Micafungin-Induced Cell Wall Damage Stimulates Morphological Changes Consistent with Microcycle Conidiation in Aspergillus nidulans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Media and Growth Conditions
2.2. Global Analysis Strain Generation, Growth Conditions, RNA Extraction, and RNA Sequencing
2.3. QRT-PCR for RNA-Sequencing Verification
2.4. Microscopy
2.5. Micafungin Concentration Experiments
2.6. Microcycle Conidiation Experiment with Wildtype and ΔmpkA
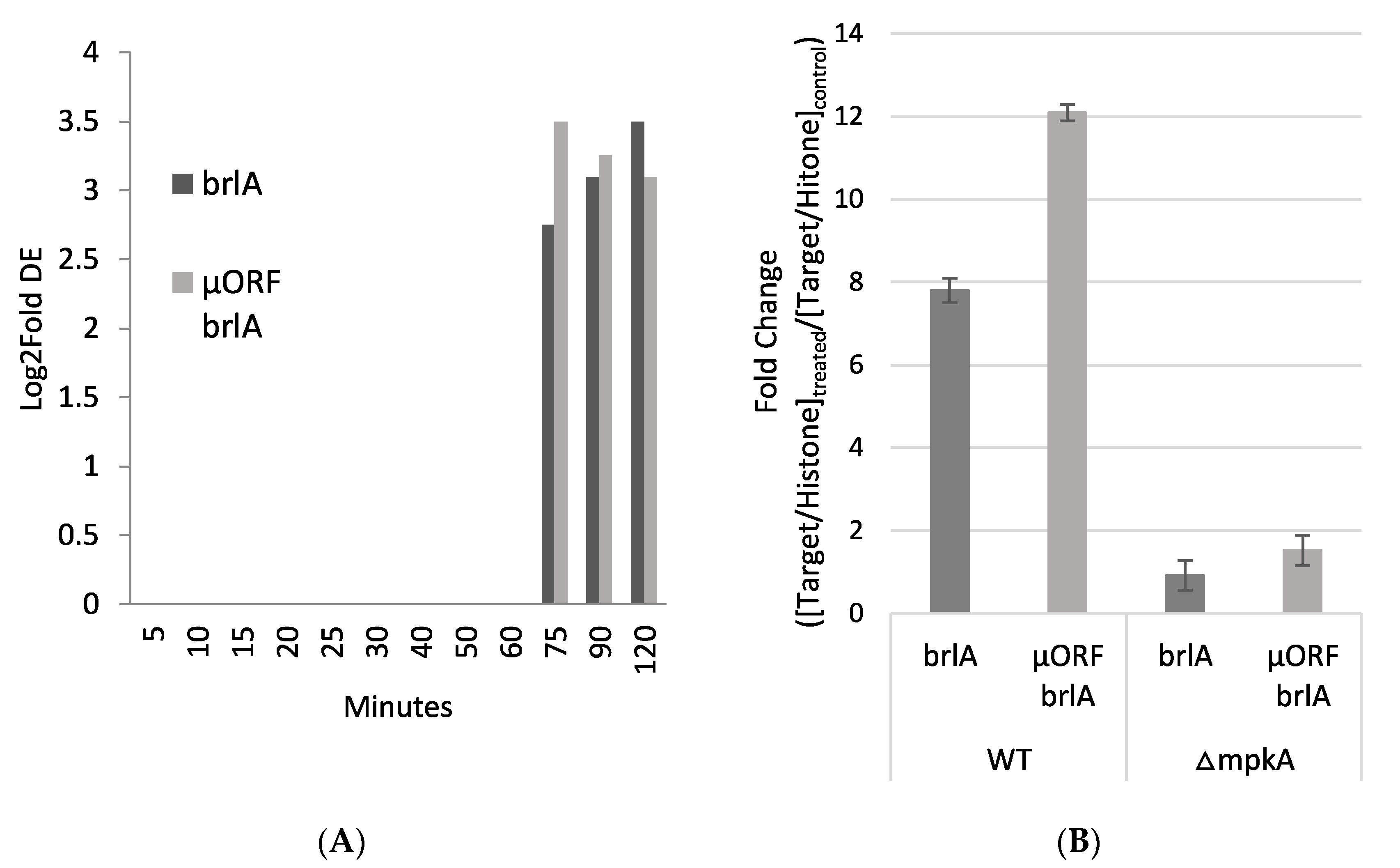
2.7. Up-Regulation of brlA Experiments
3. Results
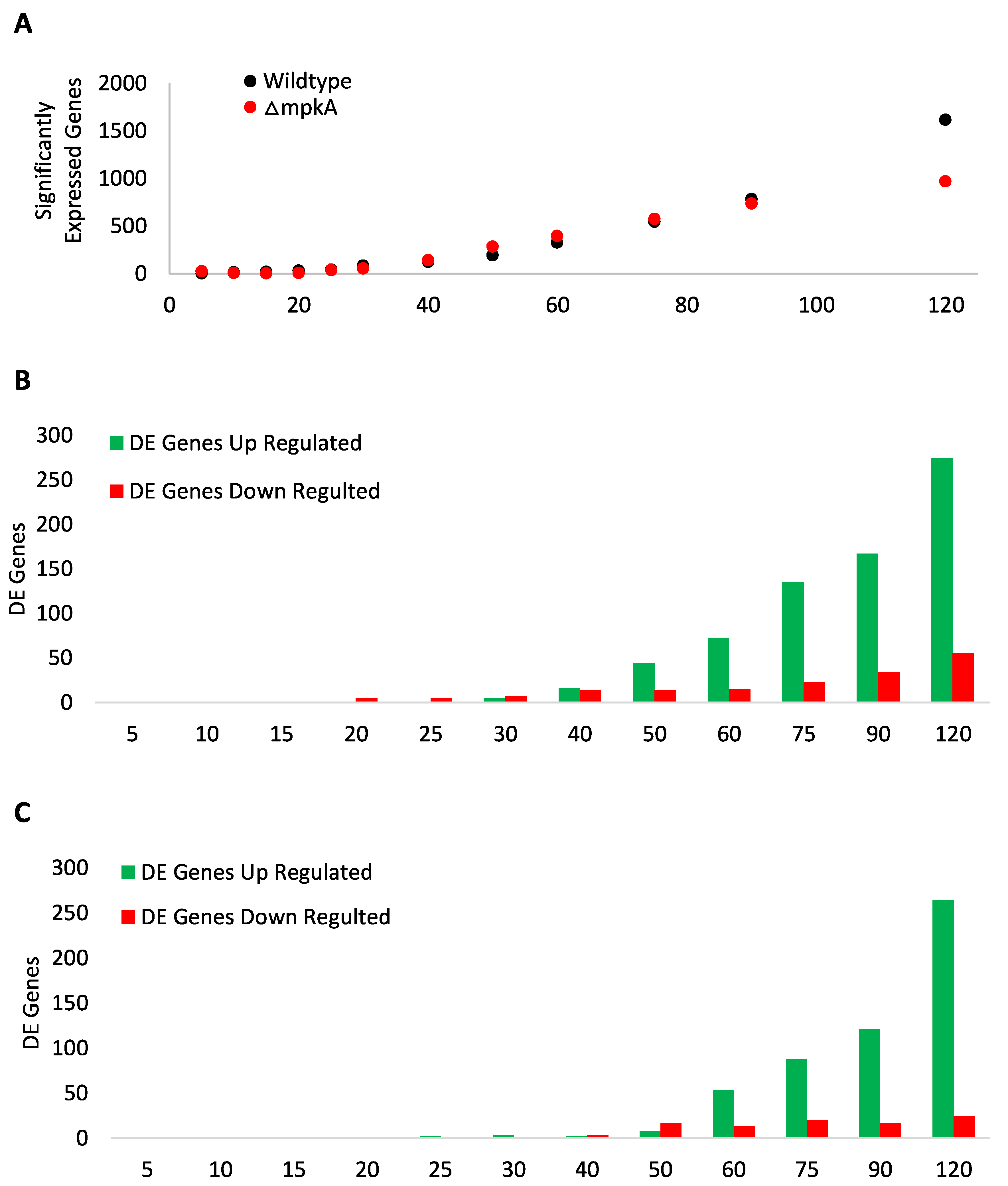
3.1. Global Transcript Expression
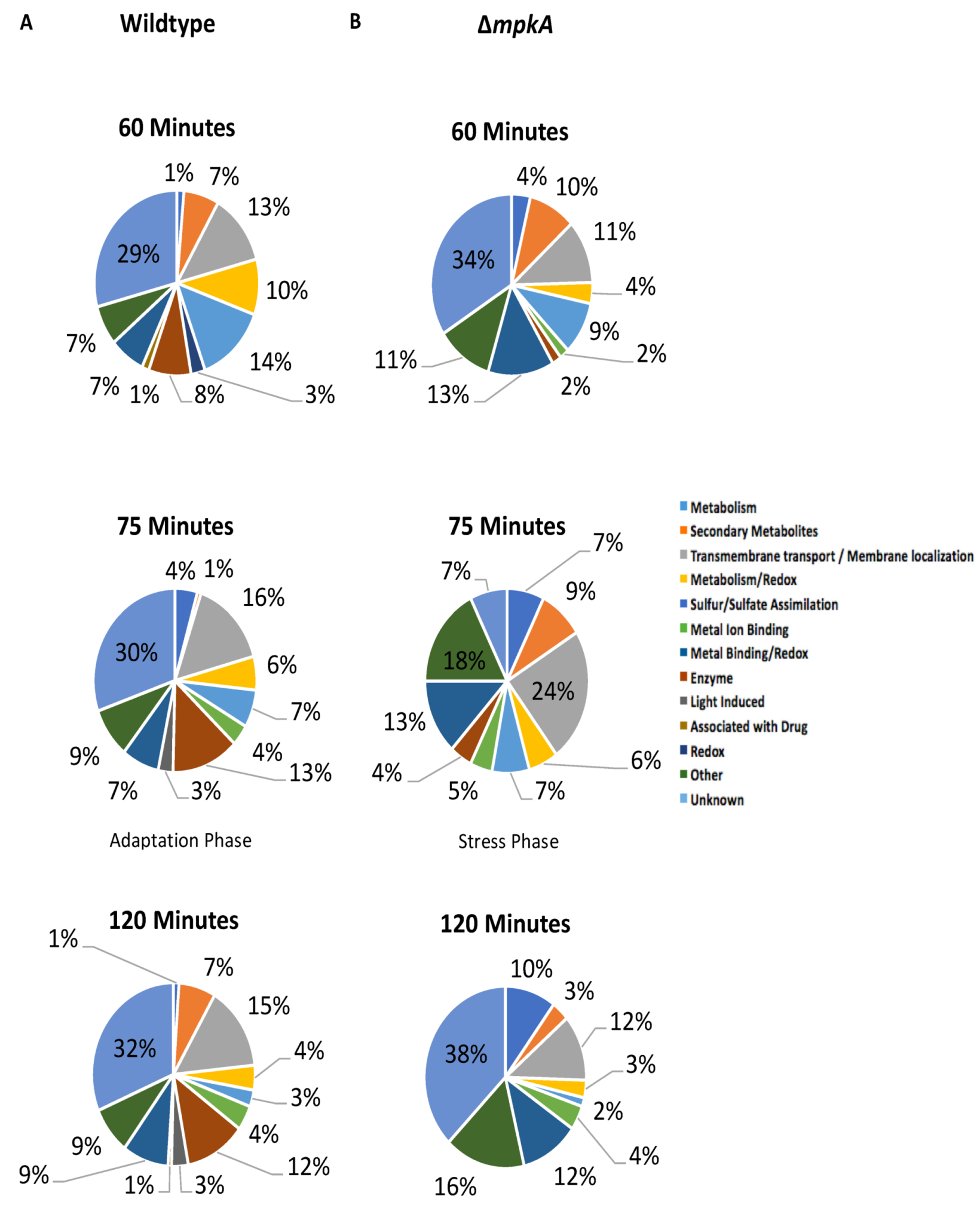
3.2. GO Term Analysis
3.3. Light Induced Gene Induction
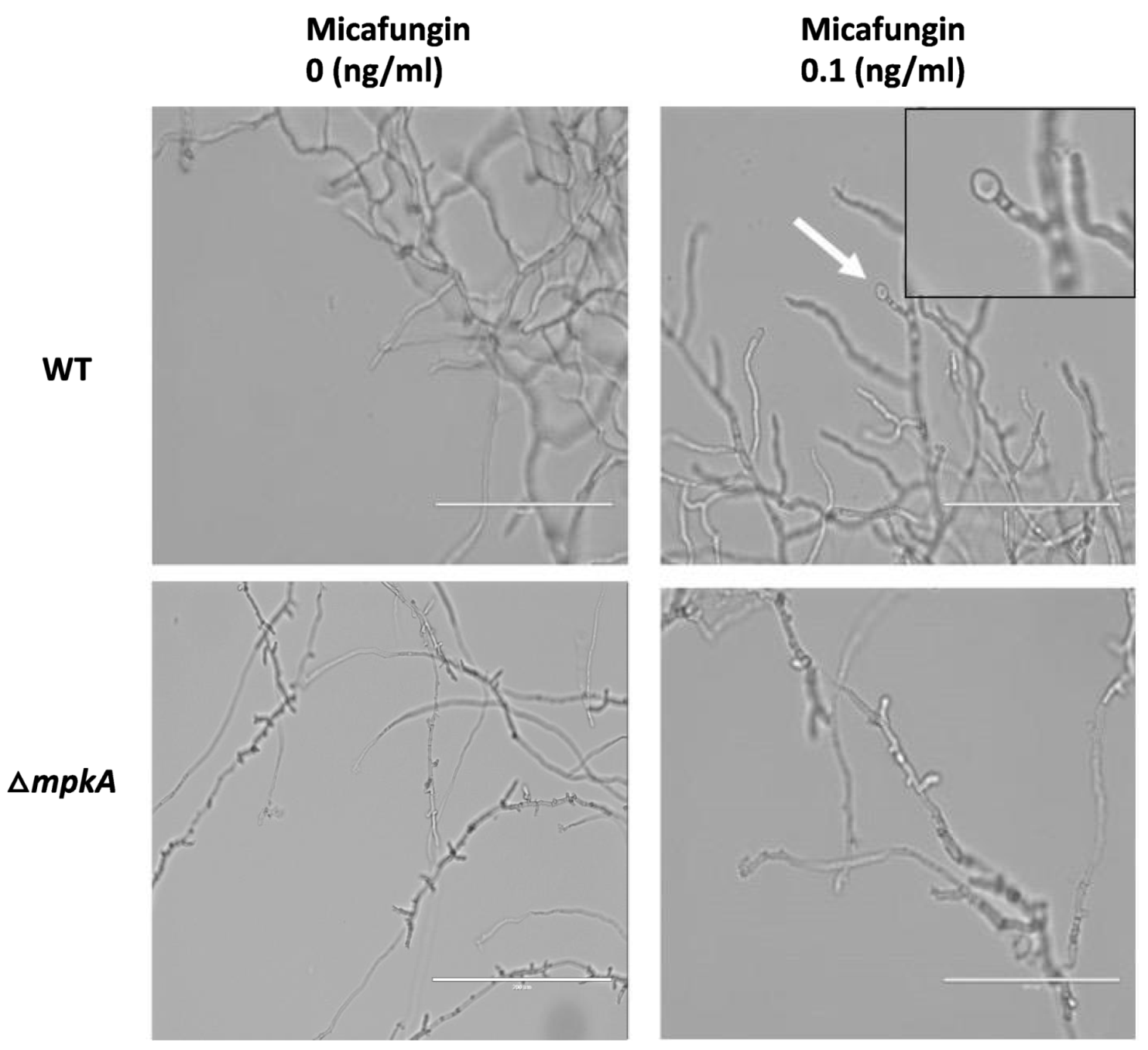
3.4. Hyphal Response to Micafungin Exposure
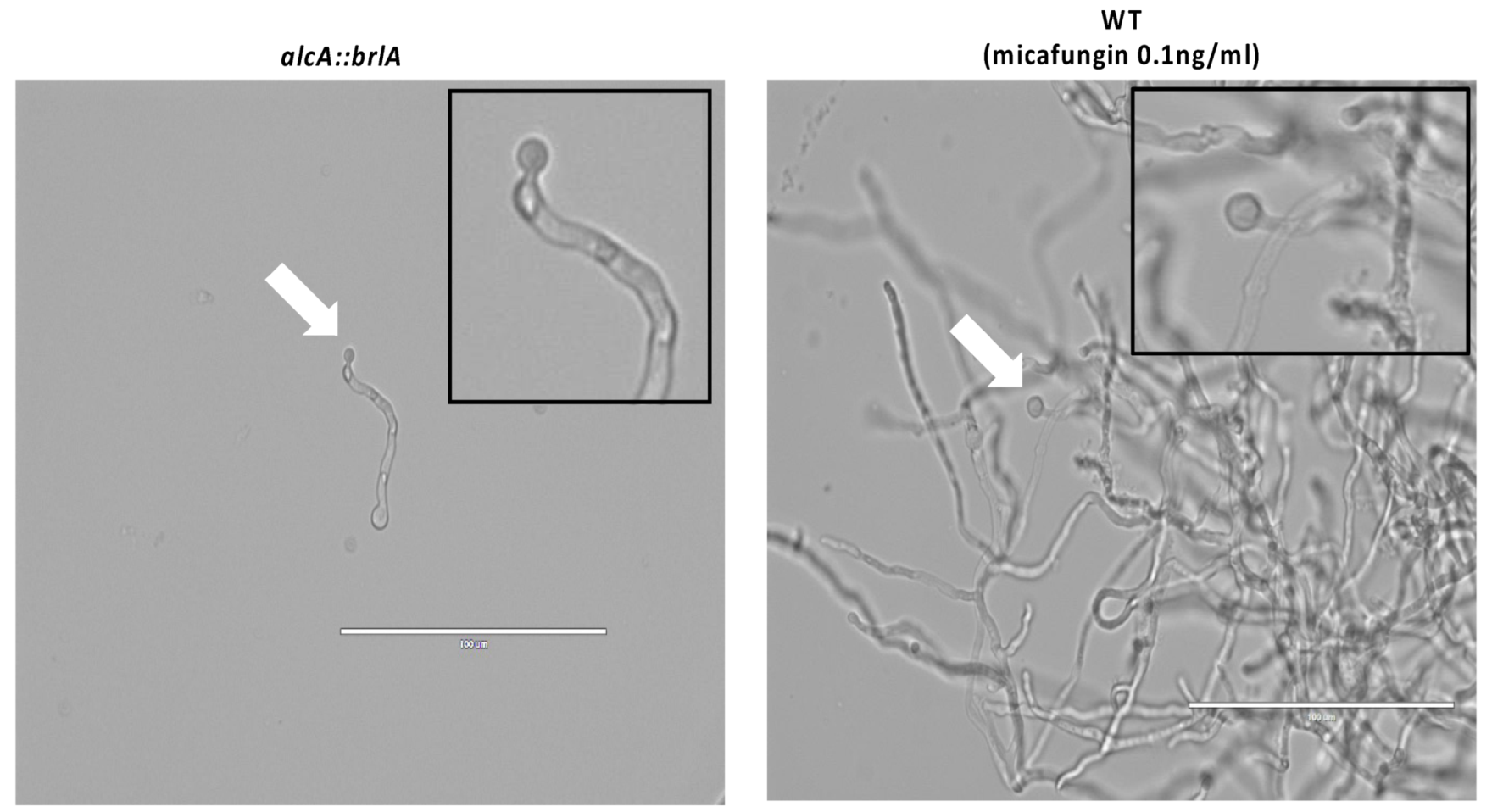
3.5. Similar Spore Morphologies upon Induced brlA Expression and Post Micafungin Exposure
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gow, N.A.; Latge, J.P.; Munro, C.A. The fungal cell wall: Structure, biosynthesis, and function. Fungal Kingd. 2017, 267–292. [Google Scholar] [CrossRef]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, T.; Mizutani, O.; Furukawa, K.; Sato, N.; Yoshimi, A.; Yamagata, Y.; Nakajima, T.; Abe, K. MpkA-dependent and-independent cell wall integrity signaling in A. nidulans. Eukaryot. Cell 2007, 6, 1497–1510. [Google Scholar] [CrossRef]
- Walker, L.A.; Gow, N.A.; Munro, C.A. Fungal echinocandin resistance. Fungal Genet. Biol. 2010, 47, 117–126. [Google Scholar] [CrossRef]
- Lima, S.L.; Colombo, A.L.; Almeida, J.N., Jr. Fungal cell wall: Emerging antifungals and drug resistance. Front. Microbiol. 2019, 10, 2573. [Google Scholar] [CrossRef]
- Chelius, C.; Huso, W.; Reese, S.; Doan, A.; Lincoln, S.; Lawson, K.; Tran, B.; Purohit, R.; Glaros, T.; Srivastava, R.; et al. Dynamic transcriptomic and phosphoproteomic analysis during cell wall stress in A. nidulans. Mol. Cell. Proteom. 2020, 19, 1310–1329. [Google Scholar] [CrossRef]
- Cole, G.T. Models of cell differentiation in conidial fungi. Microbiol. Rev. 1986, 50, 95. [Google Scholar] [CrossRef]
- Nishi, O.; Sushida, H.; Higashi, Y.; Iida, Y. Epiphytic and endophytic colonisation of tomato plants by the entomopathogenic fungus Beauveria bassiana strain GHA. Mycology 2001, 12, 39–47. [Google Scholar] [CrossRef]
- Jung, B.; Kim, S.; Lee, J. Microcyle conidiation in filamentous fungi. Mycobiology 2014, 42, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.J.; Smith, J.E. The production of conidiophores and conidia by newly germinated conidia of Aspergillus niger (microcycle conidiation). Microbiology 1971, 69, 185–197. [Google Scholar] [CrossRef]
- Son, H.; Kim, M.G.; Min, K.; Lim, J.Y.; Choi, G.J.; Kim, J.C.; Chae, S.K.; Lee, Y.W. WetA is required for conidiogenesis and conidium maturation in the ascomycete fungus Fusarium graminearum. Eukaryot. Cell 2014, 13, 87–98. [Google Scholar] [CrossRef]
- Harris, S.D.; Morrell, J.L.; Hamer, J.E. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 1994, 136, 517–532. [Google Scholar] [CrossRef] [PubMed]
- Kaminskyj, S.W. Septum position is marked at the tip of A. nidulans hyphae. Fungal Genet. Biol. 2000, 31, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Adams, T.H.; Boylan, M.T.; Timberlake, W.E. brlA is necessary and sufficient to direct conidiophore development in A. Nidulans. Cell 1988, 54, 353–362. [Google Scholar] [CrossRef]
- Si, H.; Rittenour, W.R.; Harris, S.D. Roles of A. nidulans Cdc42/Rho GTPase regulators in hyphal morphogenesis and development. Mycologia 2016, 108, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- Chelius, C.L.; Ribeiro, L.F.; Huso, W.; Kumar, J.; Lincoln, S.; Tran, B.; Goo, Y.A.; Srivastava, R.; Harris, S.D.; Marten, M.R. Phosphoproteomic and transcriptomic analyses reveal multiple functions for Aspergillus nidulans MpkA independent of cell wall stress. Fungal Genet. Biol. 2019, 125, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, M.B.; Cerqueira, G.C.; Inglis, D.O.; Skrzypek, M.S.; Binkley, J.; Chibucos, M.C.; Crabtree, J.; Howarth, C.; Orvis, J.; Shah, P.; et al. The Aspergillus Genome Database (AspGD): Recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 2002, 40, D653–D659. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, G.C.; Arnaud, M.B.; Inglis, D.O.; Skrzypek, M.S.; Binkley, G.; Simison, M.; Miyasato, S.R.; Binkley, J.; Orvis, J.; Shah, P.; et al. The Aspergillus Genome Database: Multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014, 42, D705–D710. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Navarro, J.; Greve, R.A.; Adams, T.H. Translational repression of brlA expression prevents premature development in Aspergillus. EMBO J. 1993, 12, 2449–2457. [Google Scholar] [CrossRef]
- Troppens, D.M.; Köhler, A.M.; Schlüter, R.; Hoppert, M.; Gerke, J.; Braus, G.H. Hülle Cells of A. nidulans with Nuclear Storage and Developmental Backup Functions Are Reminiscent of Multipotent Stem Cells. Mbio 2020, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Ahearn, D.G.; Price, D.; Simmons, R.B.; Mayo, A.; Zhang, S.T.; Crow, S.A., Jr. Microcycle conidiation and medusa head conidiophores of aspergilli on indoor construction materials and air filters from hospitals. Mycologia 2007, 99, 1–6. [Google Scholar] [CrossRef]
- Ruger-Herreros, C.; Rodríguez-Romero, J.; Fernández-Barranco, R.; Olmedo, M.; Fischer, R.; Corrochano, L.M.; Canovas, D. Regulation of conidiation by light in Aspergillus nidulans. Genetics 2011, 188, 809–822. [Google Scholar] [CrossRef]
- Suzuki, S.; Bayram, Ö.S.; Bayram, Ö.; Braus, G.H. conF and conJ contribute to conidia germination and stress response in the filamentous fungus Aspergillus nidulans. Fungal Genet. Biol. 2013, 56, 42–53. [Google Scholar] [CrossRef]
- Andersen, M.R.; Nielsen, J.B.; Klitgaard, A.; Petersen, L.M.; Zachariasen, M.; Hansen, T.J.; Blicher, L.H.; Gotfredsen, C.H.; Larsen, T.O.; Nielsen, K.F.; et al. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc. Natl. Acad. Sci. USA 2013, 110, E99–E107. [Google Scholar] [CrossRef]
- Valiante, V. The cell wall integrity signaling pathway and its involvement in secondary metabolite production. J. Fungi 2017, 3, 68. [Google Scholar] [CrossRef]
- Li, S.; Myung, K.; Guse, D.; Donkin, B.; Proctor, R.H.; Grayburn, W.S.; Calvo, A.M. FvVE1 regulates filamentous growth, the ratio of microconidia to macroconidia and cell wall formation in Fusarium verticillioides. Mol. Microbiol. 2006, 62, 1418–1432. [Google Scholar] [CrossRef]
- Gems, D.H.; Clutterbuck, A.J. Enhancers of conidiation mutants in Aspergillus nidulans. Genetics 1994, 137, 79–85. [Google Scholar] [CrossRef]
- Robbins, N.; Wright, G.D.; Cowen, L.E. Antifungal drugs: The current armamentarium and development of new agents. Fungal Kingd. 2016, 4, 4–5. [Google Scholar] [CrossRef] [PubMed]
- Alexander, B.D.; Johnson, M.D.; Pfeiffer, C.D.; Jiménez-Ortigosa, C.; Catania, J.; Booker, R.; Castanheira, M.; Messer, S.A.; Perlin, D.S.; Pfaller, M.A. Increasing echinocandin resistance in Candida glabrata: Clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin. Infect. Dis. 2013, 56, 1724–1732. [Google Scholar] [CrossRef] [PubMed]
- Ibe, C.; Munro, C.A. Fungal cell wall: An underexploited target for antifungal therapies. PLoS Pathog. 2001, 17, e1009470. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reese, S.; Chelius, C.; Riekhof, W.; Marten, M.R.; Harris, S.D. Micafungin-Induced Cell Wall Damage Stimulates Morphological Changes Consistent with Microcycle Conidiation in Aspergillus nidulans. J. Fungi 2021, 7, 525. https://doi.org/10.3390/jof7070525
Reese S, Chelius C, Riekhof W, Marten MR, Harris SD. Micafungin-Induced Cell Wall Damage Stimulates Morphological Changes Consistent with Microcycle Conidiation in Aspergillus nidulans. Journal of Fungi. 2021; 7(7):525. https://doi.org/10.3390/jof7070525
Chicago/Turabian StyleReese, Samantha, Cynthia Chelius, Wayne Riekhof, Mark R. Marten, and Steven D. Harris. 2021. "Micafungin-Induced Cell Wall Damage Stimulates Morphological Changes Consistent with Microcycle Conidiation in Aspergillus nidulans" Journal of Fungi 7, no. 7: 525. https://doi.org/10.3390/jof7070525





