Transcriptional Analysis of Coccidioides immitis Mycelia and Spherules by RNA Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture Conditions
2.2. RNA Extraction, Purification and Sequencing
2.3. Analysis of RNA-Seq Data
2.4. Functional Analysis of Genes
3. Results and Discussion
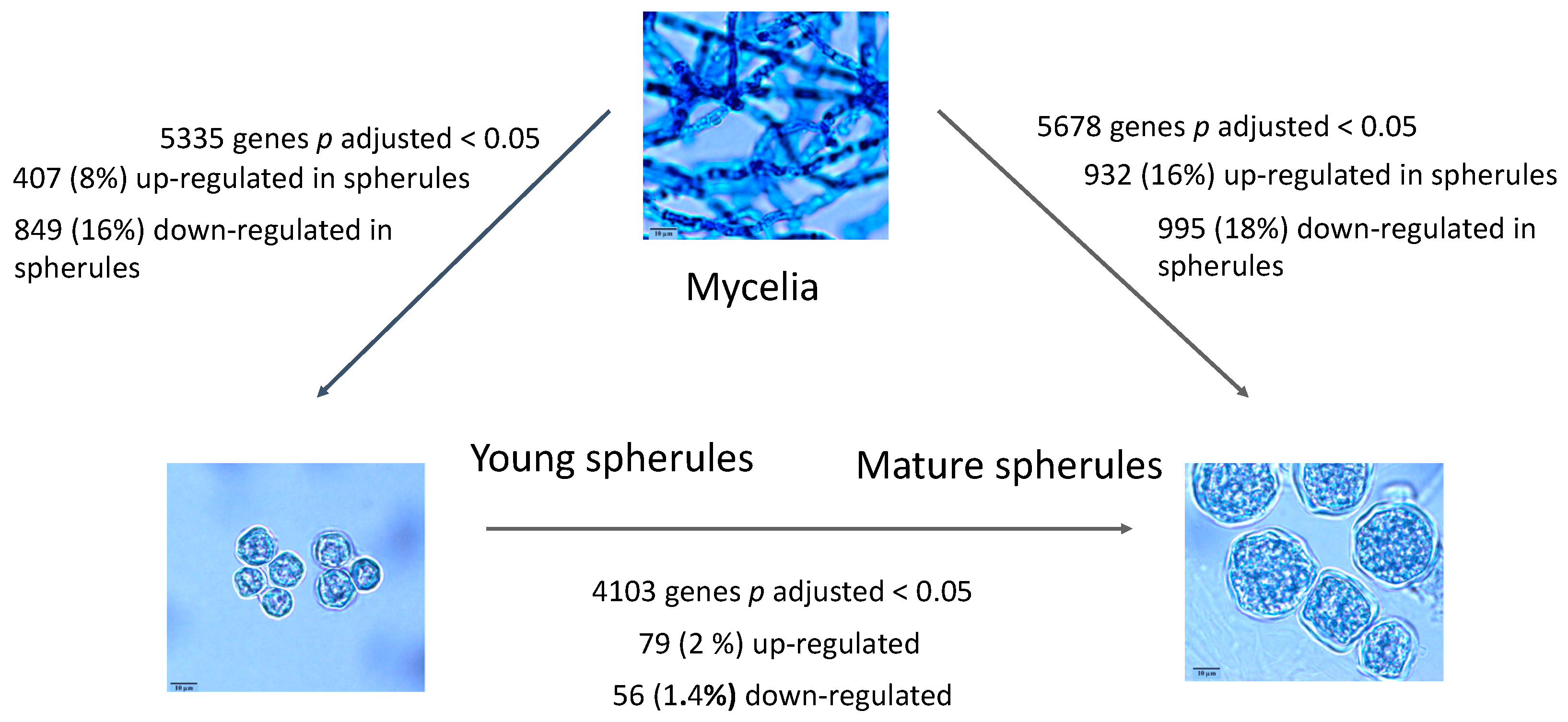
3.1. Differential Gene Expression in Young Spherules vs. Mycelia
3.2. Comparison to of Differential Gene Expression in Young Spherules to Previous Studies in Coccidioides Immitis
3.3. Differential Gene Expression in Mature Spherules vs. Mycelia
3.4. Mature Spherules Compared to Young Spherules
3.5. Expression of Transposable Elements in Young Spherules Compared to Mycelia
4. Summary
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Engelthaler, D.M.; Roe, C.C.; Hepp, C.M.; Teixeira, M.; Driebe, E.M.; Schupp, J.M.; Gade, L.; Waddell, V.; Komatsu, K.; Arathoon, E.; et al. Local Population Structure and Patterns of Western Hemisphere Dispersal for Coccidioides spp., the Fungal Cause of Valley Fever. MBio 2016, 7, e00550-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkland, T.N.; Fierer, J. Coccidioides immitis and posadasii; A review of their biology, genomics, pathogenesis, and host immunity. Virulence 2018, 9, 1426–1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, C.; Barker, B.M.; Hoover, S.; Nix, D.E.; Ampel, N.M.; Frelinger, J.A.; Orbach, M.J.; Galgiani, J.N. Recent advances in our understanding of the environmental, epidemiological, immunological, and clinical dimensions of coccidioidomycosis. Clin. Microbiol. Rev. 2013, 26, 505–525. [Google Scholar] [CrossRef] [Green Version]
- Whiston, E.; Taylor, J.W. Genomics in Coccidioides: Insights into evolution, ecology, and pathogenesis. Med. Mycol. 2014, 52, 149–155. [Google Scholar] [CrossRef]
- Converse, J.L. Effect of physico-chemical environment of spherulation of Coccidioides immitis in a chemically defined medium. J. Bacteriol. 1956, 72, 784–792. [Google Scholar] [CrossRef] [Green Version]
- Mead, H.L.; Van Dyke, M.C.C.; Barker, B.M. Proper Care and Feeding of Coccidioides: A Laboratorian’s Guide to Cultivating the Dimorphic Stages of C. immitis and C. posadasii. Curr. Protoc. Microbiol. 2020, 58, e113. [Google Scholar] [CrossRef]
- Whiston, E.; Zhang Wise, H.; Sharpton, T.J.; Jui, G.; Cole, G.T.; Taylor, J.W. Comparative transcriptomics of the saprobic and parasitic growth phases in Coccidioides spp. PLoS ONE 2012, 7, e41034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, G.T.; Kirkland, T.; Franco, M.; Zhu, S.W.; Yuan, L.; Sun, S.H.; Hearn, V.N. Immunoreactivity of a surface wall fraction produced by spherules of Coccidioides immitis. Infect. Immun. 1988, 56, 2695–2701. [Google Scholar] [CrossRef] [Green Version]
- Hung, C.Y.; Yu, J.; Seshan, K.R.; Reichard, U.; Cole, G.T. A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory fungal pathogen. Infect. Immun. 2002, 70, 3443–3456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viriyakosol, S.; Singhania, A.; Fierer, J.; Goldberg, J.; Kirkland, T.N.; Woelk, C.H. Gene expression in human fungal pathogen Coccidioides immitis changes as arthroconidia differentiate into spherules and mature. BMC Microbiol. 2013, 13, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Basenko, E.Y.; Pulman, J.A.; Shanmugasundram, A.; Harb, O.S.; Crouch, K.; Starns, D.; Warrenfeltz, S.; Aurrecoechea, C.; Stoeckert, C.J.; Kissinger, J.C.; et al. FungiDB: An Integrated Bioinformatic Resource for Fungi and Oomycetes. J. Fungi 2018, 4, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Priebe, S.; Kreisel, C.; Horn, F.; Guthke, R.; Linde, J. FungiFun2: A comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics 2015, 31, 445–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q.; et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief Bioinform. 2019, 20, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [Green Version]
- Ivey, F.D.; Magee, D.M.; Woitaske, M.D.; Johnston, S.A.; Cox, R.A. Identification of a protective antigen of Coccidioides immitis by expression library immunization. Vaccine 2003, 21, 4359–4367. [Google Scholar] [CrossRef]
- Delgado, N.; Hung, C.Y.; Tarcha, E.; Gardner, M.J.; Cole, G.T. Profiling gene expression in Coccidioides posadasii. Med. Mycol. 2004, 42. [Google Scholar] [CrossRef]
- Petito, G.; de Curcio, J.S.; Pereira, M.; Bailão, A.M.; Paccez, J.D.; Tristão, G.B.; de Morais, C.O.B.; de Souza, M.V.; de Castro Moreira Santos, A.; Fontes, W.; et al. Metabolic Adaptation of Paracoccidioides brasiliensis in Response to in vitro Copper Deprivation. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Mead, H.L.; Roe, C.C.; Higgins Keppler, E.A.; Van Dyke, M.C.C.; Laux, K.L.; Funke, A.L.; Miller, K.J.; Bean, H.D.; Sahl, J.W.; Barker, B.M. Defining Critical Genes During Spherule Remodeling and Endospore Development in the Fungal Pathogen, Coccidioides posadasii. Front. Genet. 2020, 11. [Google Scholar] [CrossRef]
- Youseff, B.H.; Holbrook, E.D.; Smolnycki, K.A.; Rappleye, C.A. Extracellular superoxide dismutase protects Histoplasma yeast cells from host-derived oxidative stress. PLoS Pathog. 2012, 8, e1002713. [Google Scholar] [CrossRef] [Green Version]
- Hwang, L.H.; Seth, E.; Gilmore, S.A.; Sil, A. SRE1 regulates iron-dependent and -independent pathways in the fungal pathogen Histoplasma capsulatum. Eukaryot. Cell 2012, 11, 16–25. [Google Scholar] [CrossRef] [Green Version]
- Gauthier, G.M.; Sullivan, T.D.; Gallardo, S.S.; Brandhorst, T.T.; Vanden Wymelenberg, A.J.; Cuomo, C.A.; Suen, G.; Currie, C.R.; Klein, B.S. SREB, a GATA transcription factor that directs disparate fates in Blastomyces dermatitidis including morphogenesis and siderophore biosynthesis. PLoS Pathog. 2010, 6, e1000846. [Google Scholar] [CrossRef]
- Mirbod-Donovan, F.; Schaller, R.; Hung, C.Y.; Xue, J.; Reichard, U.; Cole, G.T. Urease produced by Coccidioides posadasii contributes to the virulence of this respiratory pathogen. Infect. Immun. 2006, 74, 504–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wise, H.Z.; Hung, C.-Y.; Whiston, E.; Taylor, J.W.; Cole, G.T. Extracellular ammonia at sites of pulmonary infection with Coccidioides posadasii contributes to severity of the respiratory disease. Microb. Pathog. 2013, 59–60, 19–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkland, T.N. A few shared up-regulated genes may influence conidia to yeast transformation in dimorphic fungal pathogens. Med. Mycol. 2016, 54, 648–653. [Google Scholar] [CrossRef] [Green Version]
- Boyce, K.J.; McLauchlan, A.; Schreider, L.; Andrianopoulos, A. Intracellular growth is dependent on tyrosine catabolism in the dimorphic fungal pathogen Penicillium marneffei. PLoS Pathog. 2015, 11, e1004790. [Google Scholar] [CrossRef] [Green Version]
- Felipe, M.S.S.; Andrade, R.V.; Arraes, F.B.M.; Nicola, A.M.; Maranhão, A.Q.; Torres, F.A.G.; Silva-Pereira, I.; Poças-Fonseca, M.J.; Campos, E.G.; Moraes, L.M.P.; et al. Transcriptional profiles of the human pathogenic fungus Paracoccidioides brasiliensis in mycelium and yeast cells. J. Biol. Chem. 2005, 280, 24706–24714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longo, L.V.G.; Ray, S.C.; Puccia, R.; Rappleye, C.A. Characterization of the APSES-family transcriptional regulators of Histoplasma capsulatum. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrillo, A.J.; Schacht, P.; Cabrera, I.E.; Blahut, J.; Prudhomme, L.; Dietrich, S.; Bekman, T.; Mei, J.; Carrera, C.; Chen, V.; et al. Functional Profiling of Transcription Factor Genes in Neurospora crassa. G3 2017, 7, 2945–2956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webster, R.H.; Sil, A. Conserved factors Ryp2 and Ryp3 control cell morphology and infectious spore formation in the fungal pathogen Histoplasma capsulatum. Proc. Natl. Acad. Sci. USA 2008, 105, 14573–14578. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, L.; Voorhies, M.; Gilmore, S.; Beyhan, S.; Myint, A.; Sil, A. Opposing signaling pathways regulate morphology in response to temperature in the fungal pathogen Histoplasma capsulatum. PLoS Biol. 2019, 17, e3000168. [Google Scholar] [CrossRef] [Green Version]
- Bankapalli, K.; Saladi, S.; Awadia, S.S.; Goswami, A.V.; Samaddar, M.; D’Silva, P. Robust glyoxalase activity of Hsp31, a ThiJ/DJ-1/PfpI family member protein, is critical for oxidative stress resistance in Saccharomyces cerevisiae. J. Biol. Chem. 2015, 290, 26491–26507. [Google Scholar] [CrossRef] [Green Version]
- de Vries, R.P.; Riley, R.; Wiebenga, A.; Aguilar-Osorio, G.; Amillis, S.; Uchima, C.A.; Anderluh, G.; Asadollahi, M.; Askin, M.; Barry, K.; et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, G.T.; Pope, L.M.; Huppert, M.; Sun, S.H.; Starr, P. Ultrastructure and composition of conidial wall fractions of Coccidioides immitis. Exp. Mycol. 1983, 7, 297–318. [Google Scholar] [CrossRef]
- Xue, J.; Chen, X.; Selby, D.; Hung, C.Y.; Yu, J.J.; Cole, G.T. A genetically engineered live attenuated vaccine of Coccidioides posadasii protects BALB/c mice against coccidioidomycosis. Infect. Immun. 2009, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Gene ID | Product Description | Ortholog Count | Length | # TM Domains | SignalP | FC (log2 Values) | Genus-Specific |
|---|---|---|---|---|---|---|---|
| Upregulated | |||||||
| CIMG_04613 | Hypothetical protein—SOWgp | 3 | 324 | 0 | Y | 9.012 | Y |
| CIMG_10264 | Aldehyde reductase | 1176 | 314 | 0 | N | 8.116 | N |
| CIMG_09753 | ABC multidrug transporter | 569 | 1501 | 16 | N | 7.773 | N |
| CIMG_10032 | Expression library immunization antigen 1-surface protein antigen | 96 | 224 | 0 | Y | 7.092 | N |
| CIMG_10037 | Copper transporter | 121 | 193 | 2 | N | 7.072 | N |
| CIMG_01115 | Thiamine thiazole synthase, thiamine thiazole synthase, variant | 170 | 328 | 0 | N | 6.901 | N |
| CIMG_08103 | Opsin 1 | 214 | 289 | 7 | N | 6.870 | N |
| CIMG_06250 | Methyltransferase domain-containing protein | 51 | 320 | 1 | Y | 6.431 | Y |
| CIMG_11522 | Hypothetical protein—genus-specific | 2 | 85 | 1 | N | 6.376 | Y |
| CIMG_10007 | Hypothetical protein—genus-specific | 3 | 125 | 0 | N | 6.341 | Y |
| CIMG_02628 | Arp2/3 complex subunit Arc16 | 199 | 312 | 0 | N | 6.158 | N |
| CIMG_09539 | Hypothetical | 136 | 387 | 0 | N | 5.989 | N |
| CIMG_12484 | hypothetical protein—genus-specific | 1 | 124 | 0 | N | 5.977 | Y |
| CIMG_09750 | Amino acid adenylation domain-containing protein | 1262 | 8032 | 0 | N | 5.898 | N |
| CIMG_05758 | parasitic phase-specific protein PSP-1 | 458 | 279 | 7 | N | 5.752 | N |
| CIMG_11264 | Hypothetical protein | 59 | 76 | 1 | Y | 5.693 | N |
| CIMG_00534 | Glycerate kinase | 37 | 437 | 1 | N | 5.635 | N |
| CIMG_10013 | Hypothetical protein—genus-specific | 0 | 114 | 0 | N | 5.586 | Y |
| CIMG_01211 | Putative major facilitator superfamily (MFS) transporter | 11 | 553 | 13 | N | 5.294 | N |
| Downregulated | |||||||
| CIMG_03418 | CBF1-interacting co-repressor CIR domain-containing protein | 99 | 336 | 0 | N | −8.094 | N |
| CIMG_01310 | 4-hydroxyphenylpyruvate dioxygenase | 183 | 399 | 0 | N | −7.717 | N |
| CIMG_07209 | Life-span regulatory factor domain-containing protein | 56 | 151 | 0 | N | −7.491 | N |
| CIMG_03873 | Protoglobin domain-containing protein | 136 | 245 | 0 | N | −7.320 | N |
| CIMG_07928 | SAM and PH domain-containing protein | 74 | 795 | 0 | N | −6.970 | N |
| CIMG_03716 | Heat shock protein 30 | 115 | 238 | 0 | N | −6.904 | N |
| CIMG_00925 | Hypothetical protein—cell wall protein | 82 | 157 | 0 | Y | −6.903 | N |
| CIMG_00284 | Hypothetical protein | 5 | 118 | 0 | Y | −6.898 | Y |
| CIMG_09232 | Hypothetical protein | 59 | 571 | 1 | Y | −6.880 | Y |
| CIMG_00099 | C2H2 finger domain-containing protein | 116 | 595 | 0 | N | −6.868 | N |
| CIMG_08613 | Metalloproteinase 7 | 4 | 357 | 0 | Y | −6.746 | N |
| CIMG_12190 | Hypothetical protein—genus-specific | 1 | 137 | 0 | N | −6.689 | Y |
| CIMG_03848 | DnaJ domain-containing protein | 68 | 536 | 0 | N | −6.660 | N |
| CIMG_13167 | Beta-glucosidase 5 | 94 | 489 | 0 | N | −6.588 | N |
| CIMG_13284 | Hypothetical protein—genus-specific | 0 | 74 | 0 | N | −6.523 | Y |
| CIMG_04597 | GPI anchored serine-threonine rich protein | 68 | 216 | 0 | Y | −6.467 | Y |
| CIMG_00348 | Endochitinase 2 | 317 | 897 | 0 | Y | −6.445 | N |
| CIMG_10468 | Hypothetical protein—genus-specific | 3 | 132 | 0 | N | −6.370 | Y |
| CIMG_00302 | Hypothetical protein—genus-specific | 74 | 228 | 0 | Y | −6.345 | Y |
| CIMG_07925 | Hypothetical protein—genus-specific | 3 | 168 | 0 | N | −6.229 | Y |
| Group | Total | Genus-Specific | % |
|---|---|---|---|
| All genes | 9759 | 2305 | 24 |
| Upregulated | 407 | 127 | 31a |
| Downregulated | 849 | 329 | 39 a |
| Transcription Factor | Total Number | Number (Adjusted p < 0.05) | PFAM | FC (log2 Young Spherule/Mycelium) | |||
|---|---|---|---|---|---|---|---|
| >2 | >1 | <−1 | <−2 | ||||
| APSES | 5 | 3 | PF13637 | 2 | 1 | ||
| HLH transcription factor | 8 | 3 | PF00010 | 2 | 1 | ||
| C2H2 type zinc finger domain-containing protein | 29 | 21 | PF00096 | 1 | 11 | 7 | |
| fungal Zn(2)–Cys(6) binuclear cluster domain-containing | 91 | 51 | PF00172 | 4 | 16 | 17 | 7 |
| Homeobox-like domain-containing protein | 6 | 6 | PF00249 | 2 | |||
| Other Regulatory Proteins | |||||||
| ryp family | 4 | 3 | PF09729; PF11754 PF00172; PF04082 | 1 | 2 | 1 | |
| Gene ID | Product Description | Ortholog Count | Length | # TM Domains | SignalP | FC (log2 Values) |
|---|---|---|---|---|---|---|
| Upregulated | ||||||
| CIMG_03805 | Possible HSP31 | 281 | 243 | 0 | N | 4.626 |
| CIMG_12285 | hypothetical protein | 10 | 151 | 0 | N | 4.074 |
| CIMG_08736 | hypothetical protein | 5 | 156 | 0 | N | 3.705 |
| CIMG_07646 | hypothetical protein | 3 | 350 | 0 | N | 3.606 |
| CIMG_07274 | Ferritin-like domain-containing protein | 105 | 455 | 0 | Y | 3.507 |
| CIMG_13581 | hypothetical protein | 0 | 71 | 0 | N | 3.306 |
| CIMG_05456 | Possible pre-mRNA-splicing factor cef1 | 45 | 237 | 0 | N | 3.281 |
| CIMG_08524 | Dynamin family protein | 450 | 711 | 0 | N | 3.186 |
| CIMG_02168 | Possible transcription factor wer | 12 | 444 | 0 | N | 3.140 |
| CIMG_00573 | Cytochrome P450 51 | 236 | 511 | 2 | N | 3.064 |
| CIMG_08156 | Possible meiotic recombination protein dmc1/lim15 homolog | 91 | 463 | 0 | N | 3.058 |
| CIMG_04521 | DUF1399 domain-containing protein | 195 | 745 | 0 | N | 2.984 |
| CIMG_03465 | hypothetical protein | 8 | 383 | 0 | N | 2.907 |
| CIMG_06696 | Possible dual oxidase | 508 | 295 | 2 | Y | 2.902 |
| CIMG_06262 | Possible rest corepressor 1 | 37 | 185 | 0 | N | 2.900 |
| CIMG_08666 | FK506-binding protein 1 | 202 | 120 | 0 | N | 2.829 |
| CIMG_13473 | YjgH family protein | 45 | 206 | 0 | N | 2.778 |
| CIMG_04544 | Alpha-amylase AmyA | 65 | 582 | 0 | Y | 2.769 |
| CIMG_01977 | Hypothetical protein | 5 | 482 | 0 | Y | 2.740 |
| CIMG_07685 | Hypothetical protein | 8 | 105 | 0 | N | 2.711 |
| Downregulated | ||||||
| CIMG_07583 | Tyrosinase | 41 | 658 | 0 | Y | −4.248 |
| CIMG_03173 | Hypothetical protein | 5 | 189 | 1 | N | −3.863 |
| CIMG_00927 | Acyl–CoA synthetase | 137 | 693 | 1 | N | −3.204 |
| CIMG_05421 | Beta-glucan synthesis-associated protein KRE6 | 222 | 642 | 1 | N | −3.046 |
| CIMG_03257 | Cell division cycle protein Cdc20 | 172 | 599 | 0 | N | −3.030 |
| CIMG_03517 | Hypothetical protein | 85 | 2137 | 0 | N | −2.887 |
| CIMG_01729 | Aconitate hydratase, mitochondrial | 352 | 784 | 0 | N | −2.844 |
| CIMG_13686 | Hypothetical protein | 0 | 94 | 0 | N | −2.806 |
| CIMG_00254 | GTP-binding protein rhoC | 109 | 278 | 0 | N | −2.783 |
| CIMG_03888 | Betaglucosidase | 1153 | 858 | 0 | Y | −2.731 |
| CIMG_01317 | Dethiobiotin synthase | 136 | 807 | 0 | N | −2.731 |
| CIMG_01369 | Myosin rod fragments superfamily protein, putative | 1387 | 1271 | 0 | N | −2.703 |
| CIMG_08907 | Cytochrome b2 | 480 | 504 | 0 | N | −2.660 |
| CIMG_12964 | Hypothetical protein | 0 | 168 | 0 | Y | −2.615 |
| CIMG_00847 | GTP binding protein | 109 | 1469 | 0 | N | −2.609 |
| CIMG_05523 | Ca2+ regulator and membrane fusion protein Figure 1, putative | 111 | 268 | 4 | Y | −2.552 |
| CIMG_13558 | DNA replication helicase Dna2 | 158 | 1659 | 0 | N | −2.523 |
| CIMG_08747 | Putative tetratricopeptide repeat (TPR)-like superfamily protein | 232 | 702 | 0 | N | −2.493 |
| CIMG_01268 | MFS phospholipid transporter | 317 | 496 | 10 | N | −2.492 |
| CIMG_05728 | L-serine dehydratase | 173 | 426 | 0 | N | −2.475 |
| CIMG_09628 | Protein–tyrosine phosphatase | 171 | 600 | 0 | N | −2.471 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carlin, A.F.; Beyhan, S.; Peña, J.F.; Stajich, J.E.; Viriyakosol, S.; Fierer, J.; Kirkland, T.N. Transcriptional Analysis of Coccidioides immitis Mycelia and Spherules by RNA Sequencing. J. Fungi 2021, 7, 366. https://doi.org/10.3390/jof7050366
Carlin AF, Beyhan S, Peña JF, Stajich JE, Viriyakosol S, Fierer J, Kirkland TN. Transcriptional Analysis of Coccidioides immitis Mycelia and Spherules by RNA Sequencing. Journal of Fungi. 2021; 7(5):366. https://doi.org/10.3390/jof7050366
Chicago/Turabian StyleCarlin, Aaron F., Sinem Beyhan, Jesús F. Peña, Jason E. Stajich, Suganya Viriyakosol, Joshua Fierer, and Theo N. Kirkland. 2021. "Transcriptional Analysis of Coccidioides immitis Mycelia and Spherules by RNA Sequencing" Journal of Fungi 7, no. 5: 366. https://doi.org/10.3390/jof7050366
APA StyleCarlin, A. F., Beyhan, S., Peña, J. F., Stajich, J. E., Viriyakosol, S., Fierer, J., & Kirkland, T. N. (2021). Transcriptional Analysis of Coccidioides immitis Mycelia and Spherules by RNA Sequencing. Journal of Fungi, 7(5), 366. https://doi.org/10.3390/jof7050366






