Health 2007, 5, 1–38. [Google Scholar] [CrossRef]
- Stentiford, G.D.; Becnel, J.; Weiss, L.M.; Keeling, P.J.; Didier, E.S.; Williams, B.P.; Bjornson, S.; Kent, M.L.; Freeman, M.A.; Brown, M.J.F.; et al. Microsporidia—Emergent Pathogens in the Global Food Chain. Trends Parasitol 2016, 32, 336–348. [Google Scholar] [CrossRef]
- Thellier, M.; Breton, J. Enterocytozoon bieneusi in human and animals, focus on laboratory identification and molecular epidemiology. Parasite 2008, 15, 349–358. [Google Scholar] [CrossRef]
- Didier, E.S.; Weiss, L.M. Microsporidiosis: Not just in AIDS patients. Curr. Opin. Infect. Dis 2011, 24, 490–495. [Google Scholar] [CrossRef]
- Shakiba, E.; Ramazani, U.; Mardani, E.; Rahimi, Z.; Nazar, Z.M.; Najafi, F.; Moradinazar, M. Epidemiological features of HIV/AIDS in the Middle East and North Africa from 1990 to 2017. Int J. STD AIDS 2021, 956462420960632. [Google Scholar] [CrossRef]
- El Matri, A.; Ben Abdallah, T. Organ transplantation in Tunisia. Exp. Clin. Transplant. 2015, 13 (Suppl. 1), 33–36. [Google Scholar] [PubMed]
- Field, A.S.; Milner, D.A., Jr. Intestinal microsporidiosis. Clin. Lab. Med. 2015, 35, 445–459. [Google Scholar] [CrossRef]
- Heyworth, M.F. Molecular diagnosis of human microsporidian infections. Trans. R Soc. Trop. Med. Hyg. 2017, 111, 382–383. [Google Scholar] [CrossRef]
- Cannon, M.V.; Bogale, H.; Rutt, L.; Humphrys, M.; Korpe, P.; Duggal, P.; Ravel, J.; Serre, D. A high-throughput sequencing assay to comprehensively detect and characterize unicellular eukaryotes and helminths from biological and environmental samples. Microbiome 2018, 6, 195. [Google Scholar] [CrossRef]
- Hamad, I.; Abou Abdallah, R.; Ravaux, I.; Mokhtari, S.; Tissot-Dupont, H.; Michelle, C.; Stein, A.; Lagier, J.C.; Raoult, D.; Bittar, F. Metabarcoding analysis of eukaryotic microbiota in the gut of HIV-infected patients. PLoS ONE 2018, 13, e0191913. [Google Scholar] [CrossRef] [PubMed]
- Vamanu, E.; Gatea, F. Correlations between Microbiota Bioactivity and Bioavailability of Functional Compounds: A Mini-Review. Biomedicines 2020, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Weiss, L.M. Therapeutic targets for the treatment of microsporidiosis in humans. Expert Opin. Ther. Targets 2018, 22, 903–915. [Google Scholar] [CrossRef]
- Bukreyeva, I.; Angoulvant, A.; Bendib, I.; Gagnard, J.C.; Bourhis, J.H.; Dargere, S.; Bonhomme, J.; Thellier, M.; Gachot, B.; Wyplosz, B. Enterocytozoon bieneusi Microsporidiosis in Stem Cell Transplant Recipients Treated with Fumagillin. Emerg Infect. Dis 2017, 23, 1039–1041. [Google Scholar] [CrossRef]
- Molina, J.M.; Tourneur, M.; Sarfati, C.; Chevret, S.; de Gouvello, A.; Gobert, J.G.; Balkan, S.; Derouin, F. Fumagillin treatment of intestinal microsporidiosis. N. Engl. J. Med. 2002, 346, 1963–1969. [Google Scholar] [CrossRef]
- Santin, M.; Fayer, R. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: A consensus. J. Eukaryot Microbiol. 2009, 56, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Feng, Y.; Santin, M. Host Specificity of Enterocytozoon bieneusi and Public Health Implications. Trends Parasitol. 2019, 35, 436–451. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Feng, Y.; Zhang, L.; Xiao, L. Potential impacts of host specificity on zoonotic or interspecies transmission of Enterocytozoon bieneusi. Infect. Genet. Evol. 2019, 75, 104033. [Google Scholar] [CrossRef]
- Sulaiman, I.M.; Fayer, R.; Lal, A.A.; Trout, J.M.; Schaefer, F.W., 3rd; Xiao, L. Molecular characterization of microsporidia indicates that wild mammals Harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl. Environ. Microbiol. 2003, 69, 4495–4501. [Google Scholar] [CrossRef]
- Feng, Y.; Li, N.; Dearen, T.; Lobo, M.L.; Matos, O.; Cama, V.; Xiao, L. Development of a multilocus sequence typing tool for high-resolution genotyping of Enterocytozoon bieneusi. Appl Environ. Microbiol 2011, 77, 4822–4828. [Google Scholar] [CrossRef]
- Li, N.; Xiao, L.; Wang, L.; Zhao, S.; Zhao, X.; Duan, L.; Guo, M.; Liu, L.; Feng, Y. Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Negl. Trop. Dis. 2012, 6, e1809. [Google Scholar] [CrossRef] [PubMed]
- Ben Ayed, L.; Yang, W.; Widmer, G.; Cama, V.; Ortega, Y.; Xiao, L. Survey and genetic characterization of wastewater in Tunisia for Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi, Cyclospora cayetanensis and Eimeria spp. J. Water Health 2012, 10, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Weber, R.; Bryan, R.T.; Schwartz, D.A.; Owen, R.L. Human microsporidial infections. Clin. Microbiol. Rev. 1994, 7, 426–461. [Google Scholar] [CrossRef] [PubMed]
- Lono, A.; Kumar, S.; Chye, T.T. Detection of microsporidia in local HIV-positive population in Malaysia. Trans. R Soc. Trop Med. Hyg. 2011, 105, 409–413. [Google Scholar] [CrossRef]
- Raynaud, L.; Delbac, F.; Broussolle, V.; Rabodonirina, M.; Girault, V.; Wallon, M.; Cozon, G.; Vivares, C.P.; Peyron, F. Identification of Encephalitozoon intestinalis in travelers with chronic diarrhea by specific PCR amplification. J. Clin. Microbiol. 1998, 36, 37–40. [Google Scholar] [CrossRef]
- Lucas, S.B.; Papadaki, L.; Conlon, C.; Sewankambo, N.; Goodgame, R.; Serwadda, D. Diagnosis of intestinal microsporidiosis in patients with AIDS. J. Clin. Pathol. 1989, 42, 885–887. [Google Scholar] [CrossRef]
- Curry, A.; McWilliam, L.J.; Haboubi, N.Y.; Mandal, B.K. Microsporidiosis in a British patient with AIDS. J. Clin. Pathol. 1988, 41, 477–478. [Google Scholar] [CrossRef][Green Version]
- Rijpstra, A.C.; Canning, E.U.; Van Ketel, R.J.; Eeftinck Schattenkerk, J.K.; Laarman, J.J. Use of light microscopy to diagnose small-intestinal microsporidiosis in patients with AIDS. J. Infect. Dis. 1988, 157, 827–831. [Google Scholar] [CrossRef] [PubMed]
- Hocevar, S.N.; Paddock, C.D.; Spak, C.W.; Rosenblatt, R.; Diaz-Luna, H.; Castillo, I.; Luna, S.; Friedman, G.C.; Antony, S.; Stoddard, R.A.; et al. Microsporidiosis acquired through solid organ transplantation: A public health investigation. Ann. Intern. Med. 2014, 160, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.; Eichenlaub, S.; Pape, G.R.; Hoffmann, R.M. Chronic diarrhea as a result of intestinal microsposidiosis in a liver transplant recipient. Transplantation 2001, 71, 334–337. [Google Scholar] [CrossRef] [PubMed]
- Gumbo, T.; Hobbs, R.E.; Carlyn, C.; Hall, G.; Isada, C.M. Microsporidia infection in transplant patients. Transplantation 1999, 67, 482–484. [Google Scholar] [CrossRef]
- Chabchoub, N.; Abdelmalek, R.; Breton, J.; Kanoun, F.; Thellier, M.; Bouratbine, A.; Aoun, K. Genotype identification of Enterocytozoon bieneusi isolates from stool samples of HIV-infected Tunisian patients. Parasite 2012, 19, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Olano, J.P.; McBride, J.W.; Walker, D.H. Emerging pathogens: Challenges and successes of molecular diagnostics. J. Mol. Diagn. 2008, 10, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Navin, T.R.; Weber, R.; Vugia, D.J.; Rimland, D.; Roberts, J.M.; Addiss, D.G.; Visvesvara, G.S.; Wahlquist, S.P.; Hogan, S.E.; Gallagher, L.E.; et al. Declining CD4+ T-lymphocyte counts are associated with increased risk of enteric parasitosis and chronic diarrhea: Results of a 3-year longitudinal study. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1999, 20, 154–159. [Google Scholar] [CrossRef]
- Stark, D.; van Hal, S.; Barratt, J.; Ellis, J.; Marriott, D.; Harkness, J. Limited genetic diversity among genotypes of Enterocytozoon bieneusi strains isolated from HIV-infected patients from Sydney, Australia. J. Med. Microbiol. 2009, 58, 355–357. [Google Scholar] [CrossRef][Green Version]
- Wang, Z.D.; Liu, Q.; Liu, H.H.; Li, S.; Zhang, L.; Zhao, Y.K.; Zhu, X.Q. Prevalence of Cryptosporidium, microsporidia and Isospora infection in HIV-infected people: A global systematic review and meta-analysis. Parasit Vectors 2018, 11, 28. [Google Scholar] [CrossRef]
- Chabchoub, N.; Abdelmalek, R.; Mellouli, F.; Kanoun, F.; Thellier, M.; Bouratbine, A.; Aoun, K. Genetic identification of intestinal microsporidia species in immunocompromised patients in Tunisia. Am. J. Trop. Med. Hyg. 2009, 80, 24–27. [Google Scholar] [CrossRef]
- Aissa, S.; Chabchoub, N.; Abdelmalek, R.; Kanoun, F.; Goubantini, A.; Ammari, L.; Kilani, B.; Bouratbine, A.; Tiouiri-Ben Aissa, H.; Aoun, K. Asymptomatic intestinal carriage of microsporidia in HIV-positive patients in Tunisia: Prevalence, species, and pathogenesis. Med. Sante Trop 2017, 27, 281–285. [Google Scholar] [CrossRef]
- Greigert, V.; Pfaff, A.W.; Abou-Bacar, A.; Candolfi, E.; Brunet, J. Intestinal microsporidiosis in Strasbourg from 2014 to 2016: Emergence of an Enterocytozoon bieneusi genotype of Asian origin. Emerg. Microbes Infect. 2018, 7, 97. [Google Scholar] [CrossRef]
- Kicia, M.; Wesolowska, M.; Kopacz, Z.; Jakuszko, K.; Sak, B.; Kvetonova, D.; Krajewska, M.; Kvac, M. Prevalence and molecular characteristics of urinary and intestinal microsporidia infections in renal transplant recipients. Clin. Microbiol. Infect. 2016, 22, 462.e5–462.e9. [Google Scholar] [CrossRef] [PubMed]
- Desoubeaux, G.; Maakaroun-Vermesse, Z.; Lier, C.; Bailly, E.; Morio, F.; Labarthe, F.; Bernard, L.; Chandenier, J. Successful treatment with fumagillin of the first pediatric case of digestive microsporidiosis in a liver-kidney transplant. Transpl. Infect. Dis. 2013, 15, E250–E259. [Google Scholar] [CrossRef] [PubMed]
- Godron, A.; Accoceberry, I.; Couret, A.; Llanas, B.; Harambat, J. Intestinal microsporidiosis due to Enterocytozoon bieneusi in a pediatric kidney transplant recipient successfully treated with fumagillin. Transplantation 2013, 96, e66–e67. [Google Scholar] [CrossRef]
- Pomares, C.; Santin, M.; Miegeville, M.; Espern, A.; Albano, L.; Marty, P.; Morio, F. A new and highly divergent Enterocytozoon bieneusi genotype isolated from a renal transplant recipient. J. Clin. Microbiol. 2012, 50, 2176–2178. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Galvan, A.L.; Sanchez, A.M.; Valentin, M.A.; Henriques-Gil, N.; Izquierdo, F.; Fenoy, S.; del Aguila, C. First cases of microsporidiosis in transplant recipients in Spain and review of the literature. J. Clin. Microbiol. 2011, 49, 1301–1306. [Google Scholar] [CrossRef]
- Lanternier, F.; Boutboul, D.; Menotti, J.; Chandesris, M.O.; Sarfati, C.; Mamzer Bruneel, M.F.; Calmus, Y.; Mechai, F.; Viard, J.P.; Lecuit, M.; et al. Microsporidiosis in solid organ transplant recipients: Two Enterocytozoon bieneusi cases and review. Transpl. Infect. Dis. 2009, 11, 83–88. [Google Scholar] [CrossRef]
- Kicia, M.; Wesolowska, M.; Jakuszko, K.; Kopacz, Z.; Sak, B.; Kvetonova, D.; Krajewska, M.; Kvac, M. Concurrent infection of the urinary tract with Encephalitozoon cuniculi and Enterocytozoon bieneusi in a renal transplant recipient. J. Clin. Microbiol. 2014, 52, 1780–1782. [Google Scholar] [CrossRef]
- Kicia, M.; Szydlowicz, M.; Cebulski, K.; Jakuszko, K.; Piesiak, P.; Kowal, A.; Sak, B.; Krajewska, M.; Hendrich, A.B.; Kvac, M.; et al. Symptomatic respiratory Encephalitozoon cuniculi infection in renal transplant recipients. Int. J. Infect. Dis. 2018, 79, 21–25. [Google Scholar] [CrossRef]
- Kicia, M.; Sedzimirska, M.; Sak, B.; Kvac, M.; Wesolowska, M.; Hendrich, A.B.; Kopacz, Z. Respiratory microsporidiosis caused by Enterocytozoon bieneusi in an HIV-negative hematopoietic stem cell transplant recipient. Int J. Infect. Dis. 2018, 77, 26–28. [Google Scholar] [CrossRef]
- Lobo, M.L.; Xiao, L.; Antunes, F.; Matos, O. Microsporidia as emerging pathogens and the implication for public health: A 10-year study on HIV-positive and -negative patients. Int J. Parasitol. 2012, 42, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, K.; Weiss, L.M. T cell response and persistence of the microsporidia. FEMS Microbiol Rev. 2012, 36, 748–760. [Google Scholar] [CrossRef]
- Moretto, M.M.; Khan, I.A.; Weiss, L.M. Gastrointestinal cell mediated immunity and the microsporidia. PLoS Pathog. 2012, 8, e1002775. [Google Scholar] [CrossRef]
- Moretto, M.; Weiss, L.M.; Khan, I.A. Induction of a rapid and strong antigen-specific intraepithelial lymphocyte response during oral Encephalitozoon cuniculi infection. J. Immunol. 2004, 172, 4402–4409. [Google Scholar] [CrossRef]
- Khan, I.A.; Schwartzman, J.D.; Kasper, L.H.; Moretto, M. CD8+ CTLs are essential for protective immunity against Encephalitozoon cuniculi infection. J. Immunol. 1999, 162, 6086–6091. [Google Scholar]
- Han, Y.; Gao, H.; Xu, J.; Luo, J.; Han, B.; Bao, J.; Pan, G.; Li, T.; Zhou, Z. Innate and Adaptive Immune Responses Against Microsporidia Infection in Mammals. Front. Microbiol. 2020, 11, 1468. [Google Scholar] [CrossRef]
- Desoubeaux, G.; Nourrisson, C.; Moniot, M.; De Kyvon, M.A.; Bonnin, V.; De La Bretonniere, M.E.; Morange, V.; Bailly, E.; Lemaignen, A.; Morio, F.; et al. Genotyping Approach for Potential Common Source of Enterocytozoon bieneusi Infection in Hematology Unit. Emerg. Infect. Dis. 2019, 25, 1625–1631. [Google Scholar] [CrossRef]
- Cama, V.A.; Pearson, J.; Cabrera, L.; Pacheco, L.; Gilman, R.; Meyer, S.; Ortega, Y.; Xiao, L. Transmission of Enterocytozoon bieneusi between a child and guinea pigs. J. Clin. Microbiol. 2007, 45, 2708–2710. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, H.; Zhao, X.; Zhang, L.; Zhang, G.; Guo, M.; Liu, L.; Feng, Y.; Xiao, L. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J. Clin. Microbiol. 2013, 51, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Leelayoova, S.; Subrungruang, I.; Rangsin, R.; Chavalitshewinkoon-Petmitr, P.; Worapong, J.; Naaglor, T.; Mungthin, M. Transmission of Enterocytozoon bieneusi genotype a in a Thai orphanage. Am. J. Trop Med. Hyg. 2005, 73, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Pagornrat, W.; Leelayoova, S.; Rangsin, R.; Tan-Ariya, P.; Naaglor, T.; Mungthin, M. Carriage rate of Enterocytozoon bieneusi in an orphanage in Bangkok, Thailand. J. Clin. Microbiol. 2009, 47, 3739–3741. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zhou, H.; Jin, H.; Sun, L.; Li, P.; Liu, M.; Qiu, M.; Xu, L.; Li, F.; Ma, T.; et al. Genotyping of Enterocytozoon bieneusi among captive long-tailed macaques (Macaca fascicularis) in Hainan Province: High genetic diversity and zoonotic potential. Acta Trop. 2020, 201, 105211. [Google Scholar] [CrossRef] [PubMed]

| Clinical Parameters | Patients with Microsporidia (n = 18) | Patients without Microsporidia (n = 153) | p-Value a | |
|---|---|---|---|---|
| Age b | 1–15 years | 3 | 43 | >0.05 |
| 16–35 years | 8 | 37 | ||
| >35 years | 6 | 61 | ||
| Gender | Male | 7 | 92 | >0.05 |
| Female | 11 | 61 | ||
| Underlying immunodeficiency | HIV+ | 12 | 61 | <0.05 |
| Other than HIV (Bone marrow/kidney transplant, unspecified) | 6 | 92 | ||
| Diarrhea | 17 | 43 | <0.05 | |
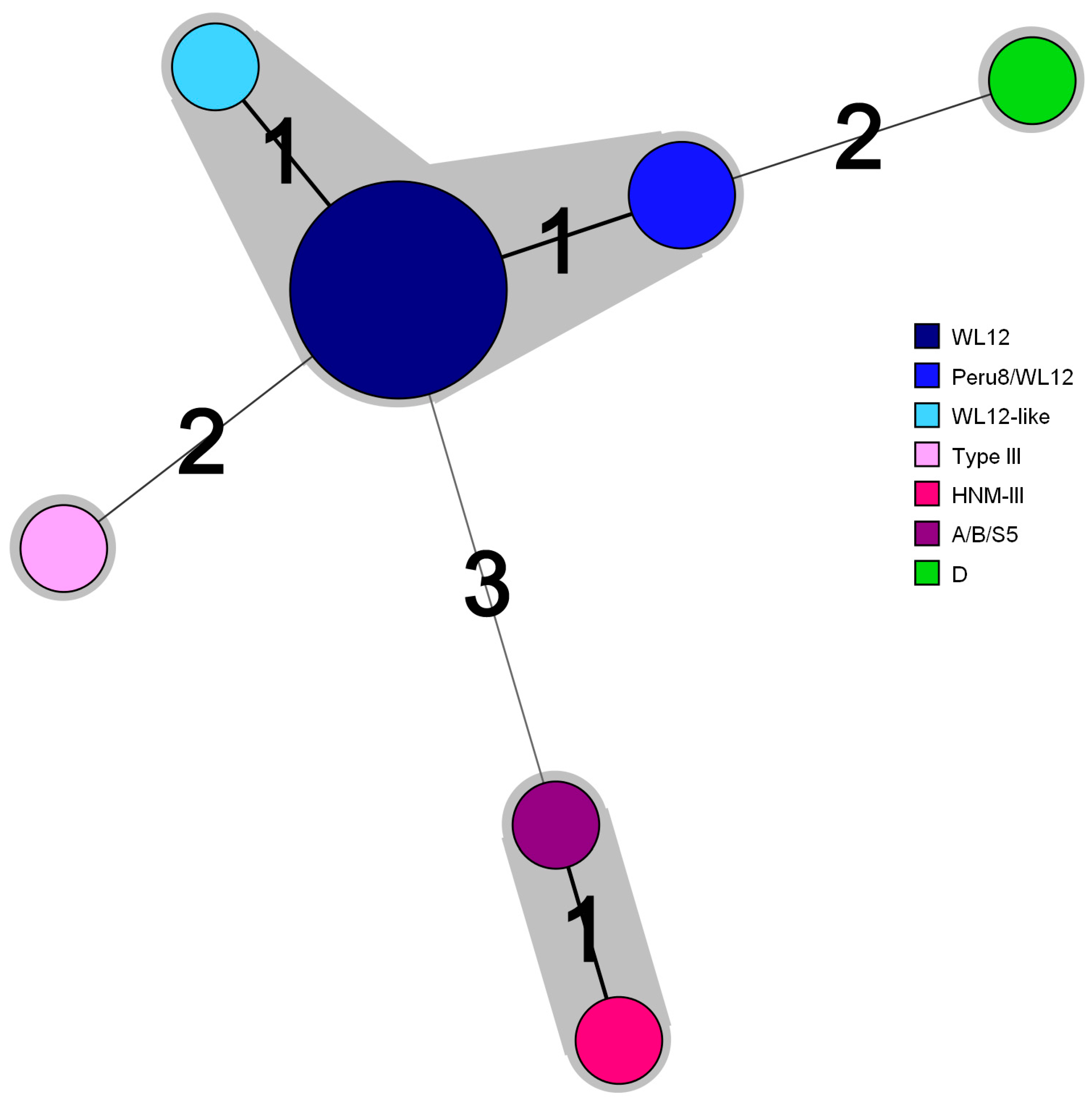
| Patient# a | Accession Number b | Underlying Disease c | ITS Genotype d | Identical or Similar Genotypes e |
|---|---|---|---|---|
| Patient 34 | MN596813 | HIV infection | HNM-III | HNM-III(C137/T): MK139696 |
| Patient 23 | MN596814 | Bone marrow transplant | WL12 | WL12: JF927952, AY237220 |
| Patient 37 | MN596815 | Kidney transplant | Type III | Type III(C97/T): AF242477 |
| Patient 55 | MN596816 | Bone marrow transplant | WL12 | WL12: JF927952, AY237220 |
| Patient 56 | MN596817 | Kidney transplant | Peru8/WL12 | Peru8(C97/T): MN747470, MH714713, MF476880, KU194602, KT267285, KJ668721, KF305584, KF261771, JX683807, JQ029733, JF927959, JF797334, AY371283, JF909995; WL12(T93/C): JF927952, AY237220 |
| Patient 74 | MN596818 | HIV infection | WL12 | WL12: JF927952, AY237220 |
| Patient 85 | MN596819 | Bone marrow transplant | WL12 | WL12: JF927952, AY237220 |
| Patient 87 | MN596820 | HIV infection | WL12 | WL12: JF927952, AY237220 |
| Patient 139 | MN596821 | HIV infection | WL12 | WL12: JF927952, AY237220 |
| Patient 152 | MN596822 | HIV infection | WL12-like | WL12(C137/G): JF927952, AY237220 |
| Patient 138 | MN596823 | HIV infection | WL12 | WL12: JF927952, AY237220 |
| Patient 11 | MN596824 | HIV infection | Peru8/WL12 | Peru8(C97/T): MN747470, MH714713, MF476880, KU194602, KT267285, KJ668721, KF305584, KF261771, JX683807, JQ029733, JF927959, JF797334, AY371283, JF909995; WL12(T93/C): JF927952, AY237220 |
| Patient 49 | MN596825 | HIV infection | WL12 | WL12: JF927952, AY237220 |
| Patient 9 | MN596826 | HIV infection | A/B/S5 | A(G31/A): MK982500, MN136776, MH500238, KU886089, KP325475, KP325476, JQ437574, AY371276, AY357185, AY168419, AF101197; B(A77/G): AF242475, AF101198, JF797332; S5(C137/T): MG458710, FJ439681, DQ885581 |
| Patient 15 | MW272504 | HIV infection | WL12 | WL12: JF927952, AY237220 |
| Patient 94 | MW272505 | HIV infection | D | D: MN704918, MN747468, MK982516, MN902234, others |
| Patient 121 | MW272506 | HIV infection | WL12 | WL12: JF927952, AY237220 |
| Number of Patients | Genotype | Nucleotide Position a | ||||||
|---|---|---|---|---|---|---|---|---|
| 31 | 93 | 97 | 117 | 134 | 137 | 178 | ||
| 10 | WL12 | G | T | T | G | C | C | G |
| 2 | Peru8/WL12 | C | ||||||
| 1 | WL12-like | G | ||||||
| 1 | HNM-III | A | C | C | T | |||
| 1 | A/B/S5 | A | C | T | ||||
| 1 | Type III | T | A | |||||
| 1 | D | C | C | T | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Messaoud, M.; Abbes, S.; Gnaien, M.; Rebai, Y.; Kallel, A.; Jemel, S.; Cherif, G.; Skhairia, M.A.; Marouen, S.; Fakhfekh, N.; et al. High Frequency of Enterocytozoon bieneusi Genotype WL12 Occurrence among Immunocompromised Patients with Intestinal Microsporidiosis. J. Fungi 2021, 7, 161. https://doi.org/10.3390/jof7030161
Messaoud M, Abbes S, Gnaien M, Rebai Y, Kallel A, Jemel S, Cherif G, Skhairia MA, Marouen S, Fakhfekh N, et al. High Frequency of Enterocytozoon bieneusi Genotype WL12 Occurrence among Immunocompromised Patients with Intestinal Microsporidiosis. Journal of Fungi. 2021; 7(3):161. https://doi.org/10.3390/jof7030161
Chicago/Turabian StyleMessaoud, Mariem, Salma Abbes, Mayssa Gnaien, Yasmine Rebai, Aicha Kallel, Sana Jemel, Ghaya Cherif, Mohamed Amine Skhairia, Sonia Marouen, Najla Fakhfekh, and et al. 2021. "High Frequency of Enterocytozoon bieneusi Genotype WL12 Occurrence among Immunocompromised Patients with Intestinal Microsporidiosis" Journal of Fungi 7, no. 3: 161. https://doi.org/10.3390/jof7030161
APA StyleMessaoud, M., Abbes, S., Gnaien, M., Rebai, Y., Kallel, A., Jemel, S., Cherif, G., Skhairia, M. A., Marouen, S., Fakhfekh, N., Mardassi, H., Belhadj, S., Znaidi, S., & Kallel, K. (2021). High Frequency of Enterocytozoon bieneusi Genotype WL12 Occurrence among Immunocompromised Patients with Intestinal Microsporidiosis. Journal of Fungi, 7(3), 161. https://doi.org/10.3390/jof7030161








