Genetic and Phenotypic Characterization of in-Host Developed Azole-Resistant Aspergillus flavus Isolates
Abstract
:1. Introduction
2. Materials and Methods
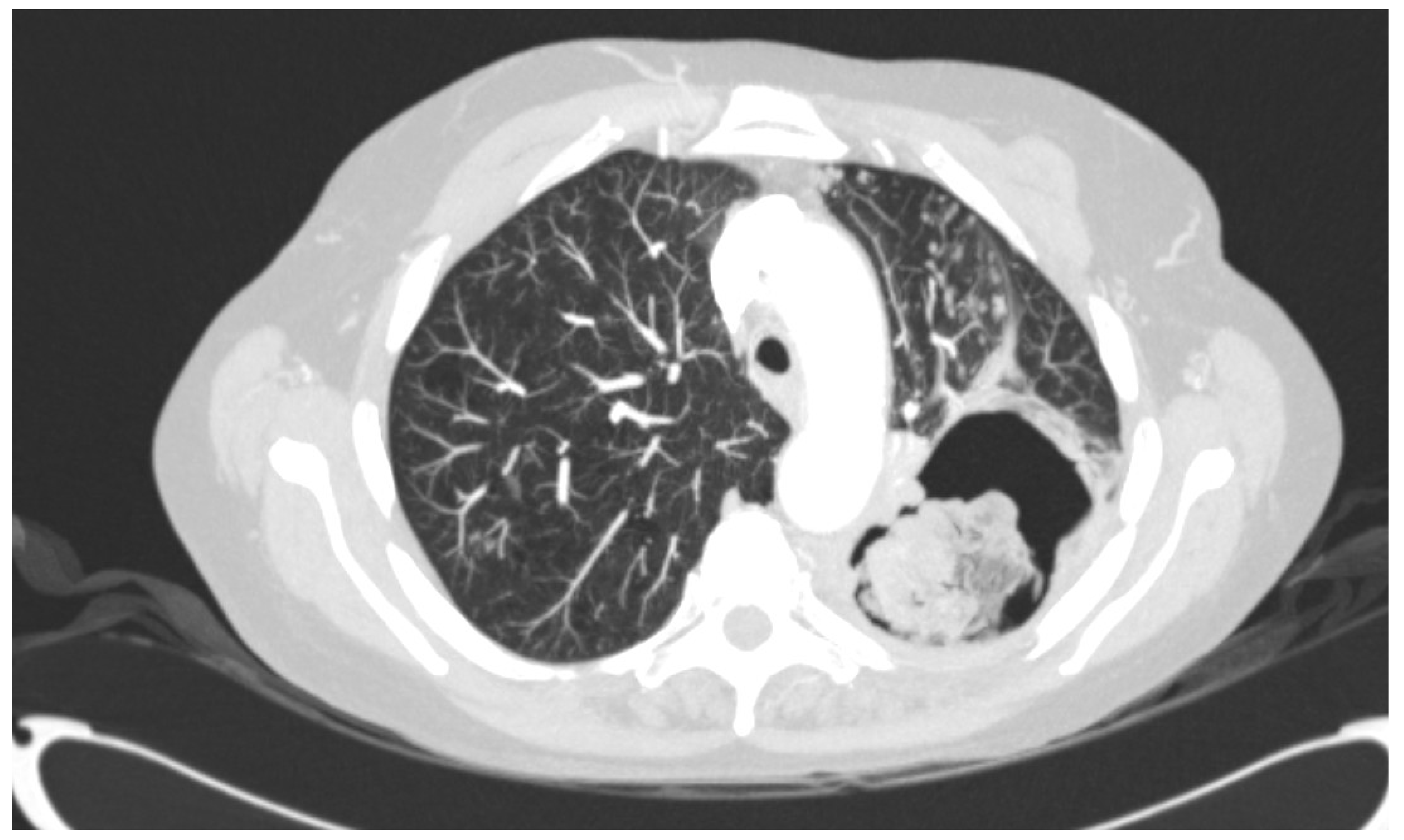
2.1. Origin of Strains and Antifungal Treatment
2.2. Whole-Genome Sequencing and Analysis
3. Results
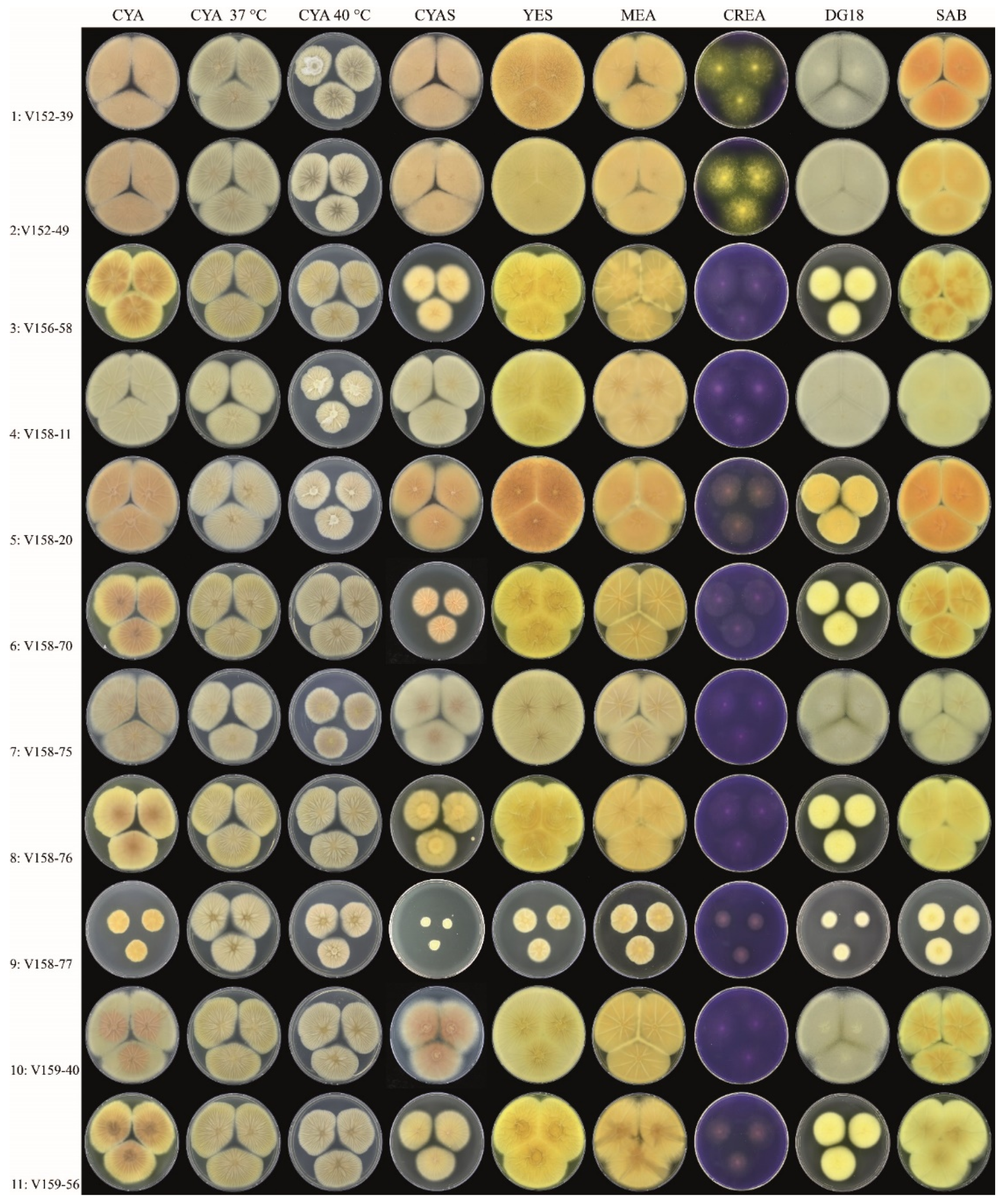
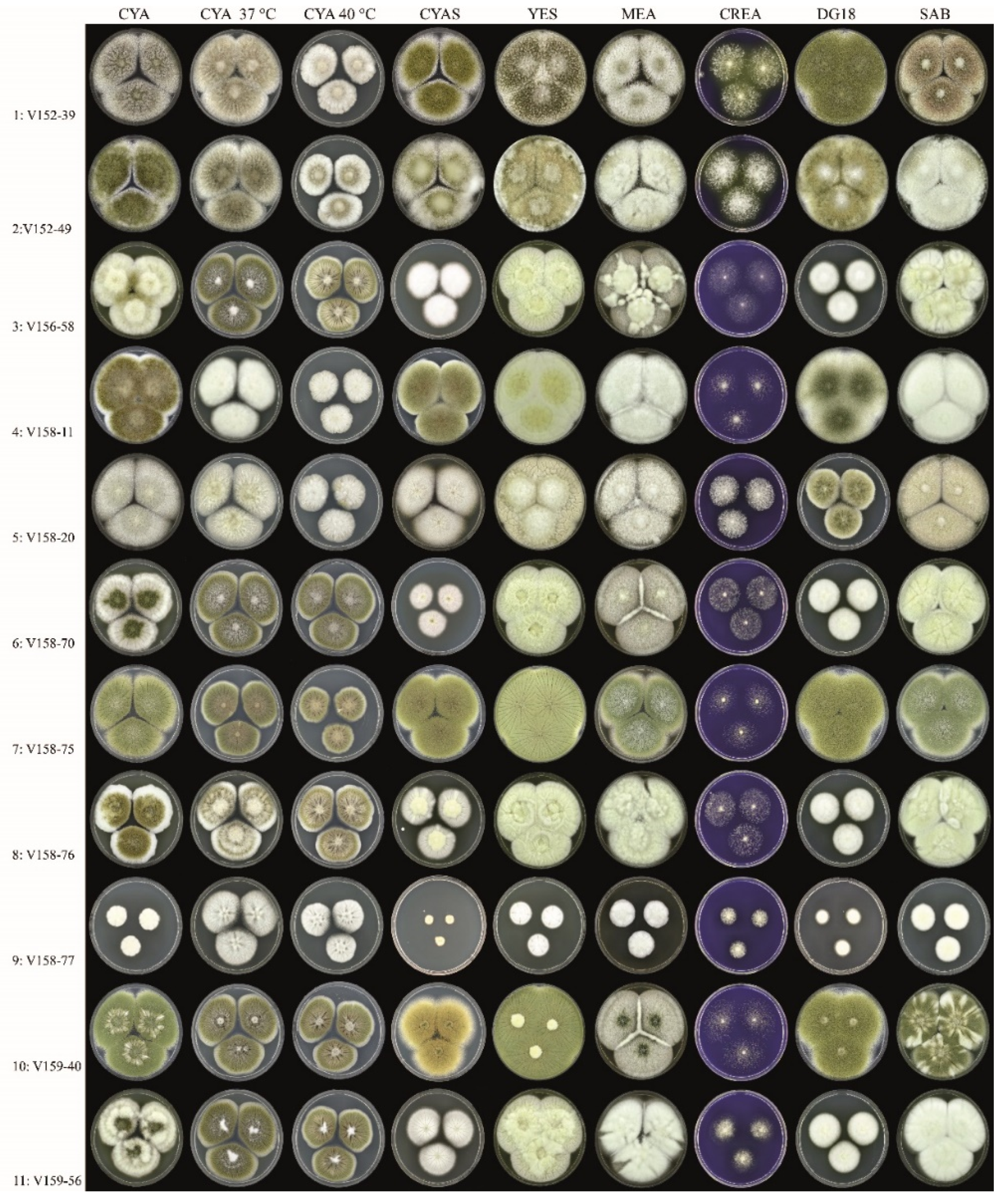
3.1. Strains, Phenotypical Analysis and Genotyping
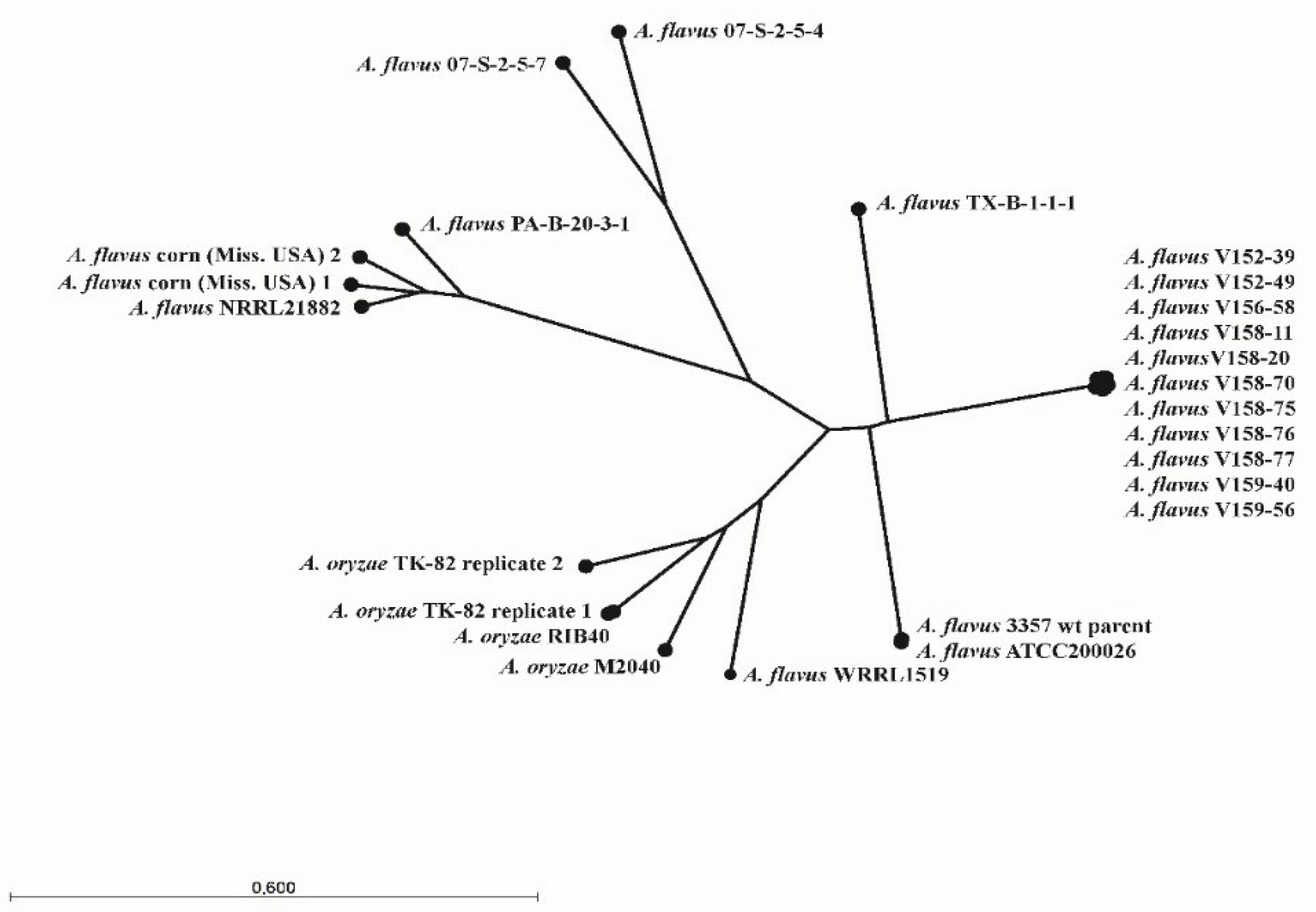
3.2. Whole-Genome Sequence Analysis
4. Discussion
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A

| Accession Number | Organism | Strain | Origin | ||
|---|---|---|---|---|---|
| 1 | SRR4142427 | A. flavus | 3357 wt parent | Unknown | USA |
| 2 | SRR5906257 | A. flavus | WRRL1519 | Almond nut | Cal. USA |
| 3 | SRR6914138 | A. flavus | ATCC200026 | Peanut | USA |
| 4 | SRR7615260 | A. flavus | NRRL21882 | Peanut | USA |
| 5 | SRR8554744 | A. flavus | from corn (Miss. USA) | Corn | Miss. USA |
| 6 | SRR8556699 | A. flavus | from corn (Miss. USA) | Corn | Miss. USA |
| 7 | SRR11596600 | A. flavus | 07-S-2-5-7 | Corn | Louis. USA |
| 8 | SRR11596612 | A. flavus | 07-S-2-5-4 | Corn | Louis. USA |
| 9 | SRR12001143 | A. flavus | TX-B-1-1-1 | Soil | USA |
| 10 | SRR12001155 | A. flavus | PA-B-20-3-1 | Soil | USA |
| 12 | SRR7615261 | A. oryzae | M2040 | Industrial strain | South Korea |
| 13 | SRR1835311 | A. oryzae | RIB40 | Industrial strain | unknown |
| 15 | DRR163532 | A. oryzae | TK-82 replicate 2 | Industrial strain | unknown |
| 16 | DRR163631 | A. oryzae | TK-82 replicate 1 | Industrial strain | unknown |
| 17 | SAMN14278081 | A. flavus | V152-39 | Clinical sample | The Netherlands |
| 18 | SAMN14278082 | A. flavus | V152-49 | Clinical sample | The Netherlands |
| 19 | SAMN14278083 | A. flavus | V156-58 | Clinical sample | The Netherlands |
| 20 | SAMN14278084 | A. flavus | V158-11 | Clinical sample | The Netherlands |
| 21 | SAMN14278085 | A. flavus | V158-20 | Clinical sample | The Netherlands |
| 22 | SAMN14278086 | A. flavus | V158-70 | Clinical sample | The Netherlands |
| 23 | SAMN14278087 | A. flavus | V158-75 | Clinical sample | The Netherlands |
| 24 | SAMN14278088 | A. flavus | V158-76 | Clinical sample | The Netherlands |
| 25 | SAMN14278089 | A. flavus | V158-77 | Clinical sample | The Netherlands |
| 26 | SAMN14278090 | A. flavus | V159-40 | Clinical sample | The Netherlands |
| 27 | SAMN14278091 | A. flavus | V159-56 | Clinical sample | The Netherlands |
| Isolate | Amino Acid Change | Colony Diameter (mm, after 7 d, 25 °C) | Unique Macromorphological Characters Compared to Isolates 1–2 a | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I997N | R52G | A221T | V99A | H25Q | P38L | A561T | Y119F | K1757R | I570T | T159I | R669Q | E35* | P445L | CYAS | DG18 | ||
| 3 | x | x | x | x | x | x | x | x | x | x | 36–38 | 32–34 | Colony diameters smaller on DG18 and CYAS, sporulation variable | ||||
| 4 | x | x | x | x | 50–53 | >60 | Colony diameters slightly smaller on CYAS; greenish-brown-colored conidia | ||||||||||
| 5 | x | x | x | x | x | x | x | x | 54–58 | 36–38 | Colony diameters smaller on DG18, slightly smaller on CYAS; sporulation generally absent or weak | ||||||
| 6 | x | x | x | x | x | x | x | x | x | 26–30 | 32–34 | Colony diameters smaller on DG18 and CYAS; sporulation variable | |||||
| 7 | x | >60 | >60 | Colony diameters similar as Isolates 1–2; abundant sporulation on all agar media, except creatine agar | |||||||||||||
| 8 | x | x | x | x | x | x | x | x | x | 36–38 | 29–32 | Colony diameters smaller on DG18 and CYAS; sporulation variable | |||||
| 9 | x | x | x | x | x | x | 8–10 | 16–18 | Colony diameters restricted on all agar media; sporulation absent | ||||||||
| 10 | x | 54–58 | >60 | Growth diameters slightly smaller on CYAS, sporulation generally moderate or good | |||||||||||||
| 11 | x | x | x | x | x | x | x | x | 35–38 | 30–33 | Colony diameters smaller on DG18 and CYAS; sporulation variable | ||||||
References
- Denning, D.W.; Cadranel, J.; Beigelman-Aubry, C.; Ader, F.; Chakrabarti, A.; Blot, S.; Ullmann, A.J.; Dimopoulos, G.; Lange, C.; European Society of Clinical Microbiology; et al. Chronic pulmonary aspergillosis: Rationale and clinical guidelines for diagnosis and management. Eur. Respir. J. 2016, 47, 45–68. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.L.; Denning, D.W. Underlying conditions in chronic pulmonary aspergillosis including simple aspergilloma. Eur. Respir. J. 2011, 37, 865–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juan Aguilar-Company; Martin, M.T.; Goterris-Bonet, L.; Martinez-Marti, A.; Sampol, J.; Roldan, E.; Almirante, B.; Ruiz-Camps, I. Chronic pulmonary aspergillosis in a tertiary care centre in Spain: A retrospective, observational study. Mycoses 2019, 62, 765–772. [Google Scholar] [CrossRef]
- Ullmann, A.J.; Aguado, J.M.; Arikan-Akdagli, S.; Denning, D.W.; Groll, A.H.; Lagrou, K.; Lass-Florl, C.; Lewis, R.E.; Munoz, P.; Verweij, P.E.; et al. Diagnosis and management of Aspergillus diseases: Executive summary of the 2017 ESCMID-ECMM-ERS guideline. Clin. Microbiol. Infect. 2018, 24 (Suppl. S1), e1–e38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verweij, P.E.; Ananda-Rajah, M.; Andes, D.; Arendrup, M.C.; Bruggemann, R.J.; Chowdhary, A.; Cornely, O.A.; Denning, D.W.; Groll, A.H.; Izumikawa, K.; et al. International expert opinion on the management of infection caused by azole-resistant Aspergillus fumigatus. Drug Resist. Updates 2015, 21–22, 30–40. [Google Scholar] [CrossRef]
- Meis, J.F.; Chowdhary, A.; Rhodes, J.L.; Fisher, M.C.; Verweij, P.E. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2016, 371. [Google Scholar] [CrossRef] [Green Version]
- Tashiro, M.; Izumikawa, K.; Hirano, K.; Ide, S.; Mihara, T.; Hosogaya, N.; Takazono, T.; Morinaga, Y.; Nakamura, S.; Kurihara, S.; et al. Correlation between triazole treatment history and susceptibility in clinically isolated Aspergillus fumigatus. Antimicrob. Agents Chemother. 2012, 56, 4870–4875. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Li, H.; Li, R.; Bu, D.; Wan, Z. Mutations in the cyp51A gene and susceptibility to itraconazole in Aspergillus fumigatus serially isolated from a patient with lung aspergilloma. J. Antimicrob. Chemother. 2005, 55, 31–37. [Google Scholar] [CrossRef]
- Verweij, P.E.; Zhang, J.; Debets, A.J.M.; Meis, J.F.; van de Veerdonk, F.L.; Schoustra, S.E.; Zwaan, B.J.; Melchers, W.J.G. In-host adaptation and acquired triazole resistance in Aspergillus fumigatus: A dilemma for clinical management. Lancet Infect. Dis. 2016, 16, e251–e260. [Google Scholar] [CrossRef]
- Howard, S.J.; Cerar, D.; Anderson, M.J.; Albarrag, A.; Fisher, M.C.; Pasqualotto, A.C.; Laverdiere, M.; Arendrup, M.C.; Perlin, D.S.; Denning, D.W. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg. Infect. Dis. 2009, 15, 1068–1076. [Google Scholar] [CrossRef]
- Chen, P.; Liu, M.; Zeng, Q.; Zhang, Z.; Liu, W.; Sang, H.; Lu, L. Uncovering new mutations conferring azole resistance in the Aspergillus fumigatus cyp51A gene. Front. Microbiol. 2019, 10, 3127. [Google Scholar] [CrossRef] [Green Version]
- Ballard, E.; Melchers, W.J.G.; Zoll, J.; Brown, A.J.P.; Verweij, P.E.; Warris, A. In-host microevolution of Aspergillus fumigatus: A phenotypic and genotypic analysis. Fungal Genet. Biol. 2018, 113, 1–13. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Cuenca-Estrella, M.; Lass-Flörl, C.; Hope, W.W.; Howard, S.J. EUCAST Definitive Document, E.Def 9.2 Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Conidia Forming Moulds; EUCAST: Växjö, Sweden, 2014. [Google Scholar]
- Rudramurthy, S.M.; de Valk, H.A.; Chakrabarti, A.; Meis, J.F.; Klaassen, C.H. High resolution genotyping of clinical Aspergillus flavus isolates from India using microsatellites. PLoS ONE 2011, 6, e16086. [Google Scholar] [CrossRef]
- Khodavaisy, S.; Badali, H.; Rezaie, S.; Nabili, M.; Moghadam, K.G.; Afhami, S.; Hagen, F.; Aala, F.; Hashemi, S.J.; Meis, J.F. Genotyping of clinical and environmental Aspergillus flavus isolates from Iran using microsatellites. Mycoses 2016, 59, 220–225. [Google Scholar] [CrossRef]
- Samson, R.A.; Houbraken, J.; Thrane, U.; Frisvad, J.C.; Andersen, B. Food and Indoor Fungi; Westerdijk Fungal Biodiversity Institute: Utrecht, The Netherlands, 2019. [Google Scholar]
- Samson, R.A.; Visagie, C.M.; Houbraken, J.; Hong, S.B.; Hubka, V.; Klaassen, C.H.; Perrone, G.; Seifert, K.A.; Susca, A.; Tanney, J.B.; et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014, 78, 141–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lestrade, P.P.A.; Meis, J.F.; Melchers, W.J.G.; Verweij, P.E. Triazole resistance in Aspergillus fumigatus: Recent insights and challenges for patient management. Clin. Microbiol. Infect. 2019, 25, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.; Kumar, R.; Kumar, N.; Masih, A.; Gupta, D.; Chowdhary, A. Investigation of multiple resistance mechanisms in voriconazole-resistant Aspergillus flavus clinical isolates from a Chest hospital surveillance in Delhi, India. Antimicrob. Agents Chemother. 2018, 62, e01928-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, M.J.; Won, E.J.; Joo, M.Y.; Park, Y.J.; Kim, S.H.; Shin, M.G.; Shin, J.H. Microsatellite typing and tesistance mechanism analysis of voriconazole-resistant Aspergillus flavus isolates in South Korean hospitals. Antimicrob. Agents Chemother. 2019, 63, e01610-18. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Sun, Y.; Chen, W.; Liu, W.; Wan, Z.; Bu, D.; Li, R. The T788G mutation in the cyp51C gene confers voriconazole resistance in Aspergillus flavus causing aspergillosis. Antimicrob. Agents Chemother. 2012, 56, 2598–2603. [Google Scholar] [CrossRef] [Green Version]
- Paul, R.A.; Rudramurthy, S.M.; Meis, J.F.; Mouton, J.W.; Chakrabarti, A. A Novel Y319H Substitution in CYP51C Associated with Azole Resistance in Aspergillus flavus. Antimicrob. Agents Chemother. 2015, 59, 6615–6619. [Google Scholar] [CrossRef] [Green Version]
- Krishnan-Natesan, S.; Chandrasekar, P.H.; Alangaden, G.J.; Manavathu, E.K. Molecular characterisation of cyp51A and cyp51B genes coding for P450 14alpha-lanosterol demethylases A (CYP51Ap) and B (CYP51Bp) from voriconazole-resistant laboratory isolates of Aspergillus flavus. Int. J. Antimicrob. Agents 2008, 32, 519–524. [Google Scholar] [CrossRef]
- Rudramurthy, S.M.; Paul, R.A.; Chakrabarti, A.; Mouton, J.W.; Meis, J.F. Invasive Aspergillosis by Aspergillus flavus: Epidemiology, Diagnosis, Antifungal Resistance, and Management. J. Fungi 2019, 5, 55. [Google Scholar] [CrossRef] [Green Version]
- Snelders, E.; Camps, S.M.; Karawajczyk, A.; Rijs, A.J.; Zoll, J.; Verweij, P.E.; Melchers, W.J. Genotype-phenotype complexity of the TR46/Y121F/T289A cyp51A azole resistance mechanism in Aspergillus fumigatus. Fungal Genet. Biol. 2015, 82, 129–135. [Google Scholar] [CrossRef]
- Lescar, J.; Meyer, I.; Akshita, K.; Srinivasaraghavan, K.; Verma, C.; Palous, M.; Mazier, D.; Datry, A.; Fekkar, A. Aspergillus fumigatus harbouring the sole Y121F mutation shows decreased susceptibility to voriconazole but maintained susceptibility to itraconazole and posaconazole. J. Antimicrob. Chemother. 2014, 69, 3244–3247. [Google Scholar] [CrossRef] [PubMed]
- Perea, S.; Lopez-Ribot, J.L.; Kirkpatrick, W.R.; McAtee, R.K.; Santillan, R.A.; Martinez, M.; Calabrese, D.; Sanglard, D.; Patterson, T.F. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 2001, 45, 2676–2684. [Google Scholar] [CrossRef] [Green Version]
- Chowdhary, A.; Prakash, A.; Sharma, C.; Kordalewska, M.; Kumar, A.; Sarma, S.; Tarai, B.; Singh, A.; Upadhyaya, G.; Upadhyay, S.; et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: Role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 2018, 73, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Singh, P.K.; de Groot, T.; Kumar, A.; Mathur, P.; Tarai, B.; Sachdeva, N.; Upadhyaya, G.; Sarma, S.; Meis, J.F.; et al. Emergence of clonal fluconazole-resistant Candida parapsilosis clinical isolates in a multicentre laboratory-based surveillance study in India. J. Antimicrob. Chemother. 2019, 74, 1260–1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sionov, E.; Chang, Y.C.; Garraffo, H.M.; Dolan, M.A.; Ghannoum, M.A.; Kwon-Chung, K.J. Identification of a Cryptococcus neoformans cytochrome P450 lanosterol 14alpha-demethylase (Erg11) residue critical for differential susceptibility between fluconazole/voriconazole and itraconazole/posaconazole. Antimicrob. Agents Chemother. 2012, 56, 1162–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheat, L.J.; Connolly, P.; Smedema, M.; Durkin, M.; Brizendine, E.; Mann, P.; Patel, R.; McNicholas, P.M.; Goldman, M. Activity of newer triazoles against Histoplasma capsulatum from patients with AIDS who failed fluconazole. J. Antimicrob. Chemother. 2006, 57, 1235–1239. [Google Scholar] [CrossRef]
- Leroux, P.; Walker, A.S. Multiple mechanisms account for resistance to sterol 14alpha-demethylation inhibitors in field isolates of Mycosphaerella graminicola. Pest Manag. Sci. 2011, 67, 44–59. [Google Scholar] [CrossRef]
- Delye, C.; Laigret, F.; Corio-Costet, M.F. A mutation in the 14 alpha-demethylase gene of Uncinula necator that correlates with resistance to a sterol biosynthesis inhibitor. Appl. Environ. Microbiol. 1997, 63, 2966–2970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hokken, M.W.J.; Zoll, J.; Coolen, J.P.M.; Zwaan, B.J.; Verweij, P.E.; Melchers, W.J.G. Phenotypic plasticity and the evolution of azole resistance in Aspergillus fumigatus; an expression profile of clinical isolates upon exposure to itraconazole. BMC Genom. 2019, 20, 28. [Google Scholar] [CrossRef]
- Losada, L.; Sugui, J.A.; Eckhaus, M.A.; Chang, Y.C.; Mounaud, S.; Figat, A.; Joardar, V.; Pakala, S.B.; Pakala, S.; Venepally, P.; et al. Genetic analysis using an isogenic mating pair of Aspergillus fumigatus identifies azole resistance genes and lack of MAT locus's role in virulence. PLoS Pathog. 2015, 11, e1004834. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Ferreira, M.E.; Malavazi, I.; Savoldi, M.; Brakhage, A.A.; Goldman, M.H.; Kim, H.S.; Nierman, W.C.; Goldman, G.H. Transcriptome analysis of Aspergillus fumigatus exposed to voriconazole. Curr. Genet. 2006, 50, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Valdes, I.D.; van den Berg, J.; Haagsman, A.; Escobar, N.; Meis, J.F.; Hagen, F.; Haas, P.J.; Houbraken, J.; Wösten, H.A.B.; de Cock, H. Comparative genotyping and phenotyping of Aspergillus fumigatus isolates from humans, dogs and the environment. BMC Microbiol. 2018, 18, 118. [Google Scholar] [CrossRef] [Green Version]
- Hokken, M.W.J.; Zwaan, B.J.; Melchers, W.J.G.; Verweij, P.E. Facilitators of adaptation and antifungal resistance mechanisms in clinically relevant fungi. Fungal Genet. Biol. 2019, 132, 103254. [Google Scholar] [CrossRef]
- Faria-Ramos, I.; Farinha, S.; Neves-Maia, J.; Tavares, P.R.; Miranda, I.M.; Estevinho, L.M.; Pina-Vaz, C.; Rodrigues, A.G. Development of cross-resistance by Aspergillus fumigatus to clinical azoles following exposure to prochloraz, an agricultural azole. BMC Microbiol. 2014, 14, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howard, S.J.; Pasqualotto, A.C.; Anderson, M.J.; Leatherbarrow, H.; Albarrag, A.M.; Harrison, E.; Gregson, L.; Bowyer, P.; Denning, D.W. Major variations in Aspergillus fumigatus arising within aspergillomas in chronic pulmonary aspergillosis. Mycoses 2013, 56, 434–441. [Google Scholar] [CrossRef]
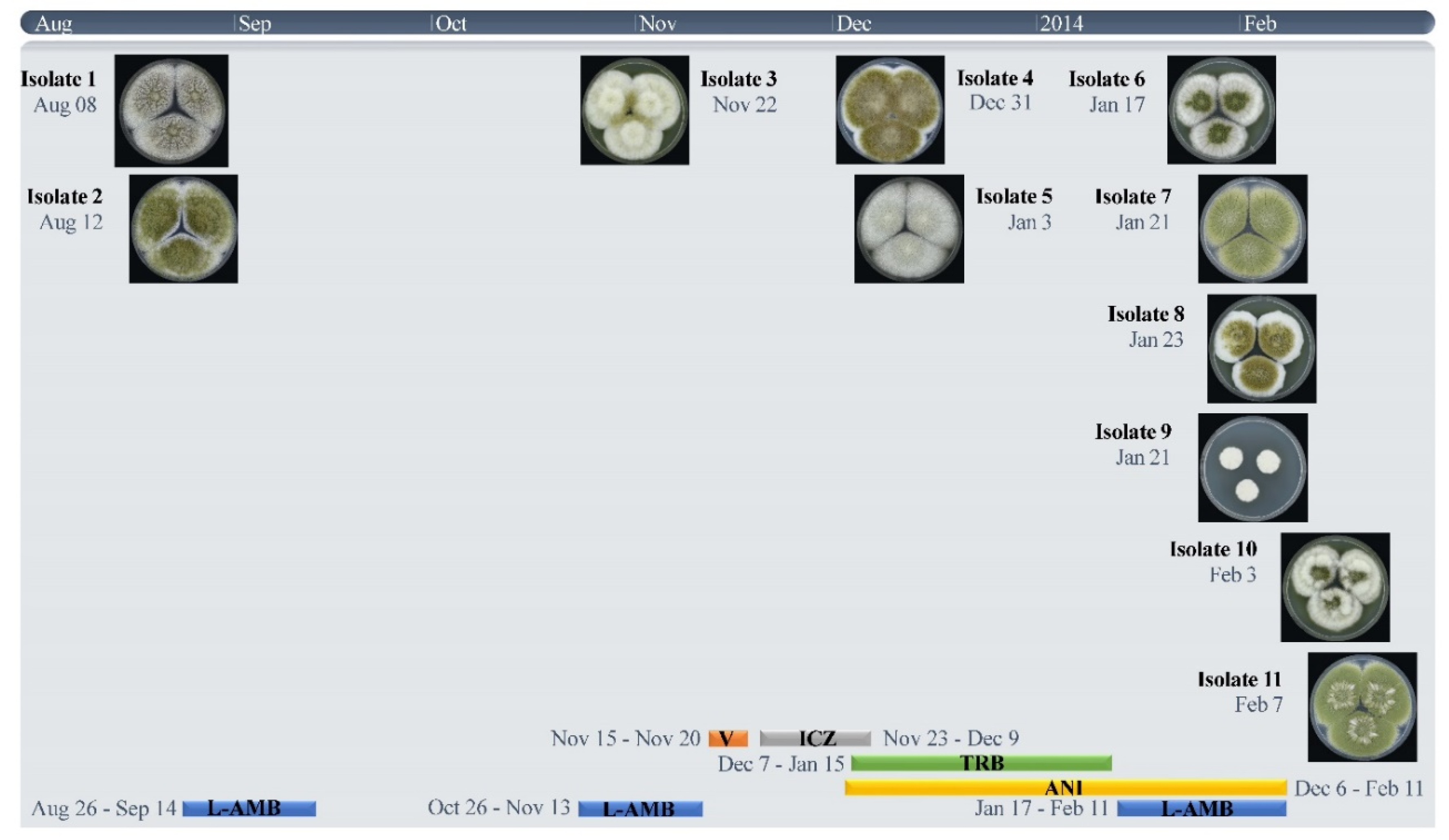
| Strain Number | Isolation Date | MIC (mg/L) | STR Numbers | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ITC | VOR | POSA | AMB | ANI c | 2 | 3 | 4 | ||
| 1 V152-39 | 8-8-2013 | 0.5 | 1 | 0.5 | 1 | 0.063 | 23-14-17 | 8-22-12 | 5-7-2011 |
| 2 V152-49 | 12-8-2013 | 1 | 4 | 0.25 | 2 | 0.031 | 23-14-17 | 8-22-12 | 5-7-2011 |
| 3 V156-58 | 22-11-2013 | 16 a | >16 | 0.5 | 2 | 0.031 | 23-14-17 | 8-22-12 | 5-7-2011 |
| 4 V158-11 | 31-12-2013 | >16 a,b | >16 b | 1 | 2 | 0.063 | 23-14-17 | 8-22-12 | 5-7-2011 |
| 5 V158-20 | 3-1-2014 | 16 a | 16 | 0.5 | 1 | 0.016 | 23-14-17 | 8-22-12 | 5-7-2011 |
| 6 V158-70 | 17-1-2014 | 2 | >16 | 0.5 | 2 | 0.016 | 23-14-17 | 8-22-12 | 5-7-2011 |
| 7 V158-75 | 21-1-2014 | >16 a | >16 | 1 | 2 | 0.125 | 23-14-17 | 8-22-12 | 5-7-2011 |
| 8 V158-76 | 23-1-2014 | >16 a | >16 | 1 | 2 | 0.125 | 23-14-17 | 8-22-12 | 5-7-2011 |
| 9 V158-77 | 21-1-2014 | 1 | 16 | 0.25 | 2 | 0.25 | 23-14-17 | 8-22-12 | 5-7-2011 |
| 10 V159-40 | 3-2-2014 | >16 a | >16 | 2 | 2 | 0.031 | 23-14-17 | 8-22-12 | 5-7-2011 |
| 11 V159-56 | 7-2-2014 | 1 | >16 | 0.5 | 2 | 0.016 | 24-14-17 | 8-22-12 | 5-7-2011 |
| Amino Acid Change | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr.-Pos. a | Allele | Isolate b | A. oryzae Gene c (RefSeq Accession Numbers) | A. flavus Gene d | Function | |||||||||
| 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||||||
| 1-498965 | T | A | I997N | I997N | I997N | I997N | I997N | AO090009000185 (XM_001816630) | AFLA_052710 | GTPase activating protein | ||||
| 1-561401 | A | G | R52G | AO090009000208 (XM_023234211) | AFLA_052490 | unknown | ||||||||
| 1-1099800 | C | T | A221T | A221T | A221T | A221T | AO090009000417 (XM_001816830) | AFLA_050600 | Aldehyde dehydrogenase | |||||
| 1-3360498 | T | C | V99A | AO090005001154 (XM_023232984) | AFLA_083950 | MSF transporter | ||||||||
| 1-3370761 | T | A | H25Q | H25Q | H25Q | H25Q | H25Q | H25Q | AO090005001151 (XM_023232999) | AFLA_083930 | DnaJ domain protein | |||
| 2-578295 | C | T | P38L | P38L | P38L | P38L | P38L | AO090001000237 (XM_001818728) | Global gene regulator VeA | |||||
| 2-579927 | G | A | A561T | AO090001000237 (XM_001818728) | Global gene regulator VeA | |||||||||
| 2-2642838 | T | A | Y119F | Y119F | Y119F | Y119F | Y119F | Y119F | Y119F | Y119F | AO090003000205 (XM_001819367) | AFLA_036130 | 14-alpha sterol demethylase | |
| 2-3253116 | T | C | K1757R | K1757R | K1757R | K1757R | K1757R | K1757R | AO090003000437 (XM_001819566) | AFLA_033810 | NPC protein An-Mlp1 | |||
| 3-851374 | A | G | I570T | I570T | I570T | I570T | I570T | I570T | AO090023000332 (XM_023235544) | AFLA_107440 | Sla1 | |||
| 4-1887254 | C | T | T159I | AO090012000739 (XM_023236608) | unknown | |||||||||
| 4-4726184 | C | T | R669Q | R669Q | R669Q | R669Q | R669Q | AO090166000062 (XM_023237067) | unknown | |||||
| 7-2588623 | C | A | E35* | E35* | E35* | E35* | E35* | E35* | AO090206000001 (XM_023233185) | AFLA_115530 | C-4 methylsterol oxidase | |||
| 8-891870 | G | A | P445L | AO090103000145 (XM_023233427) | AFLA_010830 | unknown | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buil, J.B.; Houbraken, J.; Reijers, M.H.; Zoll, J.; Sanguinetti, M.; Meis, J.F.; Verweij, P.E.; Melchers, W.J.G. Genetic and Phenotypic Characterization of in-Host Developed Azole-Resistant Aspergillus flavus Isolates. J. Fungi 2021, 7, 164. https://doi.org/10.3390/jof7030164
Buil JB, Houbraken J, Reijers MH, Zoll J, Sanguinetti M, Meis JF, Verweij PE, Melchers WJG. Genetic and Phenotypic Characterization of in-Host Developed Azole-Resistant Aspergillus flavus Isolates. Journal of Fungi. 2021; 7(3):164. https://doi.org/10.3390/jof7030164
Chicago/Turabian StyleBuil, Jochem B., Jos Houbraken, Monique H. Reijers, Jan Zoll, Maurizio Sanguinetti, Jacques F. Meis, Paul. E. Verweij, and Willem J.G. Melchers. 2021. "Genetic and Phenotypic Characterization of in-Host Developed Azole-Resistant Aspergillus flavus Isolates" Journal of Fungi 7, no. 3: 164. https://doi.org/10.3390/jof7030164







