Global Diversity and Taxonomy of Sidera (Hymenochaetales, Basidiomycota): Four New Species and Keys to Species of the Genus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Morphological Studies
2.2. DNA Extraction, PCR and Sequencing
2.3. Phylogenetic Analyses
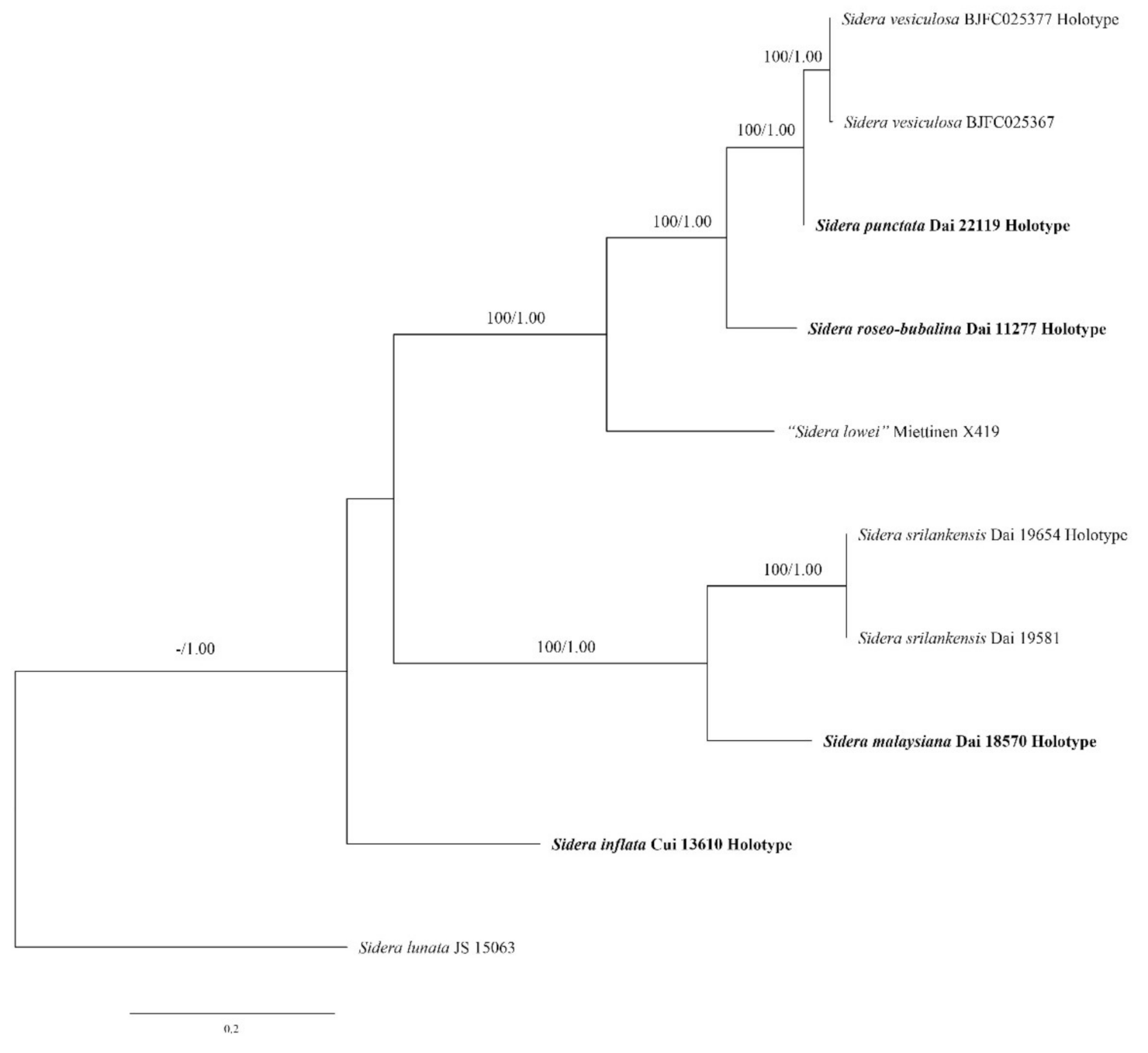
3. Results
3.1. Phylogenetic Analyses
3.2. Taxonomy
- 1.
- 2.
- 3.
- 4.
- 5.
- Sidera vulgaris sensu lato
| A key to species of Sidera worldwide | |
| 1. Hymenium grandinioid to odontioid | S. lunata |
| 1. Hymenium poroid | 2 |
| 2. Hyphal system monomitic | 3 |
| 2. Hyphal system dimitic | 6 |
| 3. Basidiospores mostly <1 μm in width | 4 |
| 3. Basidiospores mostly >1 μm in width | 5 |
| 4. Pores 7–9 per mm; basidiospores 2.9–3.7 μm long | S. vesiculosa |
| 4. Pores 6–7 per mm; basidiospores 3.9–4.5 μm long | S. roseo-bubalina |
| 5. Pores 6–8 per mm; cystidioles present, some branched | S. lowei |
| 5. Pores 8–9 per mm; cystidioles absent | S. punctata |
| 6. Basidiospores >1.5 μm in width | S. lenis |
| 6. Basidiospores <1.5 μm in width | 7 |
| 7. Skeletal hyphae becoming swollen in KOH | 8 |
| 7. Skeletal hyphae almost unchanged in KOH | 10 |
| 8. Pores 5–7 per mm; basidiospores 3.7–4.3 μm long | S. minutipora |
| 8. Pores 9–11 per mm; basidiospores 2.9–3.3 μm long | 9 |
| 9. Basidiospores allantoid, skeletal hyphae distinctly swollen in KOH | S. inflata |
| 9. Basidiospores lunate, skeletal hyphae slightly swollen in KOH | S. malaysiana |
| 10. Tramal hyphae parallel along the tubes | S. parallela |
| 10. Tramal hyphae interwoven | 11 |
| 11. Generative hyphae at dissepiments even | S. srilankensis |
| 11. Generative hyphae at dissepiments with swollen tips | 12 |
| 12. Basidiospores <3.6 μm long | S. vulgaris |
| 12. Basidiospores >3.8 μm long | 13 |
| 13. Sterile margin distinct, fimbriate; basidiospore Length/width <4 | S. minutissima |
| 13. Sterile margin indistinct to almost absent; basidiospore Length/width >4 | S. tenuis |
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miettinen, O.; Larsson, K.H. Sidera. A new genus in Hymenochaetales with poroid and hydnoid species. Mycol. Prog. 2011, 10, 131–141. [Google Scholar] [CrossRef]
- Du, R.; Wu, F.; Gate, G.M.; Dai, Y.C.; Tian, X.M. Taxonomy and phylogeny of Sidera (Hymenochaetales, Basidiomycota): Four new species and keys to species of the genus. MycoKeys 2020, 68, 115–135. [Google Scholar] [CrossRef]
- Fries, E.M. Systema Mycologicum. Berlingius Lindae. 1821, 1, 1–520. [Google Scholar]
- Rajchenberg, M. Type studies of Polyporaceae (Aphyllophorales) described by J. Rick. Nord. J. Bot. 1987, 7, 553–568. [Google Scholar] [CrossRef]
- Bourdot, H.; Galzin, A. Hyménomycètes de France. I. Hétèrobasidiés. Bull. Soc. Mycol. Fr. 1909, 25, 15–36. [Google Scholar]
- Rodway, L.; Cleland, J.B. Notes on the genus Poria No. 3. Pap. Proc. R. S. Tasman. 1929, 17, 7–24. [Google Scholar]
- Du, R.; Wang, L.; Zhou, M.; Chen, J.J. A new species of Sidera (Hymenochaetales, Basidiomycota) from tropical Asia. Phytotaxa. 2019, 387, 165–171. [Google Scholar] [CrossRef]
- Niemelä, T.; Dai, Y.C. Polypore Skeletocutis lenis and its sib S. vulgaris. Ann. Bot. Fenn. 1997, 34, 133–140. [Google Scholar]
- Niemelä, T. Polypores, lignicolous fungi. Norrlinia 2005, 13, 1–320, (In Finnish, with English summary). [Google Scholar]
- Dai, Y.C. Hymenochaetaceae (Basidiomycota) in China. Fungal Divers. 2010, 45, 131–343. [Google Scholar] [CrossRef]
- Anonymous. Flora of British Fungi. Colour Identification Chart; Her Majesty’s Stationery Office: London, UK, 1969. [Google Scholar]
- Petersen, J.H. Farvekort. The Danish Mycological Society’ s Colour-Chart; Foreningen til Svampekundskabens Fremme: Greve, Italy, 1996; pp. 1–6. [Google Scholar]
- Thiers, B. Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff; New York Botanical Garden’s Virtual Herbarium: New York, NY, USA, 2018; Available online: http://sweetgum.nybg.org/science/ih/ (accessed on 15 January 2021).
- Cui, B.K.; Li, H.J.; Ji, X.; Zhou, J.L.; Song, J.; Si, J.; Yang, Z.L.; Dai, Y.C. Species diversity, taxonomy and phylogeny of Polyporaceae (Basidiomycota) in China. Fungal Divers. 2019, 97, 137–392. [Google Scholar] [CrossRef]
- Shen, L.L.; Wang, M.; Zhou, J.L.; Xing, J.H.; Cui, B.K.; Dai, Y.C. Taxonomy and phylogeny of Postia. Multi–gene phylogeny and taxonomy of the brown–rot fungi: Postia and its related genera. Pers. Mol. Phylogeny Evol. Fungi 2019, 42, 101–126. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gefand, D.H., Sninsky, J.J., White, J.T., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Matheny, P.B.; Liu, Y.J.; Ammirati, J.F.; Hall, B.D. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am. J. Bot. 2002, 89, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Matheny, P.B. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe, Agaricales). Molec. Phylogenet. Evolut. 2005, 35, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. Bioedit: A user–friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Available online: https://www.mesquiteproject.org/ (accessed on 24 March 2021).
- Yuan, Y.; Ji, X.H.; Wu, F.; He, S.H.; Chen, J.J. Two new Gloeoporus (Polyporales, Basidiomycota) from tropical China. Nova Hedwig. 2016, 103, 169–183. [Google Scholar] [CrossRef]
- Miller, M.A.; Holder, M.T.; Vos, R.; Midford, P.E.; Liebowitz, T.; Chan, L.; Hoover, P.; Warnow, T. The CIPRES Portals. CIPRES. Available online: http://www.phylo.org/sub_sections/portal (accessed on 4 August 2009).
- Nylander, J.A.A. MrModeltest v2. Program Distributed by the Author; Evolutionary Biology Centre: Uppsala, Sweden, 2004. [Google Scholar]
- Swofford, D.L. PAUP: Phylogenetic Analysis using Parsimony Version 4.0b10; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Ronquist, F.; Teslenko, M.; Mark, P.; Avres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes3.2: Efficient Bayesian phylogenetic inference and model choice, across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A. Molecular Evolution, Phylogenetics and Epidemiology. FigTree ver. 1.4 Software. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 24 March 2021).
- Pilát, A. Polyporaceae. In Atlas des Champognons de l’s Europe; Kavina, C., Pilát, A., Eds.; Privately Published: Prague, Czech Republic, 1936–1942; pp. 1–624,374.
- Kotlaba, F.; Pouzar, Z. Type studies of polypores described by A. Pilát. Ceska Mykol. 1991, 45, 91–97. [Google Scholar]
- Murrill, W.A. Light-colored resupinate polypores 1. Mycologia 1920, 12, 77–92. [Google Scholar] [CrossRef]
- Murrill, W.A. Light-colored resupinate polypores 2. Mycologia 1920, 12, 299–308. [Google Scholar] [CrossRef]
- Pilát, A. Additamenta ad floram Sibiriae Asiaeque orientalis mycologicam. Bull. Soc. Mycol. Fr. 1933, 49, 256–339. [Google Scholar]
- Pilát, A. Additamenta ad floram Sibiriae Asiaeque orientalis mycologicam. Pars Tertia. Bull. Soc. Mycol. Fr. 1935, 51, 351–426. [Google Scholar]
- Kotlaba, F.; Pouzar, Z. Type studies of polypores described by A. Pilát-III. Ceská Mycol. 1989, 43, 36–44. [Google Scholar]
- Ryvarden, L.; Johansen, I. A Preliminary Polypore Flora of East Africa; Fungiflora: Oslo, Norway, 1980; pp. 1–636. [Google Scholar]
- Buchanan, P.K.; Ryvarden, L. An annotated checklist of polypore and polypore-like fungi recorded from New Zealand. N. Z. J. Bot. 2000, 38, 265–323. [Google Scholar] [CrossRef] [Green Version]
- Núñez, M.; Ryvarden, L. East Asian polypores 2. Synop. Fungorum. 2001, 14, 165–522. [Google Scholar]
- Ryvarden, L. Neotropical polypores 1. Synop. Fungorum. 2004, 19, 1–229. [Google Scholar]
- Dai, Y.C. Polypore diversity in China with an annotated checklist of Chinese polypores. Mycoscience 2012, 53, 49–80. [Google Scholar] [CrossRef]
- Zhou, L.W.; Nakasone, K.K.; Burdsall, H.H.; Ginns, J.; Vlasák, J.; Miettinen, O.; Spirin, V.; Niemelä, T.; Yuan, H.S.; He, S.H.; et al. Polypore diversity in North America with an annotated checklist. Mycol. Prog. 2016, 15, 771–790. [Google Scholar] [CrossRef]
- Ryvarden, L.; Melo, I. Poroid fungi of Europe, 2nd edition. In Synop. Fungorum; Ryvarden, L., Melo, I., Eds.; Fungiflora AS: Oslo, Norway, 2017; pp. 1–430. [Google Scholar]
- Decock, C.; Yombiyeni, P.; Memiaghe, H. Hymenochaetaceae from the Guineo-Congolian rainforest: Phylloporia flabelliforma sp. nov. and Phylloporia gabonensis sp. nov., two undescribed species from Gabon. Cryptogam. Mycol. 2016, 36, 449–467. [Google Scholar] [CrossRef]
- Miettinen, O.; Ryvarden, L. Polypore genera Antella, Austeria, Butyrea, Citripora, Metuloidea and Trulla (Steccherinaceae, Polyporales). Ann. Bot. Fenn. 2016, 53, 157–172. [Google Scholar] [CrossRef]
- Ji, X.H.; He, S.H.; Chen, J.J.; Si, J.; Wu, F.; Zhou, L.W.; Vlasák, J.; Tian, X.M.; Dai, Y.C. Global diversity and phylogeny of Onnia (Hymenochaetaceae) species on gymnosperms. Mycologia 2017, 109, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Spirin, V.; Vlasák, J.; Miettinen, O. Studies in the Antrodia serialis group (Polyporales, Basidiomycota). Mycologia 2017, 109, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Dai, S.J.; Vlasák, J.; Spirin, V.; Dai, Y.C. Phylogeny and global diversity of Porodaedalea, a genus of gymnosperm pathogens in the Hymenochaetales. Mycologia 2019, 111, 40–53. [Google Scholar] [CrossRef]










| Species | Growing Habit | Hymenophore | Hyphal System | Cystidioles | Skeletal Hyphae in KOH | Spores Shape | Spore Size (µm) |
|---|---|---|---|---|---|---|---|
| S. inflata | Annual | Poroid, 9–10/mm | Dimitic | Present | Swollen | Allantoid | 3–3.3 × 0.9–1.1 |
| S. lenis | Perennial | Poroid, 4–6/mm | Dimitic | Present | Swollen | Allantoid to lunate | 3.9–4.9 × 1.5–2 |
| S. lowei | Annual | Poroid, 6–8/mm | Monomitic | Present, some branched | - | Allantoid | 3.5–5 × 1–1.2 |
| S. lunata | Annual | Hydnoid, 8–9/mm | Monomitic | Present | - | Allantoid | 2.5–3.8 × 1.6–1.9 |
| S. malaysiana | Annual | Poroid, 9–11/mm | Dimitic | Present | Swollen | Lunate | 2.9–3.2 × 1–1.2 |
| S. minutipora | Annual | Poroid, 5–7/mm | Dimitic | Present | Swollen | Allantoid | 3.7–4.3 × 1–1.3 |
| S. minutissima | Annual | Poroid, 7–9/mm | Dimitic | Present | Almost unchanged | Allantoid | 3.8–4.4 × 0.9–1.3 |
| S. parallela | Annual | Poroid, 6–8/mm | Dimitic | Present | Almost unchanged | Lunate | 2.8–3.3 × 0.9–1.2 |
| S. punctata | Annual | Poroid, 8–9/mm | Monomitic | Absent | - | Allantoid to lunate | 3.8–4.8 × 1–1.3 |
| S. roseo-bubalina | Annual | Poroid, 6–7/mm | Monomitic | Present | - | Lunate | 3.9–4.5 × 0.8–1 |
| S. srilankensis | Annual | Poroid, 6–8/mm | Dimitic | Present | Almost unchanged | Lunate | 3.5–4 × 1–1.3 |
| S. tenuis | Annual | Poroid, 8–10/mm | Dimitic | Present | Almost unchanged | Allantoid | 4.2–5 × 0.8–1 |
| S. vesiculosa | Annual | Poroid, 7–9/mm | Monomitic | Present | - | Allantoid to lunate | 2.9–3.7 × 0.6–1 |
| S. vulgaris | Perennial | Poroid, 6–8/mm | Dimitic | Present, some branched | Almost unchanged | Allantoid to lunate | 2.9–3.6 × 0.9–1.4 |
| Species | Specimen No. | Locality | GenBank Accession No. | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ITS | nLSU | RPB1 | RPB2 | TEF1 | mtSSU | nSSU | |||
| Ceriporiopsis aneirina | MUAF 888 | Czech Republic | EU340895 | EU368503 | EU368504 | ||||
| Contumyces rosella | Redhead 7501 | U66452 | U66452 | ||||||
| Exidia candida | VS 8588 | Russia | KY801871 | KY801896 | KY801920 | ||||
| Exidiopsis calcea | MW 331 | Canada | AF291280 | AF291326 | |||||
| Gloeoporus dichrous | KHL 11173 | Norway | EU118627 | EU118627 | |||||
| G. hainanensis | Dai 15253 | China | KU360402 | KU360408 | |||||
| Globulicium hiemale | Hjm 19007 | Sweden | DQ873595 | DQ873595 | DQ873594 | ||||
| Hyphodermella poroides | Dai 12045 | China | KX008367 | KX011852 | |||||
| Odonticium romellii | Murdoch 38 | Finland | MF319073 | MF318929 | |||||
| Oxyporus corticola | KHL 13217 | Estonia | DQ873641 | DQ873641 | DQ873640 | ||||
| Repetobasidium conicum | KHL 12338 | USA | DQ873647 | DQ873647 | DQ873646 | ||||
| Resinicium furfuraceum | KHL 11738 | Finland | DQ873648 | DQ873648 | |||||
| Rickenella mellea | Lamoure 74 | U66438 | U66438 | ||||||
| Skvortzovia pinicola | KHL 12224 | USA | DQ873637 | DQ873637 | DQ873636 | ||||
| Sidera inflata | Cui 13610 | China | MW198480 * | MW418088 * | |||||
| S. lenis | Miettinen 11036 | Finland | FN907914 | FN907914 | |||||
| “S. lowei” | Miettinen X419 | Venezuela | FN907917 | FN907917 | |||||
| “S. lowei” | Miettinen X426 | New Zealand | FN907919 | FN907919 | |||||
| S. lunata | JS 15063 | Norway | DQ873593 | DQ873593 | |||||
| S. malaysiana | Dai 18570 | Malaysia | MW198481 * | MW192007 * | MW424992 * | ||||
| S. minutipora | Gates FF257 | Australia | FN907922 | FN907922 | |||||
| S. minutipora | Cui 16720 | Australia | MN621349 | MN621348 | MW526261 * | MW505865 * | MW446248 * | MW424986 * | MW418078 * |
| S. minutissima | Dai 19529 | Sri Lanka | MN621352 | MN621350 | MW526262 * | MW424994 * | MW418079 * | ||
| S. minutissima | Dai 18471A | China | MW198482 * | MW192008 * | |||||
| S. parallela | Cui 10346 | China | MK346145 | MW424991 * | MW418082 * | ||||
| S. parallela | Cui 10361 | China | MK346144 | ||||||
| S. punctata | Dai 22119 | China | MW418438 * | MW418437 * | MW418085 * | ||||
| S. roseo-bubalina | Dai 11277 | China | MW198483 * | MW418081 * | |||||
| S. sp. | OTU1581 | Japan | MT594995 | ||||||
| S. srilankensis | Dai 19581 | Sri Lanka | MN621345 | MN621347 | MW526265 * | MW427604 * | MW424993 * | MW418089 * | |
| S. srilankensis | Dai 19654 | Sri Lanka | MN621344 | MN621346 | MW505868 * | MW427602 * | MW424989 * | MW418087 * | |
| S. tenuis | Dai 18697 | Australia | MK331865 | MK331867 | MW526264 * | MW505866 * | MW427600 * | MW424988 * | MW418083 * |
| S. tenuis | Dai 18698 | Australia | MK331866 | MK331868 | MW505867 * | MW427601 * | MW424985 * | MW418084 * | |
| S. vesiculosa | BJFC025367 | Singapore | MH636565 | MH636567 | |||||
| S. vesiculosa | BJFC025377 | Singapore | MH636564 | MH636566 | |||||
| S. vulgaris sensu lato | Ryvarden 37198 | New Zealand | FN907918 | FN907918 | |||||
| S. vulgaris sensu lato | Dai 12730 | USA | MW198478 * | ||||||
| S. vulgaris sensu lato | Dai 21057 | Belarus | MW198484 * | MW192009 * | MW505869 * | MW427603 * | MW424987 * | MW418090 * | |
| S. vulgaris sensu lato | Dai 22151 | China | MW477794 * | MW474965 * | MW477795 * | ||||
| S. vulgaris sensu lato | Cui 11216 | China | MW198485 * | ||||||
| Skeletocutis amorpha | Miettinen 11038 | Finland | FN907913 | FN907913 | |||||
| S. chrysella | Miettinen 9472 | Finland | FN907916 | FN907916 | |||||
| S. lilacina | HHB 10522sp | USA | KY948834 | KY948894 | |||||
| S. yuchengii | FBCC 1132 | China | KY953045 | KY953045 | KY953143 | KY953109 | KY953142 | ||
| S. yunnanensis | Dai 15709 | China | KU950434 | KU950436 | MW526263 * | MW427605 * | MW424990 * | MW418080 * | |
| S. odora | L 13763sp | Canada | KY948830 | KY948893 | KY949046 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.-B.; Zhou, M.; Yuan, Y.; Dai, Y.-C. Global Diversity and Taxonomy of Sidera (Hymenochaetales, Basidiomycota): Four New Species and Keys to Species of the Genus. J. Fungi 2021, 7, 251. https://doi.org/10.3390/jof7040251
Liu Z-B, Zhou M, Yuan Y, Dai Y-C. Global Diversity and Taxonomy of Sidera (Hymenochaetales, Basidiomycota): Four New Species and Keys to Species of the Genus. Journal of Fungi. 2021; 7(4):251. https://doi.org/10.3390/jof7040251
Chicago/Turabian StyleLiu, Zhan-Bo, Meng Zhou, Yuan Yuan, and Yu-Cheng Dai. 2021. "Global Diversity and Taxonomy of Sidera (Hymenochaetales, Basidiomycota): Four New Species and Keys to Species of the Genus" Journal of Fungi 7, no. 4: 251. https://doi.org/10.3390/jof7040251









