Transcriptome Analysis of the Japanese Pine Sawyer Beetle, Monochamus alternatus, Infected with the Entomopathogenic Fungus Metarhizium anisopliae JEF-197
Abstract
1. Introduction
2. Materials and Methods
2.1. Japanese Pine Sawyer Beetles and the Entomopathogenic Fungus JEF-197
2.2. Treatment of JPS Adults with M. anisopliae JEF-197
2.3. RNA Extraction and Construction of RNA-Seq Libraries
2.4. De Novo Transcriptome Assembly and Differentially Expressed Gene (DEG) Analysis
2.5. Functional Annotation and Gene Set Enrichment Analysis
2.6. Gene Expression Validation by qRT-PCR
3. Results
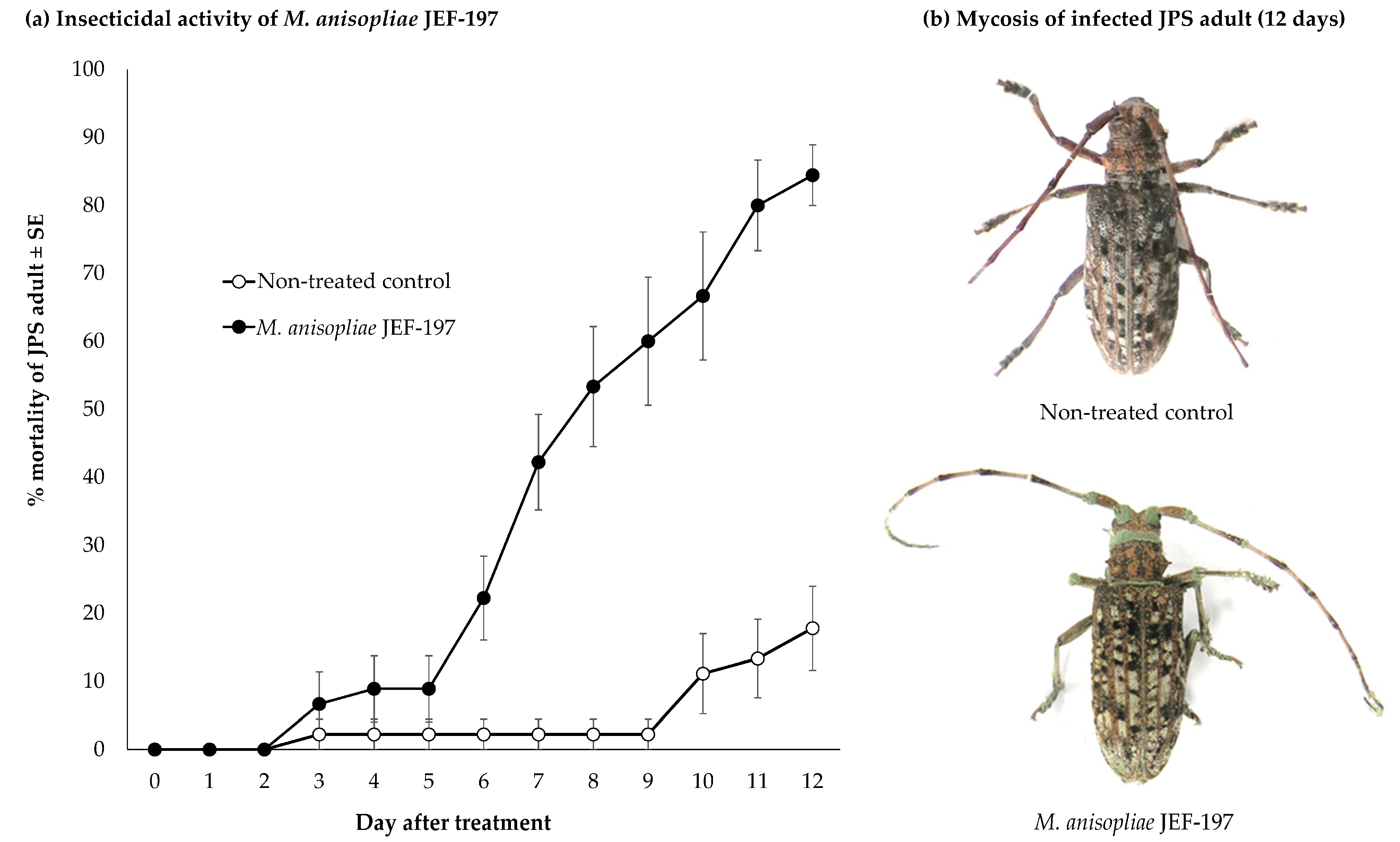
3.1. Time Course of Fungal Virulence
3.2. Construction of in Silico cDNA Libraries of M. anisopliae JEF-197 and JAPANESE Pine Sawyer Beetle
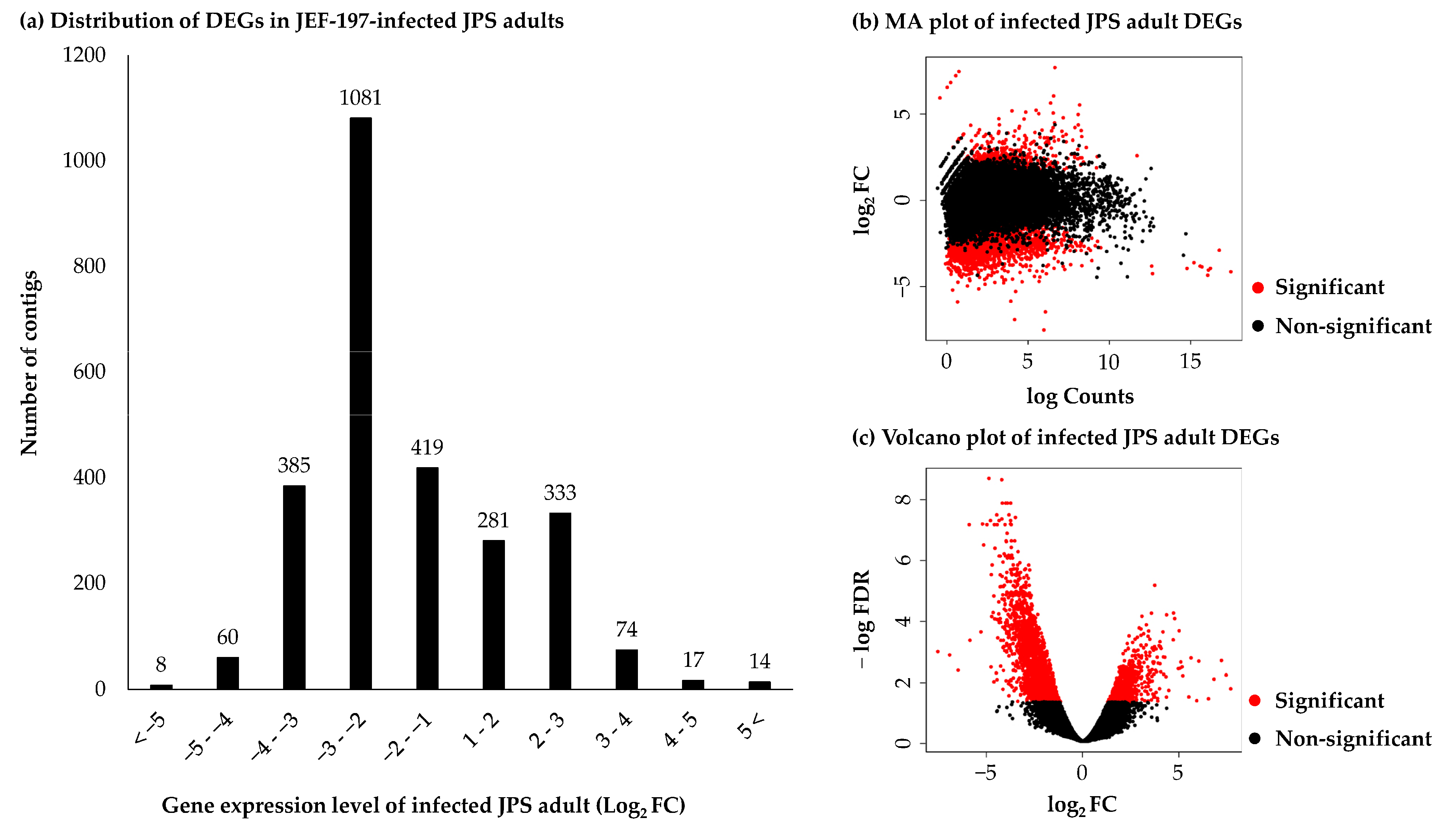
3.3. Differentially Expressed Genes in Japanese Pine Sawyer Beetle as a Result of Fungal Infection
3.4. Changes in Japanese Pine Sawyer Beetle Gene Expression after Fungal Treatment
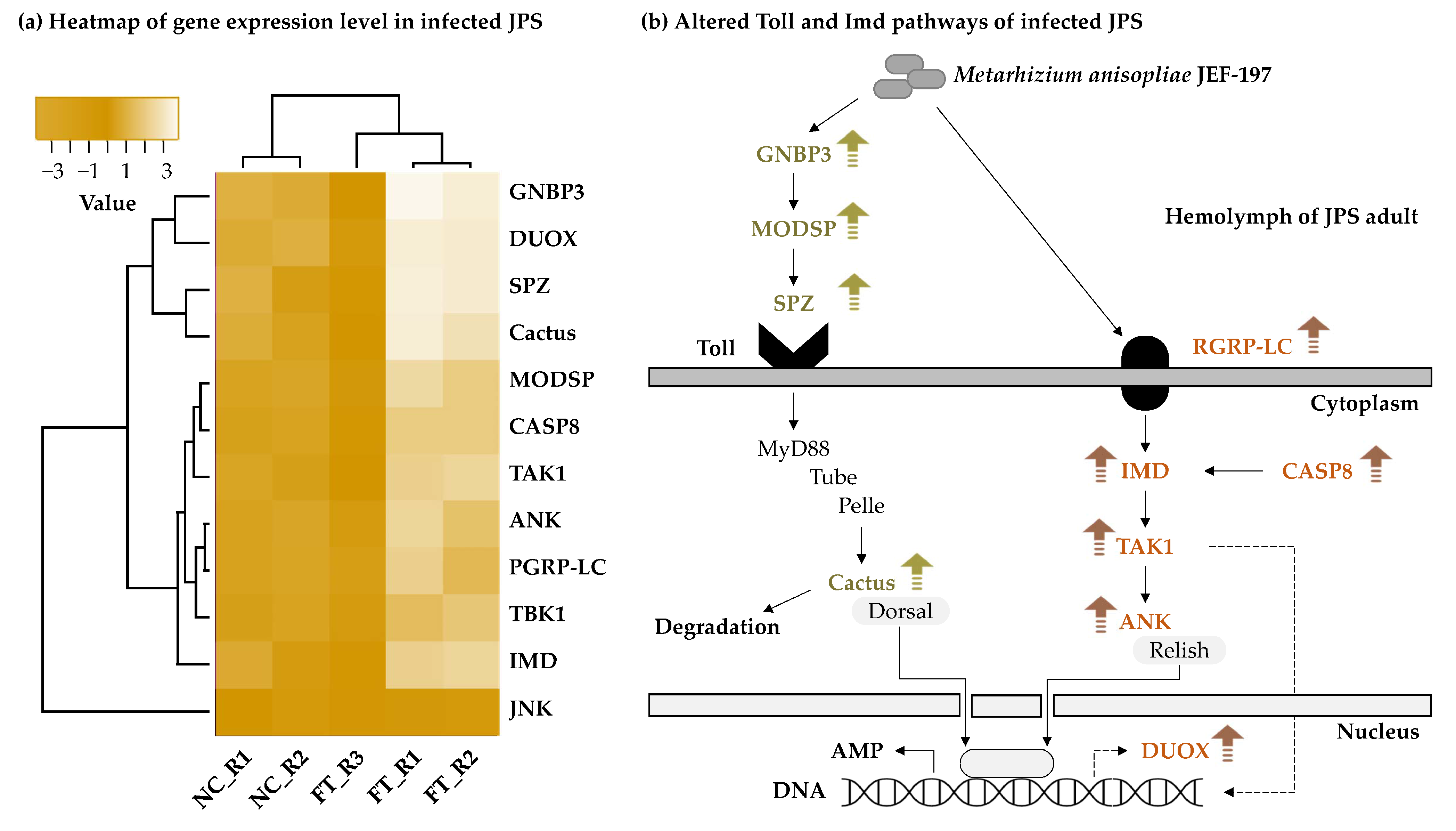
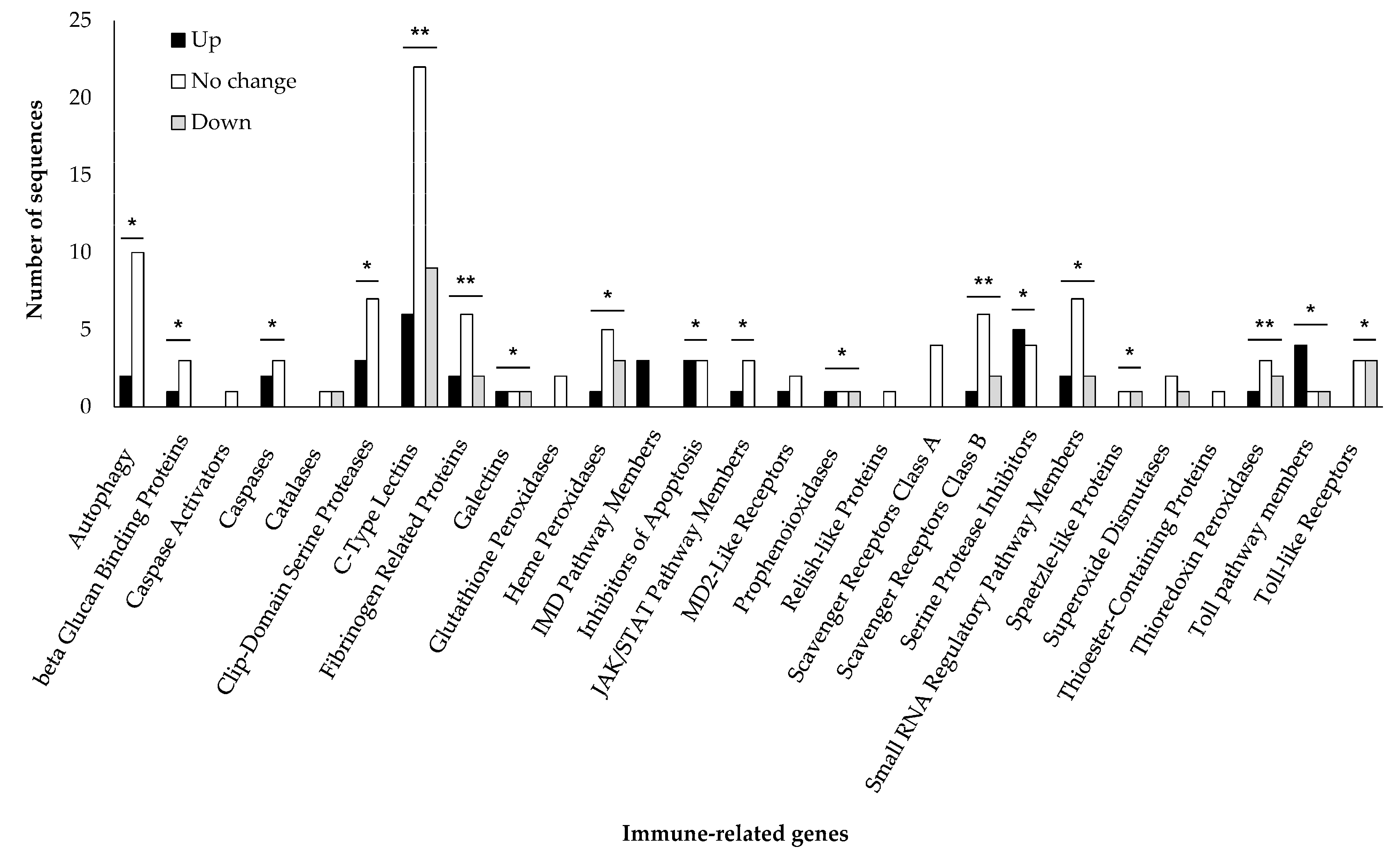
3.5. Changes in the Expression Level of Japanese Pine Sawyer Beetle Immune-Related Genes
3.6. Expression of M. anisopliae JEF-197 in Japanese Pine Sawyer Beetles
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Togashi, K. Development of Monochamus alternatus Hope (Coleoptera: Cerambycidae) in relation to oviposition time. Jpn. J. Appl. Entomol. Z. 1989, 33, 1–8. [Google Scholar] [CrossRef]
- Burgermeister, W.; Braasch, H.; Sousa, E.; Penas, A.C.; Mota, M.; Metge, K.; Bravo, M.A. First report of Bursaphelenchus xylophilus in Portugal and in Europe. Nematology 1999, 1, 727–734. [Google Scholar] [CrossRef]
- Naves, P.; Mota, M.; Pires, J.; Penas, A.C.; Sousa, E.; Bonifácio, L.; Bravo, M.A. Bursaphelenchus xylophilus (Nematoda; aphelenchoididae) associated with Monochamus galloprovincialis (Coleoptera; Cerambycidae) in Portugal. Nematology 2001, 3, 89–91. [Google Scholar] [CrossRef]
- Lee, S.; Chung, Y.; Moon, Y.; Lee, S.; Lee, D.; Choo, H.; Lee, C. Insecticidal activity and fumigation conditions of several insecticides against Japanese pine sawyer (Monochamus alternatus) larvae. J. Korean For. Soc. 2003, 92, 191–198. [Google Scholar]
- Shin, S.-C. Pine wilt disease in Korea. In Pine Wilt Disease; Springer: Tokyo, Japan, 2008; pp. 26–32. [Google Scholar] [CrossRef]
- Hemingway, J.; Ranson, H. Insecticide resistance in insect vectors of human disease. Annu. Rev. Entomol. 2000, 45, 371–391. [Google Scholar] [CrossRef]
- Whalon, M.E.; Mota-Sanchez, D.; Hollingworth, R.M. Global Pesticide Resistance in Arthropods; CABI: New York, NY, USA, 2008. [Google Scholar]
- Butt, T.M.; Jackson, C.; Magan, N. Introduction-fungal biological control agents: Progress, problems and potential. In Fungi as Biocontrol Agents: Progress, Problems and Potential; Butt, T.M., Jackson, C., Magan, N., Eds.; CABI: New York, NY, USA, 2001; pp. 1–8. [Google Scholar] [CrossRef]
- Shimazu, M. Potential of the cerambycid-parasitic type of Beauveria brongniartii (Deuteromycotina: Hyphomycetes) for microbial control of Monochamus alternatus Hope (Coleoptera: Cerambycidae). Appl. Entomol. Zool. 1994, 29, 127–130. [Google Scholar] [CrossRef][Green Version]
- Shimazu, M.; Kushida, T.; Tsuchiya, D.; Mitsuhashi, W. Microbial control of Monochamus alternatus Hope (Coleoptera: Cerambycidae) by implanting wheat-bran pellets with Beauveria bassiana in infested tree trunks. J. Jpn. For. Soc. 1992, 74, 325–330. [Google Scholar] [CrossRef][Green Version]
- Shimazu, M.; Sato, H. Effects of larval age on mortality of Monochamus alternatus Hope (Coleoptera: Cerambycidae) after application of nonwoven fabric strips with Beauveria bassiana. Appl. Entomol. Zool. 2003, 38, 1–5. [Google Scholar] [CrossRef][Green Version]
- Shimazu, M.; Tsuchiya, D.; Sato, H.; Kushida, T. Microbial control of Monochamus alternatus Hope (Coleoptera: Cerambycidae) by application of nonwoven fabric strips with Beauveria bassiana (Deuteromycotina: Hyphomycetes) on infested tree trunks. Appl. Entomol. Zool. 1995, 30, 207–213. [Google Scholar] [CrossRef]
- Francardi, V.; Rumine, P.; De Silva, J. On microbial control of Monochamus galloprovincialis (Olivier)(Coleoptera Cerambycidae) by means of Beauveria bassiana (Bals.) Vuillemin (Deuteromycotina Hyphomycetes). Redia 2003, 86, 129–132. [Google Scholar]
- Kim, J.C.; Lee, S.J.; Kim, S.; Lee, M.R.; Baek, S.; Park, S.E.; Kim, J.; Shin, T.Y.; Kim, J.S. Management of pine wilt disease vectoring Monochamus alternatus adults using spray and soil application of Metarhizium anisopliae JEF isolates. J. Asia Pac. Entomol. 2020, 23, 224–233. [Google Scholar] [CrossRef]
- Kim, J.C.; Baek, S.; Park, S.E.; Kim, S.; Lee, M.R.; Jo, M.; Im, J.S.; Ha, P.; Kim, J.S.; Shin, T.Y. Colonization of Metarhizium anisopliae on the surface of pine tree logs: A promising biocontrol strategy for the Japanese pine sawyer, Monochamus alternatus. Fungal Biol. 2020, 124, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Prior, C.; Lomer, C.; Herren, H.; Paraiso, A.; Kooyman, C.; Smit, J. The IIBC/IITA/DFPV collaborative research programme on the biological control of locusts and grasshoppers. In Proceedings of the Biological Control of Locusts and Grasshoppers: Proceedings of a Workshop Held at the International Institute of Tropical Agriculture, Cotonou, Republic of Benin, 29 April–1 May 1991; pp. 8–18. [Google Scholar]
- St Leger, R.; Joshi, L.; Bidochka, M.J.; Roberts, D.W. Construction of an improved mycoinsecticide overexpressing a toxic protease. Proc. Natl. Acad. Sci. USA 1996, 93, 6349–6354. [Google Scholar] [CrossRef]
- Gillespie and, J.P.; Kanost, M.R.; Trenczek, T. Biological mediators of insect immunity. Annu. Rev. Entomol. 1997, 42, 611–643. [Google Scholar] [CrossRef] [PubMed]
- Lowenberger, C. Innate immune response of Aedes aegypti. Insect Biochem. Molec. 2001, 31, 219–229. [Google Scholar] [CrossRef]
- Muta, T.; Iwanaga, S. The role of hemolymph coagulation in innate immunity. Curr. Opin. Immunol. 1996, 8, 41–47. [Google Scholar] [CrossRef]
- Schmidt, O.; Theopold, U.; Strand, M. Innate immunity and its evasion and suppression by hymenopteran endoparasitoids. BioEssays 2001, 23, 344–351. [Google Scholar] [CrossRef]
- Strand, M.R.; Pech, L.L. Immunological basis for compatibility in parasitoid-host relationships. Annu. Rev. Entomol. 1995, 40, 31–56. [Google Scholar] [CrossRef]
- Gross, J.; Müller, C.; Vilcinskas, A.; Hilker, M. Antimicrobial Activity of Exocrine Glandular Secretions, Hemolymph, and Larval Regurgitate of the Mustard Leaf Beetle Phaedon cochleariae. J. Invertebr. Pathol. 1998, 72, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Boucias, D.; Latgé, J. Nonspecific induction of germination of Conidiobolus obscurus and Nomuraea rileyi with host and non-host cuticle extracts. J. Invertebr. Pathol. 1988, 51, 168–171. [Google Scholar] [CrossRef]
- Holder, D.J.; Keyhani, N.O. Adhesion of the entomopathogenic fungus Beauveria (Cordyceps) bassiana to substrata. Appl. Environ. Microb. 2005, 71, 5260–5266. [Google Scholar] [CrossRef]
- Wraight, S.; Butt, T.; Galaini-Wraight, S.; Allee, L.; Soper, R.; Roberts, D.W. Germination and infection processes of the entomophthoralean fungus Erynia radicans on the potato leafhopper, Empoasca fabae. J. Invertebr. Pathol. 1990, 56, 157–174. [Google Scholar] [CrossRef]
- Zipfel, P.F.; Skerka, C.; Kupka, D.; Luo, S. Immune escape of the human facultative pathogenic yeast Candida albicans: The many faces of the Candida Pra1 protein. Int. J. Med. Microbiol. Suppl. 2011, 301, 423–430. [Google Scholar] [CrossRef]
- Brey, P.T.; Lee, W.-J.; Yamakawa, M.; Koizumi, Y.; Perrot, S.; Francois, M.; Ashida, M. Role of the integument in insect immunity: Epicuticular abrasion and induction of cecropin synthesis in cuticular epithelial cells. Proc. Natl. Acad. Sci. USA 1993, 90, 6275–6279. [Google Scholar] [CrossRef] [PubMed]
- Butt, T.M.; Hoch, H.C.; Staples, R.C.; Leger, R.J.S. Use of fluorochromes in the study of fungal cytology and differentiation. Exp. Mycol. 1989, 13, 303–320. [Google Scholar] [CrossRef]
- Santi, L.; da Silva, W.O.B.; Berger, M.; Guimarães, J.A.; Schrank, A.; Vainstein, M.H. Conidial surface proteins of Metarhizium anisopliae: Source of activities related with toxic effects, host penetration and pathogenesis. Toxicon 2010, 55, 874–880. [Google Scholar] [CrossRef] [PubMed]
- Schrank, A.; Vainstein, M.H. Metarhizium anisopliae enzymes and toxins. Toxicon 2010, 56, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Birzele, F.; Schaub, J.; Rust, W.; Clemens, C.; Baum, P.; Kaufmann, H.; Weith, A.; Schulz, T.W.; Hildebrandt, T. Into the unknown: Expression profiling without genome sequence information in CHO by next generation sequencing. Nucleic Acids Res. 2010, 38, 3999–4010. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Qin, Z.; Zhang, X. Opportunities and methods for studying alternative splicing in cancer with RNA-Seq. Cancer Lett. 2013, 340, 179–191. [Google Scholar] [CrossRef] [PubMed]
- McGettigan, P.A. Transcriptomics in the RNA-seq era. Curr. Opin. Chem. Biol. 2013, 17, 4–11. [Google Scholar] [CrossRef]
- Lin, T.; Cai, Z.; Wu, H. Transcriptome analysis of the Japanese pine sawyer beetle, Monochamus alternatus (Coleoptera: Cerambycidae) by high-throughput Illumina sequencing. J. Asia Pac. Entomol. 2015, 18, 439–445. [Google Scholar] [CrossRef]
- Zhou, J.; Yu, H.Y.; Zhang, W.; Ahmad, F.; Hu, S.N.; Zhao, L.L.; Zou, Z.; Sun, J.H. Comparative analysis of the Monochamus alternatus immune system. Insect Sci. 2018, 25, 581–603. [Google Scholar] [CrossRef]
- Wu, S.; Zhu, X.; Liu, Z.; Shao, E.; Rebeca, C.-L.; Guo, Y.; Xiong, Y.; Mou, Y.; Xu, R.; Hu, X. Identification of genes relevant to pesticides and biology from global transcriptome data of Monochamus alternatus Hope (Coleoptera: Cerambycidae) larvae. PLoS ONE 2016, 11, e0147855. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, D.-Z.; Min, S.-F.; Mi, F.; Zhou, S.-S.; Wang, M.-Q. Analysis of chemosensory gene families in the beetle Monochamus alternatus and its parasitoid Dastarcus helophoroides. Comp. Biochem. Physiol. Part. D Genom. Proteom. 2014, 11, 1–8. [Google Scholar] [CrossRef]
- Zhang, W.; Meng, J.; Ning, J.; Qin, P.; Zhou, J.; Zou, Z.; Wang, Y.; Jiang, H.; Ahmad, F.; Zhao, L. Differential immune responses of Monochamus alternatus against symbiotic and entomopathogenic fungi. Sci. China Life Sci. 2017, 60, 902–910. [Google Scholar] [CrossRef][Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 16 April 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. g: Profiler—A web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef]
- Oosthuizen, J.L.; Gomez, P.; Ruan, J.; Hackett, T.L.; Moore, M.M.; Knight, D.A.; Tebbutt, S.J. Dual organism transcriptomics of airway epithelial cells interacting with conidia of Aspergillus fumigatus. PLoS ONE 2011, 6, e20527. [Google Scholar] [CrossRef] [PubMed]
- Westermann, A.J.; Gorski, S.A.; Vogel, J. Dual RNA-seq of pathogen and host. Nat. Rev. Microbiol. 2012, 10, 618–630. [Google Scholar] [CrossRef]
- Heinz, E.; Hacker, C.; Dean, P.; Mifsud, J.; Goldberg, A.V.; Williams, T.A.; Nakjang, S.; Gregory, A.; Hirt, R.P.; Lucocq, J.M. Plasma membrane-located purine nucleotide transport proteins are key components for host exploitation by microsporidian intracellular parasites. PLoS Pathog. 2014, 10, e1004547. [Google Scholar] [CrossRef] [PubMed]
- Cossart, P.; Roy, C.R. Manipulation of host membrane machinery by bacterial pathogens. Curr. Opin. Cell Biol. 2010, 22, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Alix, E.; Mukherjee, S.; Roy, C.R. Subversion of membrane transport pathways by vacuolar pathogens. J. Cell Biol. 2011, 195, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Tanji, T.; Ip, Y.T. Regulators of the Toll and Imd pathways in the Drosophila innate immune response. Trends Immunol. 2005, 26, 193–198. [Google Scholar] [CrossRef]
- Lee, S.H.; Carpenter, J.F.; Chang, B.S.; Randolph, T.W.; Kim, Y.S. Effects of solutes on solubilization and refolding of proteins from inclusion bodies with high hydrostatic pressure. Protein Sci. 2006, 15, 304–313. [Google Scholar] [CrossRef]
- Ma, C.; Kanost, M.R. A β1, 3-glucan recognition protein from an insect, Manduca sexta, agglutinates microorganisms and activates the phenoloxidase cascade. J. Biol. Chem. 2000, 275, 7505–7514. [Google Scholar] [CrossRef]
- Ochiai, M.; Ashida, M. A pattern-recognition protein for β-1, 3-glucan: The binding domain and the cDNA cloning of β-1, 3-glucan recognition protein from the silkworm, Bombyx mori. J. Biol. Chem. 2000, 275, 4995–5002. [Google Scholar] [CrossRef] [PubMed]
- Buchon, N.; Poidevin, M.; Kwon, H.-M.; Guillou, A.; Sottas, V.; Lee, B.-L.; Lemaitre, B. A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc. Natl. Acad. Sci. USA 2009, 106, 12442–12447. [Google Scholar] [CrossRef] [PubMed]
- Gobert, V.; Gottar, M.; Matskevich, A.A.; Rutschmann, S.; Royet, J.; Belvin, M.; Hoffmann, J.A.; Ferrandon, D. Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science 2003, 302, 2126–2130. [Google Scholar] [CrossRef]
- Ligoxygakis, P.; Pelte, N.; Hoffmann, J.A.; Reichhart, J.-M. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science 2002, 297, 114–116. [Google Scholar] [CrossRef]
- Hoffmann, J.A.; Reichhart, J.-M. Drosophila innate immunity: An evolutionary perspective. Nat. Immunol. 2002, 3, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, J.A. The immune response of Drosophila. Nature 2003, 426, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Hedengren-Olcott, M.; Olcott, M.C.; Mooney, D.T.; Ekengren, S.; Geller, B.L.; Taylor, B.J. Differential activation of the NF-κB-like factors Relish and Dif in Drosophila melanogaster by fungi and Gram-positive bacteria. J. Biol. Chem. 2004, 279, 21121–21127. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.L.; Dunlap, C.A.; Muturi, E.J.; Barletta, A.B.; Rooney, A.P. Entomopathogenic fungal infection leads to temporospatial modulation of the mosquito immune system. PLoS Negl. Trop. Dis. 2018, 12, e0006433. [Google Scholar] [CrossRef] [PubMed]
- Hedengren, M.; Dushay, M.S.; Ando, I.; Ekengren, S.; Wihlborg, M.; Hultmark, D. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol. Cell 1999, 4, 827–837. [Google Scholar] [CrossRef]
- Stöven, S.; Silverman, N.; Junell, A.; Hedengren-Olcott, M.; Erturk, D.; Engström, Y.; Maniatis, T.; Hultmark, D. Caspase-mediated processing of the Drosophila NF-κB factor Relish. Proc. Natl. Acad. Sci. USA 2003, 100, 5991–5996. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.-H.; Ha, E.-M.; Lee, W.-J. Innate immunity and gut–microbe mutualism in Drosophila. Dev. Comp. Immunol. 2010, 34, 369–376. [Google Scholar] [CrossRef]
- Bae, Y.S.; Choi, M.K.; Lee, W.-J. Dual oxidase in mucosal immunity and host–microbe homeostasis. Trends Immunol. 2010, 31, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H.; Loh, K.M.; Nusse, R. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 2014, 346. [Google Scholar] [CrossRef] [PubMed]
- Wodarz, A.; Nusse, R. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 1998, 14, 59–88. [Google Scholar] [CrossRef] [PubMed]
- Artavanis-Tsakonas, S.; Rand, M.D.; Lake, R.J. Notch signaling: Cell fate control and signal integration in development. Science 1999, 284, 770–776. [Google Scholar] [CrossRef] [PubMed]
- Xin, C.; Yang, J.; Mao, Y.; Chen, W.; Wang, Z.; Song, Z. GATA-type transcription factor MrNsdD regulates dimorphic transition, conidiation, virulence and microsclerotium formation in the entomopathogenic fungus Metarhizium rileyi. Microb. Biotechnol. 2020, 13, 1489–1501. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Yin, Y.P.; Song, J.Z.; Hu, S.J.; Cheng, W.; Qiu, L. A p53-like transcription factor, BbTFO1, contributes to virulence and oxidative and thermal stress tolerances in the insect pathogenic fungus, Beauveria bassiana. PLoS ONE 2021, 16, e0249350. [Google Scholar] [CrossRef]
- Zhao, X.; Luo, T.; Huang, S.; Peng, N.; Yin, Y.; Luo, Z.; Zhang, Y. A novel transcription factor negatively regulates antioxidant response, cell wall integrity and virulence in the fungal insect pathogen, Beauveria bassiana. Environ. Microbiol. 2021. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, T.; Qiao, L.; Zhu, J.; Fan, J.; Zhang, T.; Wang, Z.-x.; Li, W.; Chen, A.; Huang, B. DNA methyltransferases contribute to the fungal development, stress tolerance and virulence of the entomopathogenic fungus Metarhizium robertsii. Appl. Microbiol. Biotechnol. 2017, 101, 4215–4226. [Google Scholar] [CrossRef]
- Lei, B.; Zhou, N.; Guo, Y.; Zhao, W.; Tan, Y.-W.; Yu, Y.; Lu, H. Septin ring assembly is regulated by Spt20, a structural subunit of the SAGA complex. J. Cell Sci. 2014, 127, 4024–4036. [Google Scholar] [CrossRef]
- Anderson, R.M.; Sinclair, D.A. Yeast RecQ Helicases: Clues to DNA Repair, Genome Stability and Aging. In Molecular Mechanisms of Werner’s Syndrome; Lebel, M., Ed.; Springer Science & Business Media: Berlin, Germany, 2004; pp. 78–104. [Google Scholar]
- Saier, M.H., Jr. Molecular phylogeny as a basis for the classification of transport proteins from bacteria, archaea and eukarya. Adv. Mircrob. Physiol. 1998, 40, 81–136. [Google Scholar] [CrossRef]
- Chabane, S.; Gachet, E.; Képès, F. Over-expression of the yeast BFR2 gene partially suppresses the growth defects induced by Brefeldin A and by four ER-to-Golgi mutations. Curr. Genet. 1998, 33, 21–28. [Google Scholar] [CrossRef]
- Rangel, D.E.; Braga, G.U.; Flint, S.D.; Anderson, A.J.; Roberts, D.W. Variations in UV-B tolerance and germination speed of Metarhizium anisopliae conidia produced on insects and artificial substrates. J. Invertebr. Pathol. 2004, 87, 77–83. [Google Scholar] [CrossRef]
- Garre, V.; Tenberge, K.B.; Eising, R. Secretion of a fungal extracellular catalase by Claviceps purpurea during infection of rye: Putative role in pathogenicity and suppression of host defense. Phytopathology 1998, 88, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Shang, F.; Taylor, A. Ubiquitin–proteasome pathway and cellular responses to oxidative stress. Free Radic. Biol. Med. 2011, 51, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-X.; Zhou, X.-Z.; Meng, H.-M.; Liu, Y.-J.; Zhou, Q.; Huang, B. Comparative transcriptomic analysis of the heat stress response in the filamentous fungus Metarhizium anisopliae using RNA-Seq. Appl. Microbiol. Biotechnol. 2014, 98, 5589–5597. [Google Scholar] [CrossRef]
- Diz, A.P.; Martínez-fernández, M.; Rolán-alvarez, E. Proteomics in evolutionary ecology: Linking the genotype with the phenotype. Mol. Ecol. 2012, 21, 1060–1080. [Google Scholar] [CrossRef] [PubMed]
- Nesvizhskii, A.I. Proteogenomics: Concepts, applications and computational strategies. Nat. Methods 2014, 11, 1114. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Casado, G.; Covey, P.A.; Bedinger, P.A.; Mueller, L.A.; Thannhauser, T.W.; Zhang, S.; Fei, Z.; Giovannoni, J.J.; Rose, J.K. Enabling proteomic studies with RNA-Seq: The proteome of tomato pollen as a test case. Proteomics 2012, 12, 761–774. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-C.; Lee, M.-R.; Kim, S.; Park, S.-E.; Lee, S.-J.; Shin, T.-Y.; Kim, W.-J.; Kim, J. Transcriptome Analysis of the Japanese Pine Sawyer Beetle, Monochamus alternatus, Infected with the Entomopathogenic Fungus Metarhizium anisopliae JEF-197. J. Fungi 2021, 7, 373. https://doi.org/10.3390/jof7050373
Kim J-C, Lee M-R, Kim S, Park S-E, Lee S-J, Shin T-Y, Kim W-J, Kim J. Transcriptome Analysis of the Japanese Pine Sawyer Beetle, Monochamus alternatus, Infected with the Entomopathogenic Fungus Metarhizium anisopliae JEF-197. Journal of Fungi. 2021; 7(5):373. https://doi.org/10.3390/jof7050373
Chicago/Turabian StyleKim, Jong-Cheol, Mi-Rong Lee, Sihyeon Kim, So-Eun Park, Se-Jin Lee, Tae-Young Shin, Woo-Jin Kim, and Jaesu Kim. 2021. "Transcriptome Analysis of the Japanese Pine Sawyer Beetle, Monochamus alternatus, Infected with the Entomopathogenic Fungus Metarhizium anisopliae JEF-197" Journal of Fungi 7, no. 5: 373. https://doi.org/10.3390/jof7050373
APA StyleKim, J.-C., Lee, M.-R., Kim, S., Park, S.-E., Lee, S.-J., Shin, T.-Y., Kim, W.-J., & Kim, J. (2021). Transcriptome Analysis of the Japanese Pine Sawyer Beetle, Monochamus alternatus, Infected with the Entomopathogenic Fungus Metarhizium anisopliae JEF-197. Journal of Fungi, 7(5), 373. https://doi.org/10.3390/jof7050373






