Exploring the Diversity of Fungal DyPs in Mangrove Soils to Produce and Characterize Novel Biocatalysts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains for Cloning and Heterologous Expression
2.2. Sediment Sampling, RNA Extraction, and cDNA Synthesis
2.3. Probe Design, cDNA Capture by Hybridization, and High-Throughput Sequencing of Fungal DyP cDNA
2.4. Bioinformatic Analysis and Statistics
2.5. Cloning and Expression of DyP-encoding cDNA
2.6. Production and Purification of Recombinant DyP
2.7. Structural Analysis
2.8. Standard Conditions for Peroxidase Activity
2.9. Influence of Temperature and pH on DyP Activity and Enzyme Stability
2.10. Effect of Hydrogen Peroxide and Sea Salt on DyP Activity
2.11. Substrate Specificity and Kinetics
2.12. Decolorization Properties
3. Results
3.1. Diversity and Capture of Fungal DyP Encoding cDNAs
3.2. Phylogenetic Analysis
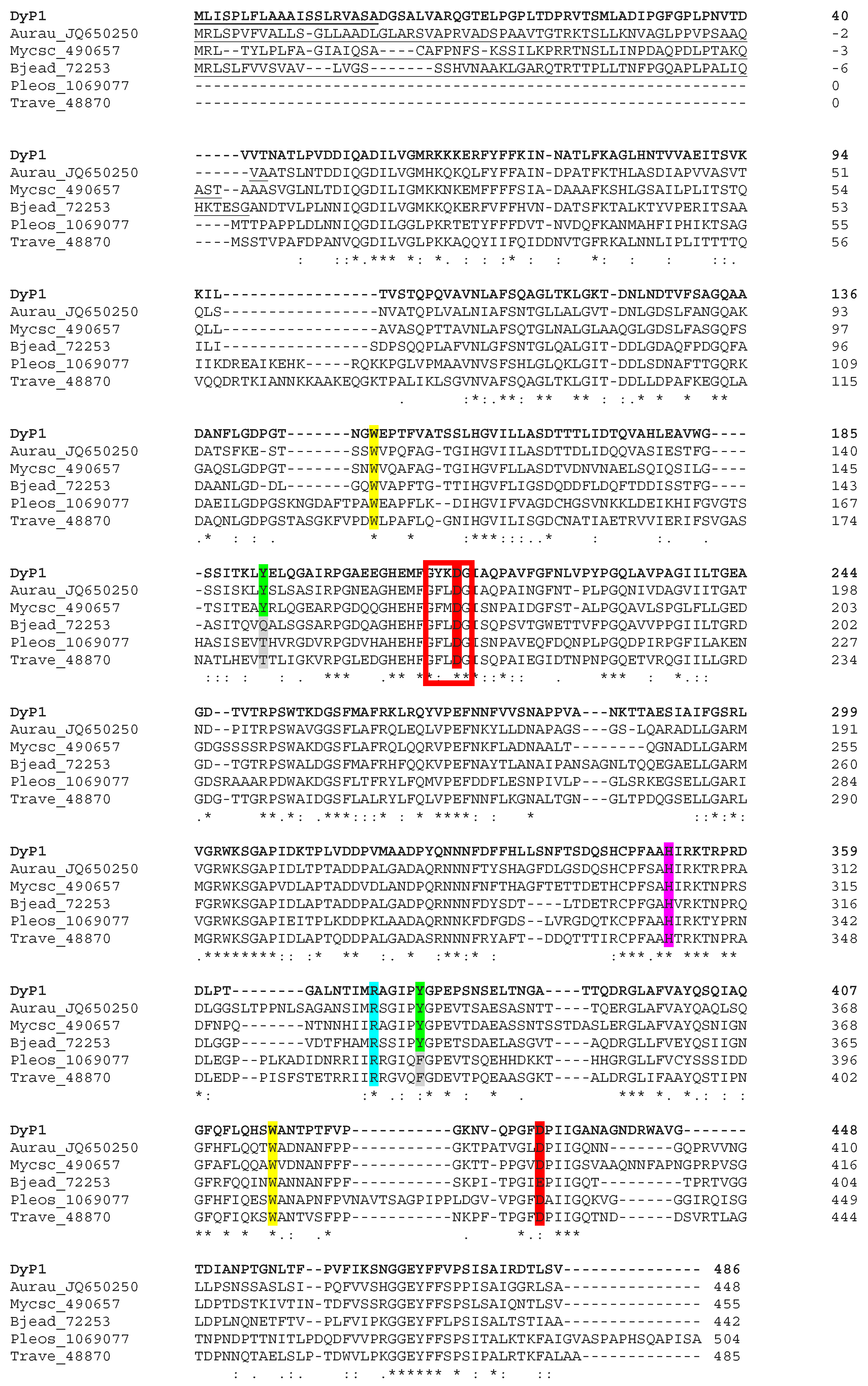
3.3. Structural Analysis
3.4. Heterologous Production and Purification of the Recombinant DyP1
3.5. Catalytic Properties
3.6. Enzyme Activity and Stability at Different pH and Temperature
3.7. Decolorization of Industrial Dyes
3.8. Effect of Hydrogen Peroxide on DyP1 Activity
3.9. Influence of Sea Salt on DyP1 Activity and Surface Charge of the Recombinant DyP1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Thatoi, H.; Behera, B.C.; Mishra, R.R. Ecological role and biotechnological potential of mangrove fungi: A review. Mycology 2013, 4, 54–71. [Google Scholar] [CrossRef]
- Kathiresan, K.; Bingham, B.L. Biology of mangroves and mangrove ecosystems. Adv. Mar. Biol. 2001, 40, 84–254. [Google Scholar]
- Luis, P.; Saint-Genis, G.; Vallon, L.; Bourgeois, C.; Bruto, M.; Marchand, C.; Record, E.; Hugoni, M. Contrasted ecological niches shape fungal and prokaryotic community structure in mangroves sediments. Environ. Microbiol. 2019, 21, 1407–1424. [Google Scholar] [CrossRef] [PubMed]
- Latha, R.; Mitra, S. Mangrove fungi in India. Curr. Sci. 2004, 86, 1586. [Google Scholar]
- Kida, M.; Fujitake, N. Organic Carbon Stabilization Mechanisms in Mangrove Soils: A Review. Forests 2020, 11, 981. [Google Scholar] [CrossRef]
- Chen, G.; Azkab, M.H.; Chmura, G.L.; Chen, S.; Sastrosuwondo, P.; Ma, Z.; IWayan, E.D.; Xijie, Y.; Chen, B. Mangroves as a major source of soil carbon storage in adjacent seagrass meadows. Sci. Rep. 2017, 7, 42406. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Lee, S.Y. Ecology of mangrove fungi and their role in nutrient cycling—What gaps occur in our knowledge. Hydrobiologia 1995, 295, 107–118. [Google Scholar] [CrossRef]
- López-Mondéjar, R.; Zühlke, D.; Becher, D.; Riedel, K.; Baldrian, P. Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci. Rep. 2016, 6, 25279. [Google Scholar] [CrossRef]
- Datta, R.; Kelkar, A.; Baraniya, D.; Molaei, A.; Moulick, A.; Meena, R.S.; Formanek, P. Enzymatic degradation of lignin in soil: A review. Sustainability 2017, 9, 1163. [Google Scholar] [CrossRef] [Green Version]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 2013, 6, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Arfi, Y.; Chevret, D.; Henrissat, B.; Berrin, J.G.; Levasseur, A.; Record, E. Characterization of salt-adapted secreted lignocellulolytic enzymes from the mangrove fungus Pestalotiopsis sp. Nat. Commun. 2013, 4, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Ben Ali, W.; Navarro, D.; Kumar, A.; Drula, E.; Turbé-Doan, A.; Correia, L.O.; Baumberger, S.; Bertrand, E.; Faulds, C.B.; Henrissat, B.; et al. Characterization of the CAZy Repertoire from the Marine-Derived Fungus Stemphylium lucomagnoense in Relation to Saline Conditions. Mar. Drugs 2020, 18, 461. [Google Scholar] [CrossRef]
- Bucher, V.V.C.; Pointing, S.B.; Hyde, K.D.; Reddy, C.A. Production of Wood Decay Enzymes, Loss of Mass, and Lignin Solubilization in Wood by Diverse Tropical Freshwater Fungi. Microb. Ecol. 2004, 48, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Ben Ali, W.; Ayed, A.B.; Turbé-Doan, A.; Bertrand, E.; Mathieu, Y.; Faulds, C.B.; Lomascolo, A.; Sciara, G.; Record, E.; Mechichi, T. Enzyme Properties of a Laccase Obtained from the Transcriptome of the Marine-Derived Fungus Stemphylium lucomagnoense. Int. J. Mol. Sci. 2020, 21, 8402. [Google Scholar] [CrossRef]
- Kamei, I.; Daikoku, C.; Tsutsumi, Y.; Kondo, R. Saline-dependent regulation of manganese peroxidase genes in the hypersaline-tolerant white rot fungus Phlebia sp. strain MG-60. Appl. Environ. Microb. 2008, 74, 2709–2716. [Google Scholar] [CrossRef] [Green Version]
- Zámocký, M.; Hofbauer, S.; Schaffner, I.; Gasselhuber, B.; Nicolussi, A.; Soudi, M.; Pirker, K.F.; Furtmüller, P.G.; Obinger, C. Independent evolution of four heme peroxidase superfamilies. Arch. Biochem. Biophys. 2015, 574, 108–119. [Google Scholar] [CrossRef]
- Acharya, G.; Kaur, G.; Subramanian, S. Evolutionary relationships between heme-binding ferredoxin α + β barrels. BMC Bioinform. 2016, 17, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celis, A.I.; DuBois, J.L. Substrate, product, and cofactor: The extraordinarily flexible relationship between the CDE superfamily and heme. Arch. Biochem. Biophys. 2015, 574, 3–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, T.; Sugano, Y. A structural and functional perspective of DyP-type peroxidase family. Arch. Biochem. Biophys. 2015, 574, 49–55. [Google Scholar] [CrossRef]
- Linde, D.; Pogni, R.; Cañellas, M.; Lucas, F.; Guallar, V.; Baratto, M.C.; Sinicropi, A.; Sáez-Jiménez, V.; Coscolín, C.; Romero, A.; et al. Catalytic surface radical in dye-decolorizing peroxidase: A computational, spectroscopic and directed mutagenesis study. Biochem. J. 2015, 466, 253–262. [Google Scholar] [CrossRef]
- Sciara, G.; Kendrew, S.G.; Miele, A.E.; Marsh, N.G.; Federici, L.; Malatesta, F.; Giuliana, S.; Carmelinda, S.; Vallone, B. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. EMBO J. 2003, 22, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Thompson, T.B.; Katayama, K.; Watanabe, K.; Hutchinson, C.R.; Rayment, I. Structural and Functional Analysis of Tetracenomycin F2 Cyclase fromStreptomyces glaucescens. J. Biol. Chem. 2004, 279, 37956–37963. [Google Scholar] [CrossRef] [Green Version]
- Goblirsch, B.; Kurker, R.C.; Streit, B.R.; Wilmot, C.M.; DuBois, J.L. Chlorite dismutases, DyPs, and EfeB: 3 microbial heme enzyme families comprise the CDE structural superfamily. J. Mol. Biol. 2011, 408, 379–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugano, Y. DyP-type peroxidases comprise a novel heme peroxidase family. Cell Mol. Life Sci. 2009, 66, 1387–1403. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, K.; Nishihashi, Y.; Narioka, T.; Yoshida, T.; Morita, M.; Sugano, Y. Characterization of a novel DyP-type peroxidase from Streptomyces avermitilis. J. Biosci. Bioeng. 2017, 123, 425–430. [Google Scholar] [CrossRef]
- Gomi, N.; Yoshida, S.; Matsumoto, K.; Okudomi, M.; Konno, H.; Hisabori, T.; Sugano, Y. Degradation of the synthetic dye amaranth by the fungus Bjerkandera adusta Dec 1: Inference of the degradation pathway from an analysis of decolorized products. Biodegradation 2011, 22, 1239–1245. [Google Scholar] [CrossRef]
- Faraco, V.; Piscitelli, A.; Sannia, G.; Giardina, P. Identification of a new member of the dye-decolorizing peroxidase family from Pleurotus ostreatus. World J. Microbiol. Biotechnol. 2007, 23, 889–893. [Google Scholar] [CrossRef]
- Liers, C.; Bobeth, C.; Pecyna, M.; Ullrich, R.; Hofrichter, M. DyP-like peroxidases of the jelly fungus Auricularia auricula-judae oxidize nonphenolic lignin model compounds and high-redox potential dyes. Appl. Microbiol. Biotechnol. 2010, 85, 1869–1879. [Google Scholar] [CrossRef]
- Linde, D.; Coscolin, C.; Liers, C.; Hofrichter, M.; Martínez, A.T.; Ruiz-Dueñas, F.J. Heterologous expression and physicochemical characterization of a fungal dye-decolorizing peroxidase from Auricularia auricula-judae. Protein Expr. Purif. 2014, 103, 28–37. [Google Scholar] [CrossRef]
- Amara, S.; Perrot, T.; Navarro, D.; Deroy, A.; Benkhelfallah, A.; Chalak, A.; Daou, M.; Chevret, D.; Faulds, C.B.; Berrin, J.G.; et al. Enzyme activities of two recombinant heme-containing peroxidases, TvDyP and TvVP2, identified from the secretome of Trametes versicolor. Appl. Environ. Microbiol. 2018, 84, e02826-17. [Google Scholar] [CrossRef] [Green Version]
- Hofrichter, M.; Ullrich, R.; Pecyna, M.J.; Liers, C.; Lundell, T. New and classic families of secreted fungal heme peroxidases. Appl. Microbiol. Biotechnol. 2010, 87, 871–897. [Google Scholar] [CrossRef]
- Brenelli, L.B.; Persinoti, G.F.; Cairo, J.P.L.F.; Liberato, M.V.; Gonçalves, T.A.; Otero, I.V.R.; Mainardi, P.H.; Felby, C.; Sette, L.D.; Squina, F.M. Novel redox-active enzymes for ligninolytic applications revealed from multiomics analyses of Peniophora sp. CBMAI 1063, a laccase hyper-producer strain. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Wikee, S.; Hatton, J.; Turbé-Doan, A.; Mathieu, Y.; Daou, M.; Lomascolo, A.; Kumar, A.; Lymyong, A.; Sciara, G.; Faulds, C.B.; et al. Characterization and Dye Decolorization Potential of Two Laccases from the Marine-Derived Fungus Pestalotiopsis sp. Int. J. Mol. Sci. 2019, 20, 1864. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.Y.; Xue, D.S.; Feng, X.Y.; Yao, S.J. Screening and production of ligninolytic enzyme by a marine-derived fungal Pestalotiopsis sp. J63. Appl. Biochem. Biotechnol. 2011, 165, 1754–1769. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, Z.; Lin, Y.; Ng, T.B.; Ye, X.; Lin, J. Immobilized Cerrena sp. laccase: Preparation, thermal inactivation, and operational stability in malachite green decolorization. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-a sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D. The Condensed Protocols from Molecular Cloning: A Laboratory Manual, 1st ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2006. [Google Scholar]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef]
- Savelli, B.; Li, Q.; Webber, M.; Jemmat, A.; Robitaille, A.; Zamocky, M.; Mathé, C.; Dunand, C. RedoxiBase: A database for ROS homeostasis regulated proteins. Redox Biol. 2019, 26, 101247. [Google Scholar] [CrossRef] [PubMed]
- Parisot, N.; Denonfoux, J.; Dugat-Bony, E.; Peyret, P.; Peyretaillade, E. KASpOD-a web service for highly specific and explorative oligonucleotide design. Bioinformatics 2012, 28, 3161–3162. [Google Scholar] [CrossRef] [PubMed]
- Bragalini, C.; Ribière, C.; Parisot, N.; Vallon, L.; Prudent, E.; Peyretaillade, E.; Girlanda, M.; Peyret, P.; Marmeisse, R.; Luis, P. Solution hybrid selection capture for the recovery of functional full-length eukaryotic cDNAs from complex environmental samples. DNA Res. 2014, 21, 685–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kellner, H.; Luis, P.; Pecyna, M.J.; Barbi, F.; Kapturska, D.; Krüger, D.; Zak, D.R.; Marmeisse, R.; Vandenbol, M.; Hofrichter, M. Widespread occurrence of expressed fungal secretory peroxidases in forest soils. PLoS ONE 2014, 9, e95557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahé, F.; Rognes, T.; Quince, C.; De Vargas, C. Swarm: Robust and fast clustering method for amplicon-based studies. Peer J. 2014, 2, e593. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open-source tool for metagenomics. Peer J. 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Escudie, F.; Auer, L.; Bernard, M.; Mariadassou, M.; Hernadez-Raquet, M.; Pascal, G. FROGS: Find, rapidly, OTUs with Galaxy solution. Bioinformatics 2018, 34, 1287–1294. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Leung, H.C.; Yiu, S.M.; Chin, F.Y. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- De Castro, E.; Sigrist, C.J.A.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef]
- Sigrist, C.J.A.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Fueyo, E.; Linde, D.; Almendral, D.; Lopez-Lucendo, M.F.; Ruiz Dueñas, F.J.; Martínez, A.T. Description of the first fungal dye-decolorizing peroxidase oxidizing manganese (II). Appl. Microbiol. Biotechnol. 2015, 99, 8927–8942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolwek, J.; Behrens, C.; Linke, D.; Krings, U.; Berger, R.G. Cell-free one-pot conversion of (+) -valencene to (+) -nootkatone by a unique dye-decolorizing peroxidase combined with a laccase from Funalia trogii. J. Ind. Microbiol. Biotechnol. 2018, 45, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Kung, C.P.; Wu, Y.R.; Chuang, H.W. Expression of a dye-decolorizing peroxidase results in hypersensitive response to cadmium stress through reducing the ROS signal in Arabidopsis. Environ. Exp. Bot. 2014, 101, 47. [Google Scholar] [CrossRef]
- Linde, D.; Ruiz-Dueñas, F.J.; Fernández-Fueyo, E.; Guallar, V.; Hammel, K.E.; Pogni, R.; Martínez, A.T. Basidiomycete DyPs: Genomic diversity, structural-functional aspects, reaction mechanism and environmental significance. Arch. Biochem. Biophys. 2015, 574, 66–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johjima, T.; Ohkuma, M.; Kudo, T. Isolation and cDNA cloning of novel hydrogen peroxide-dependent phenol oxidase from the basidiomycete Termitomyces albuminosus. Appl. Microbiol. Biotechnol. 2003, 61, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Scheibner, M.; Hulsdau, B.; Zelena, K.; Nimtz, M.; de Boer, L.; Berger, R.G.; Zorn, H. Novel peroxidases of Marasmius scorodonius degrade b-carotene. Appl. Microbiol. Biotechnol. 2008, 77, 1241–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelena, K.; Zorn, H.; Nimtz, M.; Berger, R.G. Heterologous expression of the msp2 gene from Marasmius scorodonius. Arch. Microbiol. 2009, 191, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Salvachúa, D.; Prieto, A.; Martínez, Á.T.; Martínez, M.J. Characterization of a novel dye-decolorizing peroxidase (DyP)-type enzyme from Irpex lacteus and its application in enzymatic hydrolysis of wheat straw. Appl. Environ. Microbiol. 2013, 79, 4316–4324. [Google Scholar] [CrossRef] [Green Version]
- Lauber, C.; Schwarz, T.; Quoc, K.N.; Lorenz, P.; Lochnit, G.; Zorn, H. Identification, heterologous expression and characterization of a dye-decolorizing peroxidase of Pleurotus sapidus. AMB Express 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colpa, D.I.; Fraaije, M.W.; van Bloois, E. DyP-type peroxidases: A promising and versatile class of enzymes. J. Ind. Microb. Biotechnol. 2014, 41, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sáez-Jiménez, V.; Baratto, M.C.; Pogni, R.; Rencoret, J.; Gutiérrez, A.; Santos, J.I.; Martínez, A.T.; Ruiz-Dueñas, F.J. Demonstration of lignin-to-peroxidase direct electron transfer: A transient-state kinetics, directed mutagenesis, EPR and NMR study. J. Biol. Chem. 2015, 290, 23201–23213. [Google Scholar] [CrossRef] [Green Version]
- Zerva, A.; Christakopoulos, P.; Topakas, E. Characterization and application of a novel class II thermophilic peroxidase from Myceliophthora thermophila in biosynthesis of polycatechol. Enzym. Microb. Technol. 2015, 75, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Chalak, A.; Villares, A.; Moreau, C.; Haon, M.; Grisel, S.; d’Orlando, A.; Herpoël-Gimbert, I.; Labourel, A.; Cathala, B.; Berrin, J.G. Influence of the carbohydrate-binding module on the activity of a fungal AA9 lytic polysaccharide monooxygenase on cellulosic substrates. Biotechnol. Biofuels 2019, 12, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valderrama, B.; Ayala, M.; Vazquez-Duhalt, R. Suicide inactivation of peroxidases and the challenge of engineering more robust enzymes. Chem Biol. 2002, 9, 555–565. [Google Scholar] [CrossRef] [Green Version]
- Bouillon, S.; Borges, A.; Castañeda-Moya, E.; Diele, K.; Dittmar, T.; saraDuke, N.; Kristensen, E.; Lee, S.; Marchand, C.; Middelburg, J.; et al. Mangrove Production and Carbon sinks: A revision of global budget estimates. Glob. Biogeochem. Cycles 2008, 22, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Saravanakumar, K.; Anburaj, R.; Gomathi, V.; Kandasamy, K. Ecology of soil microbes in a tropical mangrove forest of south east coast of India. Biocatal Agric. Biotechnol. 2016, 8, 73–85. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The Carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [Green Version]
- Barbi, F.; Prudent, E.; Vallon, L.; Buée, M.; Dubost, A.; Legout, A.; Marneisse, R.; Fraissinet-Tachet, L.; Luis, P. Tree species select diverse soil fungal communities expressing different sets of lignocellulolytic enzyme-encoding genes. Soil Biol. Biochem. 2016, 100, 149–159. [Google Scholar] [CrossRef]
- Deborde, J.; Marchand, C.; Molnar, N.; Patrona, L.D.; Meziane, T. Concentrations and fractionation of carbon, iron, sulfur, nitrogen and phosphorus in mangrove sediments along an intertidal gradient (semi-arid climate, New Caledonia). J. Mar. Sci. Eng. 2015, 3, 52–72. [Google Scholar] [CrossRef]
- Molnar, N.; Marchand, C.; Deborde, J.; Patrona, L.C.; Meziane, T. Seasonal pattern of the biogeochemical properties of mangrove sediments receiving shrimp farm effluents (New Caledonia). J. Aquacult Res. Dev. 2014, 05, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Leopold, A.; Marchand, C.; Renchon, A.; Deborde, J.; Quiniou, T.; Allenbach, M. Net ecosystem CO2 exchange in the “Coeur de Voh” mangrove, New Caledonia: Effects of water stress on mangrove productivity in a semi-arid climate. Agric. For. Meteorol. 2016, 223, 217–232. [Google Scholar] [CrossRef]
- Alongi, D.M.; Clough, B.F.; Robertson, A.I. Nutrientuse efficiency in arid-zone forests of the mangroves Rhizophora stylosa and Avicennia marina. Aquat. Bot. 2005, 82, 121–131. [Google Scholar] [CrossRef]
- Thiem, D.; Gołębiewski, M.; Hulisz, P.; Piernik, A.; Hrynkiewicz, K. How does salinity shape bacterial and fungal microbiomes of Alnus glutinosa roots? Front. Microbiol. 2018, 9, 651. [Google Scholar] [CrossRef]
- Ruiz-Dueñas, F.J.; Pogni, R.; Morales, M.; Giansanti, S.; Mate, M.J.; Romero, A.; Martínez, M.J.; Basosi, R.; Martínez, A.T. Protein radicals in fungal versatile peroxidase: Catalytic tryptophan radical in both Compound I and Compound II and studies on W164Y, W164H and W164S variants. J. Biol. Chem. 2009, 284, 7986–7994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.T.; Doyle, W.A.; Dorlet, P.; Ivancich, A. Spectroscopic evidence for an engineered, catalytically active Trp radical that creates the unique reactivity of lignin peroxidase. Proc. Nat. Acad. Sci. USA 2009, 106, 16084–16089. [Google Scholar] [CrossRef] [Green Version]
- Miki, Y.; Calviño, F.R.; Pogni, R.; Giansanti, S.; Ruiz-Dueñas, F.J.; Martínez, M.J.; Basosi, R.; Romero, A.; Martínez, A.T. Crystallographic, kinetic, and spectroscopic study of the first ligninolytic peroxidase presenting a catalytic tyrosine. J. Biol. Chem. 2011, 286, 15525–15534. [Google Scholar] [CrossRef] [Green Version]
- Adnan, L.A.; Yusoff, A.R.M.; Hadibarata, T.; Khudhair, A.B. Biodegradation of bis-azo dye Reactive Black 5 by white-rot fungus Trametes gibbosa sp. WRF 3 and its metabolite characterization. Water Air Soil Pollut. 2014, 225, 2119. [Google Scholar] [CrossRef]
- Hadibarata, T.; Adnan, L.A.; Yusoff, A.R.M.; Yuniarto, A.; Zubir, M.M.F.A.; Khudhair, A.B.; Teh, Z.C.; Naser, M.A. Microbial decolorization of an azo dye reactive black 5 using white-rot fungus Pleurotus eryngii F032. Water Air Soil Pollut. 2013, 224, 1595. [Google Scholar] [CrossRef]
- Kim, S.J.; Ishikawa, K.; Hirai, M.; Shoda, M. Characteristics of a newly isolated fungus, Geotrichum candidum Dec 1, which decolorizes various dyes. J. Ferment Bioeng. 1995, 79, 601–607. [Google Scholar] [CrossRef]
- Hanapi, S.Z.; Abdelgalil, S.A.; Hatti-Kaul, R.; Aziz, R.; El Enshasy, H.A. Isolation of a new efficient dye decolorizing white rot fungus Cerrena Sp. WICC F39. J. Sci. Ind. Res. 2018, 77, 399–404. [Google Scholar]
- Park, C.; Lim, J.S.; Lee, Y.; Lee, B.; Kim, S.W.; Lee, J.; Kim, S. Optimization and morphology for decolorization of reactive black 5 by Funalia trogii. Enzym. Microb. Technol. 2007, 40, 1758–1764. [Google Scholar] [CrossRef]
- Husain, Q. Immobilized Peroxidase Catalyzed Decolorization and Degradation of Industrially Important Dyes from Polluted Water Salt-responsive lytic polysaccharide monooxygenases from the mangrove fungus Pestalotiopsis sp. NCi6. Biocatal. Springer Cham. 2019, 139–166. [Google Scholar] [CrossRef]
- Patel, I.; Kracher, D.; Ma, S.; Garajova, S.; Haon, M.; Faulds, C.B.; Berrin, J.G.; Ludwig, R.; Record, E. Salt-responsive lytic polysaccharide monooxygenases from the mangrove fungus Pestalotiopsis sp. NCi6. Biotechnol. Biofuels 2016, 9, 108. [Google Scholar] [CrossRef]
- Kern, M.; McGeehan, J.E.; Streeter, S.D.; Martin, R.N.A.; Besser, K.; Elias, L.; Eborall, W.; Malyon, G.P.; Payne, C.M.; Himmel, M.E.; et al. Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance. Proc. Nat. Acad Sci. USA 2013, 110, 10189–10194. [Google Scholar] [CrossRef] [Green Version]
- Paul, S.; Bag, S.K.; Das, S.; Harvill, E.T.; Dutta, C. Molecular signature of hypersaline adaptation: Insights from genome and proteome composition of halophilic prokaryotes. Genome Biol. 2008, 9, R70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanyi, J.K. Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol. Rev. 1974, 38, 272–290. [Google Scholar] [CrossRef] [PubMed]





| Df | F | P | R2 | |
|---|---|---|---|---|
| Variable | ||||
| Tree species | 1 | 3.011 | 0.032 | 0.062 |
| Sediment depth | 1 | 3.453 | 0.021 | 0.072 |
| Season | 1 | 8.579 | 5.8 × 10−5 | 0.178 |
| Interaction | ||||
| Tree × Depth | 1 | 6.789 | 7.1 × 10−4 | 0.141 |
| Tree × Season | 1 | 2.462 | 0.062 | 0.051 |
| Depth ×Season | 1 | 2.926 | 0.036 | 0.061 |
| Tree × Depth × Season | 1 | 4.946 | 0.005 | 0.102 |
| Residuals | 16 |
| DyP1 | |
|---|---|
| E. glandulosa DyP A0A165BX62 | 59% (459) |
| P. indica DyP G4TL25 | 57% (460) |
| E. glandulosa DyP A0A165FCE7 | 56% (460) |
| E. glandulosa DyP A0A165G2C1 | 54% (498) |
| E. glandulosa DyP A0A165GZG2 | 54% (469) |
| S. stellatus DyP A0A0C9U2H4 | 54% (465) |
| S. vermifera DyP A0A0C3B0S6 | 53% (460) |
| E. glandulosa DyP A0A166ARP7 | 52% (505) |
| A. auricula-judae DyPI2DBY1 | 51% (505) |
| S. stellatus DyP A0A0C9UT91 | 51% (505) |
| S. stellatus DyP A0A0C9VF44 | 49% (472) |
| S. stellatus DyP A0A0C9VPJ8 | 47% (506) |
| Purification Step | Volume (mL) | Total Activity (U mL−1) | Protein (mg) | Specific Activity (U mg−1) | Yield (%) | Purification (Fold) |
|---|---|---|---|---|---|---|
| Culture medium | 500 | 376 | 4682 | 0.08 | 100 | 1 |
| IMAC | 25 | 164 | 63.3 | 2.59 | 43.6 | 32.4 |
| Substrate | Parameters | |||
|---|---|---|---|---|
| Km (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) | pH | |
| ABTS | 0.651 +/− 0.081 | 0.322 | 0.49 | 3 |
| RB19 | 1.497 +/− 0.878 | 3.34 | 2.23 | 2.6 |
| DMP | 0 | 0 | 0 | 2.6–7 |
| Mn2+ | 0 | 0 | 0 | 2.6–6 |
| VA | 0 | 0 | 0 | 2.6–7 |
| Dye | DyP1 | TvDyP1 |
|---|---|---|
| AB | 18.8 +/− 0.008 | 75.0 +/− 0.007 |
| BB | − | − |
| RB5 | 32.3 +/− 0.009 | − |
| DB79 | 5.2 +/− 0.005 | − |
| VG | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Ayed, A.; Saint-Genis, G.; Vallon, L.; Linde, D.; Turbé-Doan, A.; Haon, M.; Daou, M.; Bertrand, E.; Faulds, C.B.; Sciara, G.; et al. Exploring the Diversity of Fungal DyPs in Mangrove Soils to Produce and Characterize Novel Biocatalysts. J. Fungi 2021, 7, 321. https://doi.org/10.3390/jof7050321
Ben Ayed A, Saint-Genis G, Vallon L, Linde D, Turbé-Doan A, Haon M, Daou M, Bertrand E, Faulds CB, Sciara G, et al. Exploring the Diversity of Fungal DyPs in Mangrove Soils to Produce and Characterize Novel Biocatalysts. Journal of Fungi. 2021; 7(5):321. https://doi.org/10.3390/jof7050321
Chicago/Turabian StyleBen Ayed, Amal, Geoffroy Saint-Genis, Laurent Vallon, Dolores Linde, Annick Turbé-Doan, Mireille Haon, Marianne Daou, Emmanuel Bertrand, Craig B. Faulds, Giuliano Sciara, and et al. 2021. "Exploring the Diversity of Fungal DyPs in Mangrove Soils to Produce and Characterize Novel Biocatalysts" Journal of Fungi 7, no. 5: 321. https://doi.org/10.3390/jof7050321
APA StyleBen Ayed, A., Saint-Genis, G., Vallon, L., Linde, D., Turbé-Doan, A., Haon, M., Daou, M., Bertrand, E., Faulds, C. B., Sciara, G., Adamo, M., Marmeisse, R., Comtet-Marre, S., Peyret, P., Abrouk, D., Ruiz-Dueñas, F. J., Marchand, C., Hugoni, M., Luis, P., ... Record, E. (2021). Exploring the Diversity of Fungal DyPs in Mangrove Soils to Produce and Characterize Novel Biocatalysts. Journal of Fungi, 7(5), 321. https://doi.org/10.3390/jof7050321









