The Vacuolar Morphogenesis Protein Vam6-Like Protein Vlp1 Is Required for Pathogenicity of Cryptococcus neoformans
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Generation of vlp1Δ Mutants and Their Complemented and Overexpressed Strains
2.3. Melanin Production and Capsule Formation Assay
2.4. Virulence Studies
2.5. Fungal Burdens and Histopathology in Infected Organs
2.6. Serum Treatment and Cryptococcus–Macrophage Interaction Assay
2.7. Blood Analysis
2.8. Ethics Statement
3. Results
3.1. Identification of Vlp1 in C. neoformans
3.2. Disruption, Complementation and Overexpression of VLP1
3.3. Disruption of VLP1 Affects Melanin Production and Capsule Formation in C. neoformans
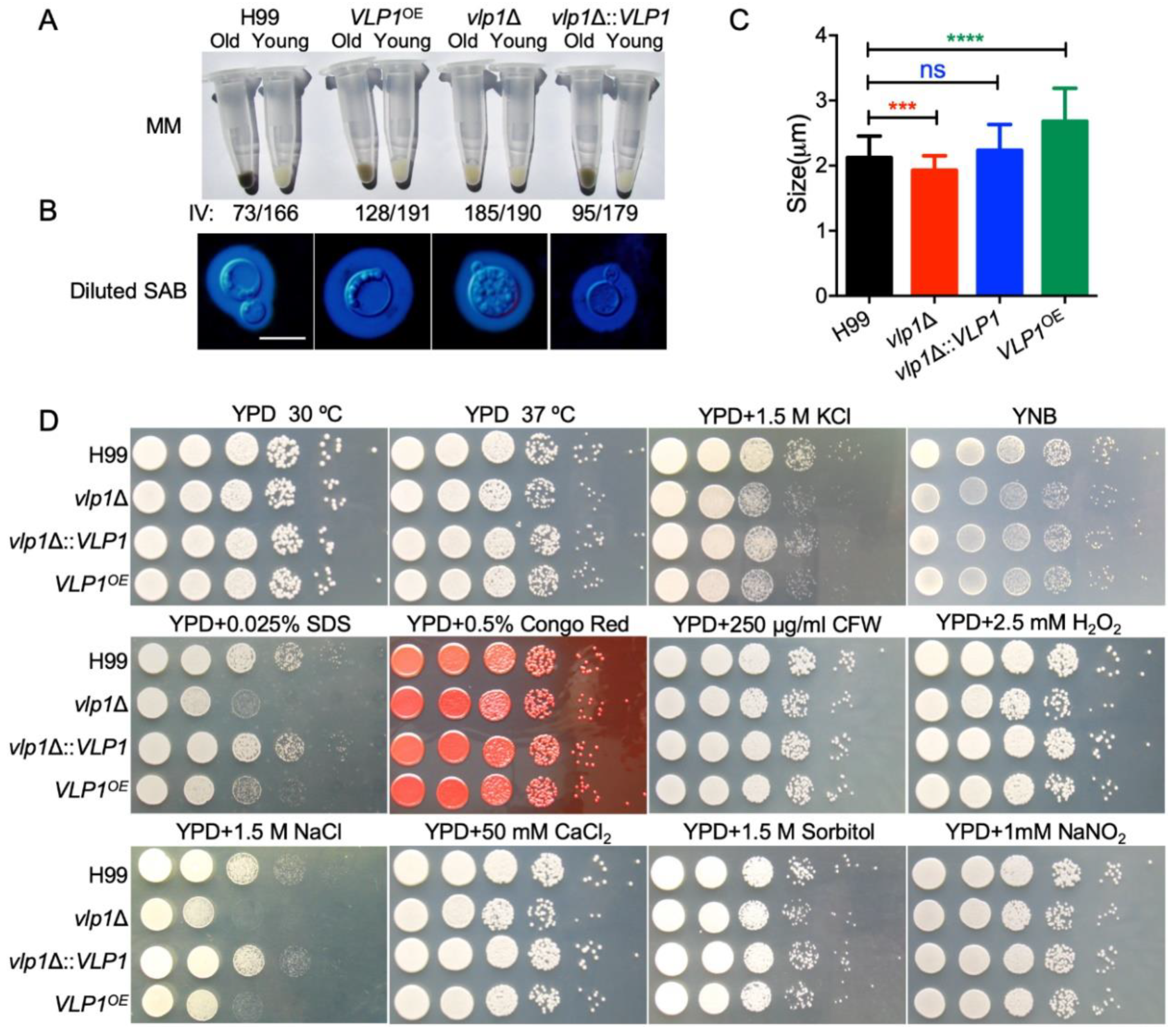
3.4. Vlp1 Is Required for Membrane Integrity
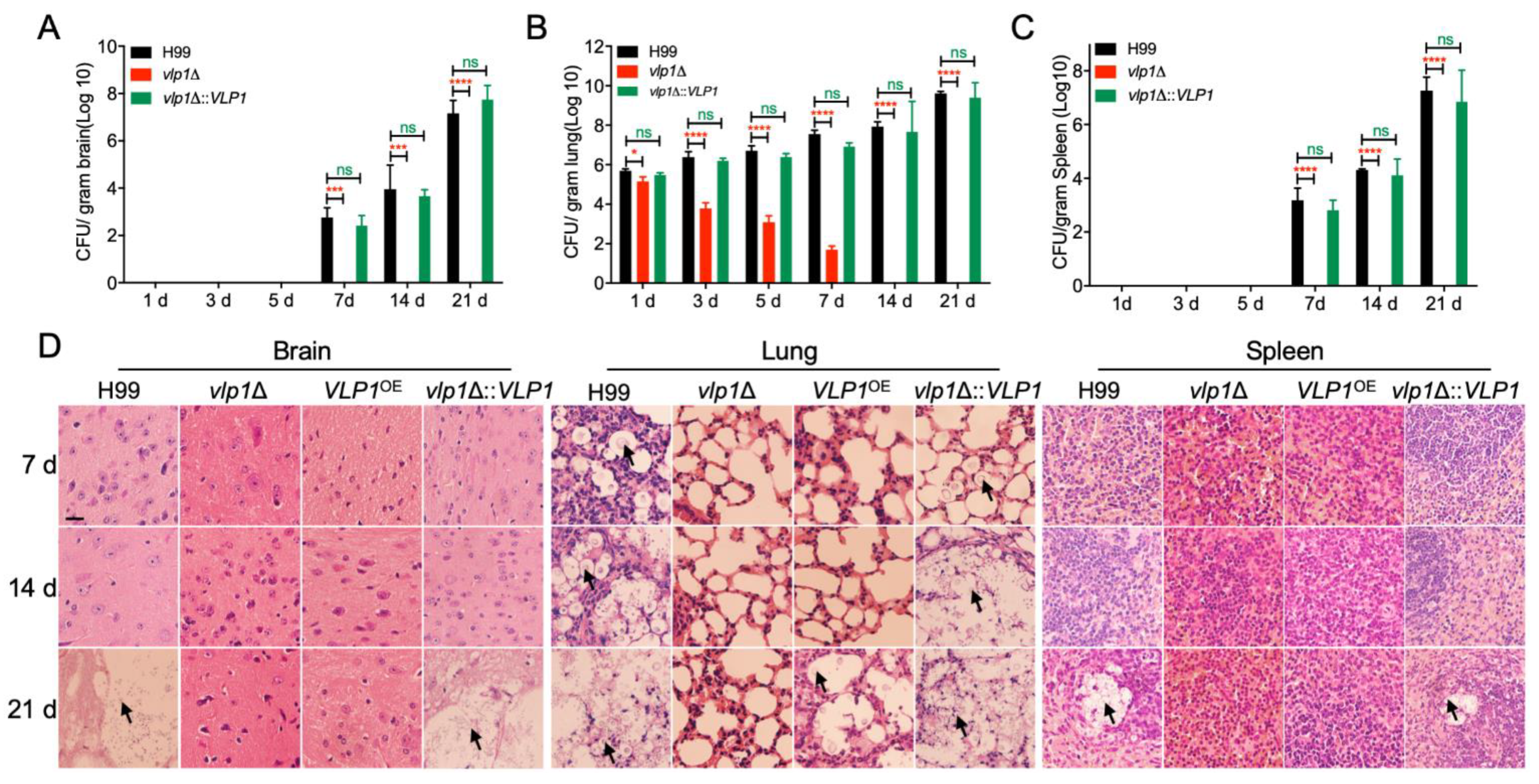
3.5. Vlp1 Is Essential for Fungal Infection
3.6. Vlp1 Is Essential for Progression of Fungal Infection
3.7. Vlp1 Is Important for Proliferation inside Macrophage and Survival in the Host Complement System
3.8. Vlp1 Is Not Required to Stimulate the Immune Response in Blood
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Casadevall, A.; Perfect., J.R. Cryptococcus Neoformans; ASM Press: Washington, DC, USA, 1998. [Google Scholar]
- Kronstad, J.W.; Attarian, R.; Cadieux, B.; Choi, J.; D’Souza, C.A.; Griffiths, E.J.; Geddes, J.M.; Hu, G.; Jung, W.H.; Kretschmer, M.; et al. Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat. Rev. Microbiol. 2011, 9, 193–203. [Google Scholar] [CrossRef]
- Erwig, L.P.; Gow, N.A. Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 2016, 14, 163–176. [Google Scholar] [CrossRef]
- Rajasingham, R.; Smith, R.M.; Park, B.J.; Jarvis, J.N.; Govender, N.P.; Chiller, T.M.; Denning, D.W.; Loyse, A.; Boulware, D.R. Global burden of disease of HIV-associated cryptococcal meningitis: An updated analysis. Lancet Infect. Dis. 2017, 17, 873–881. [Google Scholar] [CrossRef]
- Iyer, K.R.; Revie, N.M.; Fu, C.; Robbins, N.; Cowen, L.E. Treatment strategies for cryptococcal infection: Challenges, advances and future outlook. Nat. Rev. Microbiol. 2021. [Google Scholar] [CrossRef]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef] [PubMed]
- Lecker, S.H.; Goldberg, A.L.; Mitch, W.E. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. 2006, 17, 1807–1819. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.M.; Busino, L. The Biology of F-box Proteins: The SCF Family of E3 Ubiquitin Ligases. Adv. Exp. Med. Biol. 2020, 1217, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Jonkers, W.; Rep, M. Lessons from fungal F-box proteins. Eukaryot. Cell 2009, 8, 677–695. [Google Scholar] [CrossRef]
- Liu, T.B.; Xue, C. The Ubiquitin-Proteasome System and F-box Proteins in Pathogenic Fungi. Mycobiology 2011, 39, 243–248. [Google Scholar] [CrossRef]
- Liu, T.B.; Wang, Y.; Stukes, S.; Chen, Q.; Casadevall, A.; Xue, C. The F-Box protein Fbp1 regulates sexual reproduction and virulence in Cryptococcus neoformans. Eukaryot. Cell 2011, 10, 791–802. [Google Scholar] [CrossRef]
- Liu, T.B.; Xue, C. Fbp1-mediated ubiquitin-proteasome pathway controls Cryptococcus neoformans virulence by regulating fungal intracellular growth in macrophages. Infect. Immun. 2014, 82, 557–568. [Google Scholar] [CrossRef]
- Masso-Silva, J.; Espinosa, V.; Liu, T.B.; Wang, Y.; Xue, C.; Rivera, A. The F-Box Protein Fbp1 Shapes the Immunogenic Potential of Cryptococcus neoformans. mBio 2018, 9. [Google Scholar] [CrossRef]
- Han, L.T.; Wu, L.; Liu, T.B. A Predicted Mannoprotein Cmp1 Regulates Fungal Virulence in Cryptococcus neoformans. Pathogens 2020, 9, 881. [Google Scholar] [CrossRef]
- Orner, E.P.; Bhattacharya, S.; Kalenja, K.; Hayden, D.; Del Poeta, M.; Fries, B.C. Cell Wall-Associated Virulence Factors Contribute to Increased Resilience of Old Cryptococcus neoformans Cells. Front. Microbiol. 2019, 10, 2513. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza, O.; Casadevall, A. Experimental modulation of capsule size in Cryptococcus neoformans. Biol. Proced. Online 2004, 6, 10–15. [Google Scholar] [CrossRef]
- Perfect, J.R.; Ketabchi, N.; Cox, G.M.; Ingram, C.W.; Beiser, C.L. Karyotyping of Cryptococcus neoformans as an epidemiological tool. J. Clin. Microbiol. 1993, 31, 3305–3309. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.; Cox, G.M.; Wang, P.; Toffaletti, D.L.; Perfect, J.R.; Heitman, J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect. Immun. 2003, 71, 4831–4841. [Google Scholar] [CrossRef] [PubMed]
- Price, M.S.; Nichols, C.B.; Alspaugh, J.A. The Cryptococcus neoformans Rho-GDP dissociation inhibitor mediates intracellular survival and virulence. Infect. Immun. 2008, 76, 5729–5737. [Google Scholar] [CrossRef]
- Fan, C.L.; Han, L.T.; Jiang, S.T.; Chang, A.N.; Zhou, Z.Y.; Liu, T.B. The Cys2His2 zinc finger protein Zfp1 regulates sexual reproduction and virulence in Cryptococcus neoformans. Fungal Genet. Biol. 2019, 124, 59–72. [Google Scholar] [CrossRef]
- Fraser, J.A.; Subaran, R.L.; Nichols, C.B.; Heitman, J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: Implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 2003, 2, 1036–1045. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.M.; Mukherjee, J.; Cole, G.T.; Casadevall, A.; Perfect, J.R. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 2000, 68, 443–448. [Google Scholar] [CrossRef]
- Kingsbury, J.M.; McCusker, J.H. Fungal homoserine kinase (thr1Delta) mutants are attenuated in virulence and die rapidly upon threonine starvation and serum incubation. Eukaryot. Cell 2010, 9, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Basenko, E.Y.; Pulman, J.A.; Shanmugasundram, A.; Harb, O.S.; Crouch, K.; Starns, D.; Warrenfeltz, S.; Aurrecoechea, C.; Stoeckert, C.J., Jr.; Kissinger, J.C.; et al. FungiDB: An Integrated Bioinformatic Resource for Fungi and Oomycetes. J. Fungi (Basel) 2018, 4, 39. [Google Scholar] [CrossRef]
- Raymond, C.K.; Howald-Stevenson, I.; Vater, C.A.; Stevens, T.H. Morphological classification of the yeast vacuolar protein sorting mutants: Evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 1992, 3, 1389–1402. [Google Scholar] [CrossRef]
- Nakamura, N.; Hirata, A.; Ohsumi, Y.; Wada, Y. Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membranes and involved in the vacuolar assembly in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1997, 272, 11344–11349. [Google Scholar] [CrossRef]
- Wurmser, A.E.; Sato, T.K.; Emr, S.D. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 2000, 151, 551–562. [Google Scholar] [CrossRef]
- Bermas, A.; Geddes-McAlister, J. Combatting the evolution of antifungal resistance in Cryptococcus neoformans. Mol. Microbiol. 2020, 114, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Sinha, H.; David, L.; Pascon, R.C.; Clauder-Munster, S.; Krishnakumar, S.; Nguyen, M.; Shi, G.; Dean, J.; Davis, R.W.; Oefner, P.J.; et al. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics 2008, 180, 1661–1670. [Google Scholar] [CrossRef]
- Kozel, T.R. Virulence factors of Cryptococcus neoformans. Trends Microbiol. 1995, 3, 295–299. [Google Scholar] [CrossRef]
- Kronstad, J.; Jung, W.H.; Hu, G. Beyond the big three: Systematic analysis of virulence factors in Cryptococcus neoformans. Cell Host Microbe 2008, 4, 308–310. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza, O. Basic principles of the virulence of Cryptococcus. Virulence 2019, 10, 490–501. [Google Scholar] [CrossRef]
- D’Souza, C.A.; Alspaugh, J.A.; Yue, C.; Harashima, T.; Cox, G.M.; Perfect, J.R.; Heitman, J. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 2001, 21, 3179–3191. [Google Scholar] [CrossRef] [PubMed]
- Gerik, K.J.; Bhimireddy, S.R.; Ryerse, J.S.; Specht, C.A.; Lodge, J.K. PKC1 is essential for protection against both oxidative and nitrosative stresses, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans. Eukaryot. Cell 2008, 7, 1685–1698. [Google Scholar] [CrossRef]
- Bahn, Y.S.; Kojima, K.; Cox, G.M.; Heitman, J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell 2005, 16, 2285–2300. [Google Scholar] [CrossRef] [PubMed]



| Strains and Plasmids | Genotype or Properties | Source or Reference |
|---|---|---|
| C. neoformans | ||
| H99 | MATα | Perfect et al., 1993 [17] |
| KN99a | MATa | Nielsen et al., 2003 [18] |
| TBL131 | MATα vlp1Δ::NEO | In this study |
| TBL268 | MATa vlp1Δ::NEO | In this study |
| TBL278 | MATα vlp1Δ::NEO GFP-VLP1::NAT | In this study |
| TBL342 | MATa vlp1Δ::NEO GFP-VLP1::NAT | In this study |
| TBL350 | MATα vlp1Δ::NEO VLP1-HA::NAT | In this study |
| TBL351 | MATavlp1Δ::NEO VLP1-HA::NAT | In this study |
| TBL381 | MATα vlp1Δ::NEO VLP1-HA::NAT | In this study |
| TBL382 | MATavlp1Δ::NEO VLP1::NAT | In this study |
| Plasmids | ||
| pCN19 | Ampr Plasmid harboring GFP under histone H3 promoter | Price et al., 2008 [19] |
| pTBL1 | Ampr Plasmid harboring NAT marker | Fan et al., 2019 [20] |
| pTBL20 | Ampr Vector for PACTIN-ZFP1HA-NAT for ZFP1 overexpression | Fan et al., 2019 [20] |
| pTBL155 | Ampr Vector for PH3-GFP-VLP1 for VLP1 localization | In this study |
| pTBL187 | Ampr Vector for PH3-GFP-VLP1 for VLP1 localization | In this study |
| pTBL208 | Ampr Vector for PACTIN-VLP1-NAT for VLP1 overexpression | In this study |
| pTBL212 | Ampr Vector for PVLP1-VLP1-NAT for VLP1 complementation | In this study |
| Primers | Targeted Genes | Sequence (5′-3′) |
|---|---|---|
| TL17 | M13F | GTAAAACGACGGCCAG |
| TL18 | M13R | CAGGAAACAGCTATGAC |
| TL19 | NEO split F | GGGCGCCCGGTTCTTTTTGTCA |
| TL20 | NEO split R | TTGGTGGTCGAATGGGCAGGTAGC |
| TL59 | NEO R4 | TGTGGATGCTGGCGGAGGATA |
| TL217 | GAPDH Q-PCR F1 | TGAGAAGGACCCTGCCAACA |
| TL218 | GAPDH Q-PCR R1 | ACTCCGGCTTGTAGGCATCAA |
| TL327 | VLP1 KO F1 | GGTCAGGCGTGGAAGCGTCATACA |
| TL328 | VLP1 KO R1 | CTGGCCGTCGTTTTACGCTGCAACAAGTCGCGTCATTTAC |
| TL329 | VLP1 KO F2 | GTCATAGCTGTTTCCTGTCCGCAATGCCACAAGAGACTG |
| TL330 | VLP1 KO R2 | GAGACCCGGGGCCTAATACCTAAT |
| TL331 | VLP1 KO F3 | GCTCACCCCGAAAACAGATACAGG |
| TL332 | VLP1 KO R3 | AAGGCGGGGAGGGAGGATTC |
| TL333 | VLP1 KO F4 | AGCTGTGCCCCATCTTTTTAGTTA |
| TL971 | VLP1 CDS F1 | GACGAGCTGTAcGGATCCATGGCGACCGTCGAGTGCGTGCTG (BamHI) |
| TL972 | VLP1 CDS R1 | CTGGCGGCCGTTACTAGTCTATTCTCTTGAAGCGGCCAAGTT(SpeI) |
| TL1016 | VLP1 Comp F1 | GATATCGAATTCCTGCAGCCCGGGGGATCCGGAAAAGTTAAAAGTCATTGGCAGTT (BamHI) |
| TL1017 | VLP1 Comp R1 | CGGTGGCGGCCGCTCTAGAACTAGTGGATCTTGGAAAAGAAAAGGAAAGTGAAATC (BamHI) |
| TL1018 | VLP1-HA F1 | CGCCCAACATGTCTGGATCCATGGCGACCGTCGAGTGCGTGCTG (BamHI) |
| TL1019 | VLP1-HA R1 | ACGTCGTATGGGTAGGATCCTTCTCTTGAAGCGGCCAAGTTTGC (BamHI) |
| TL1281 | VLP1 Q-PCR F1 | GAAAGGGAGGAAAGAGGAAGTG |
| TL1282 | VLP1 Q-PCR R1 | TATTCTCTTGAAGCGGCCAAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, C.-L.; Liu, T.-B. The Vacuolar Morphogenesis Protein Vam6-Like Protein Vlp1 Is Required for Pathogenicity of Cryptococcus neoformans. J. Fungi 2021, 7, 418. https://doi.org/10.3390/jof7060418
Fan C-L, Liu T-B. The Vacuolar Morphogenesis Protein Vam6-Like Protein Vlp1 Is Required for Pathogenicity of Cryptococcus neoformans. Journal of Fungi. 2021; 7(6):418. https://doi.org/10.3390/jof7060418
Chicago/Turabian StyleFan, Cheng-Li, and Tong-Bao Liu. 2021. "The Vacuolar Morphogenesis Protein Vam6-Like Protein Vlp1 Is Required for Pathogenicity of Cryptococcus neoformans" Journal of Fungi 7, no. 6: 418. https://doi.org/10.3390/jof7060418
APA StyleFan, C.-L., & Liu, T.-B. (2021). The Vacuolar Morphogenesis Protein Vam6-Like Protein Vlp1 Is Required for Pathogenicity of Cryptococcus neoformans. Journal of Fungi, 7(6), 418. https://doi.org/10.3390/jof7060418







