Use of Wheat Straw for Value-Added Product Xylanase by Penicillium chrysogenum Strain A3 DSM105774
Abstract
1. Introduction
2. Materials and Methods
2.1. Wheat Straw
2.2. Reagents and Chemicals
2.3. Media
2.4. Isolation of Xylanase-Producing Fungal Strains
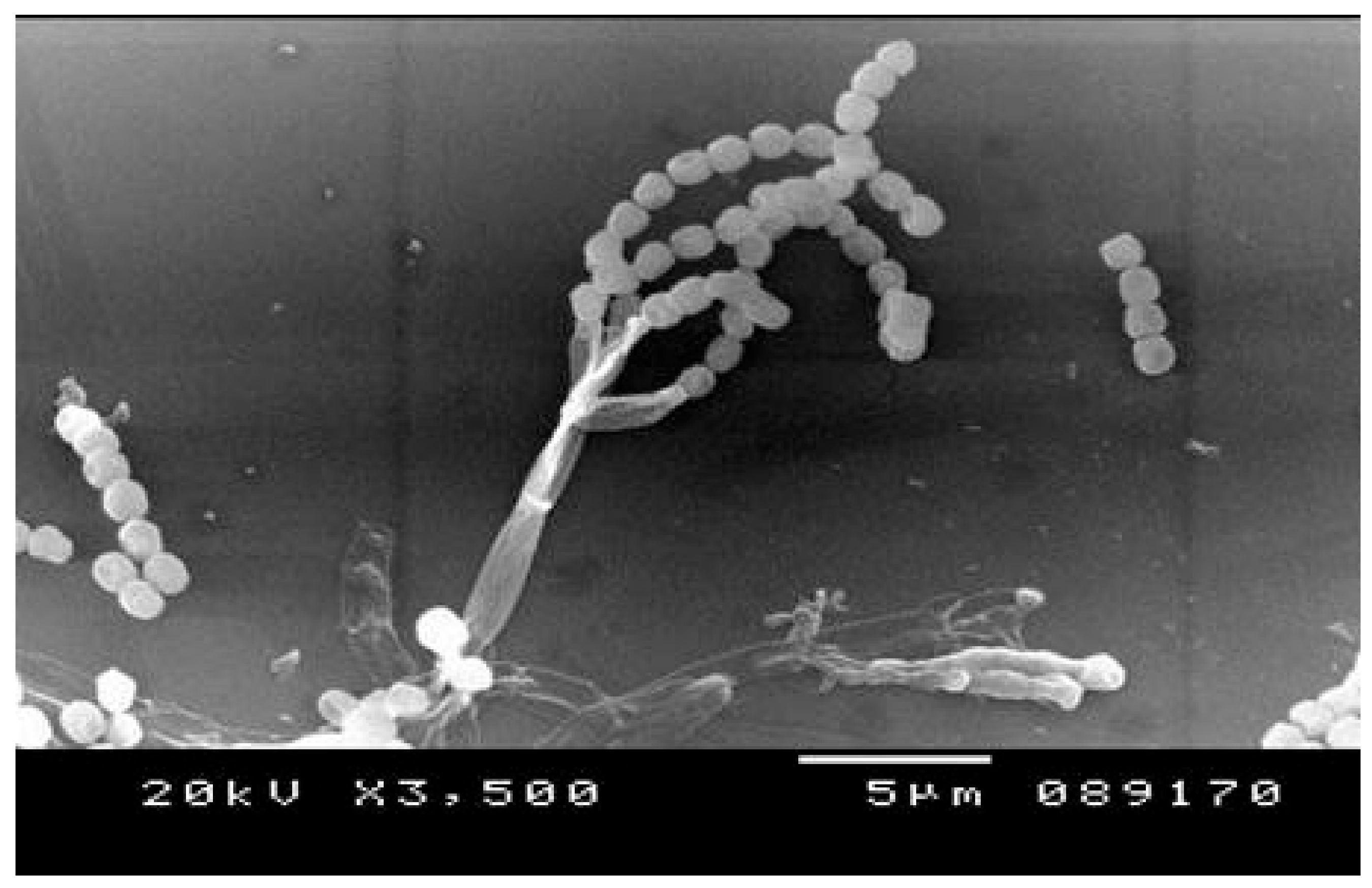
2.5. Scnnaing Electron Microscopy
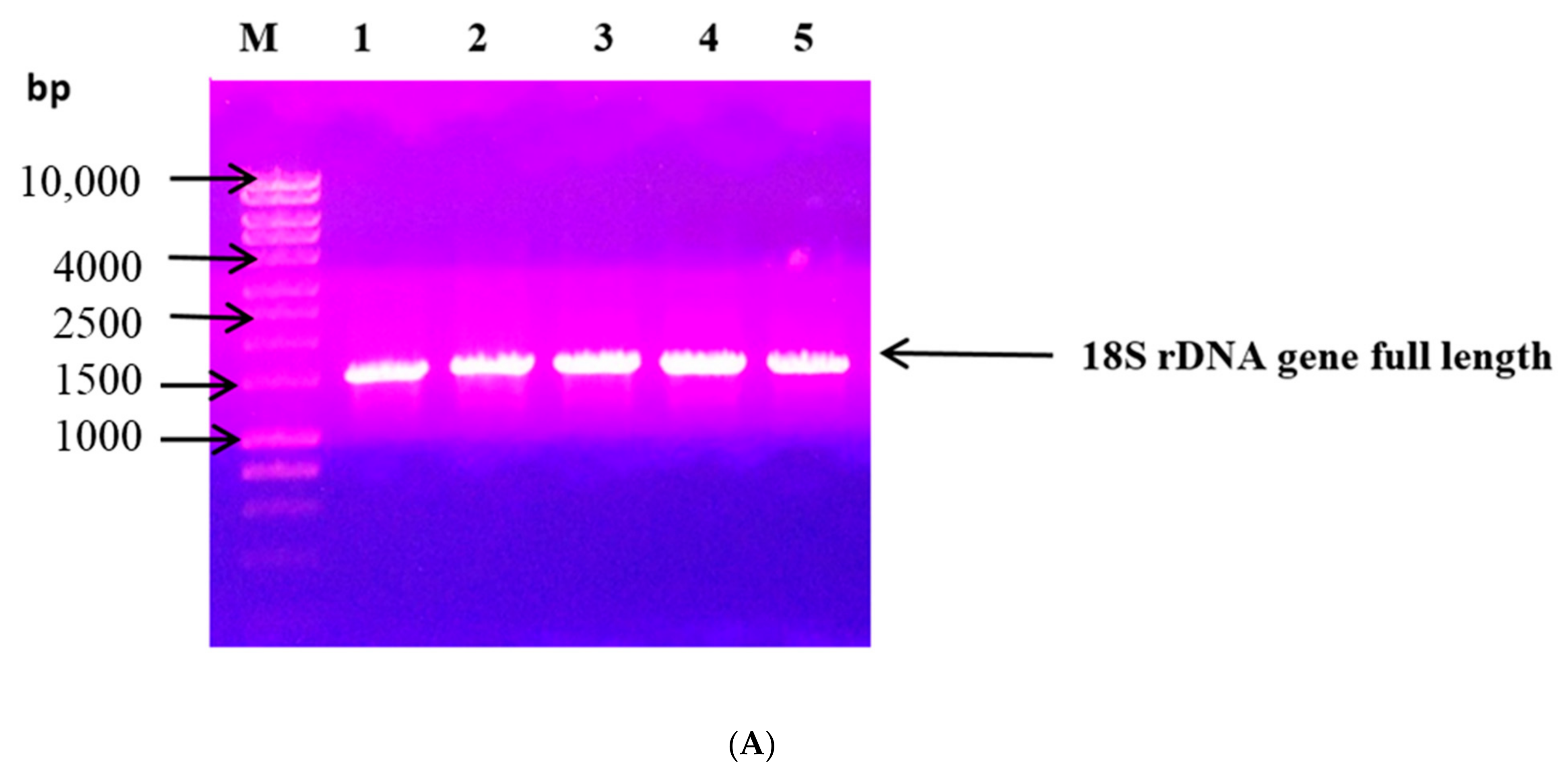
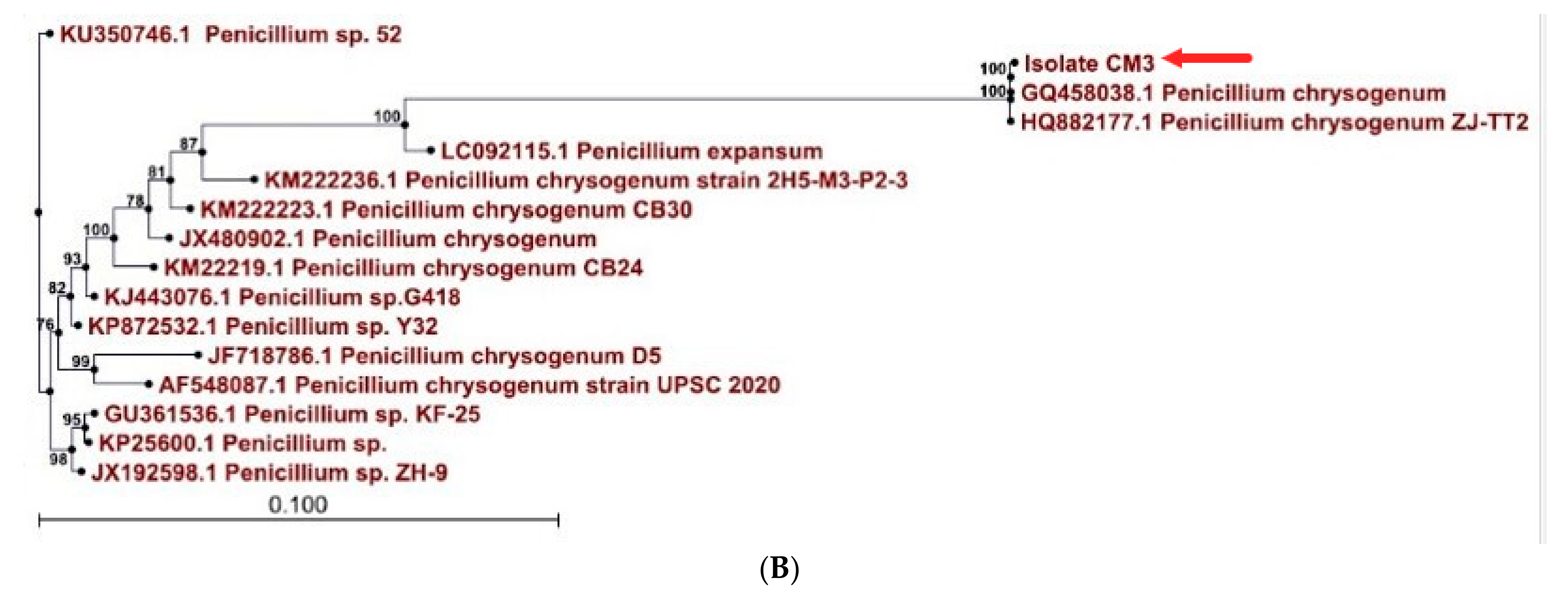
2.6. 18S rDNA Sequencing Approach
2.7. Xylanase Assay
2.8. Optimization of Xylanase Production
2.9. One Factor at a Time (OFAT)
2.10. Plackett–Burman Design (PBD)
2.11. Box–Behnken Design (BBD)
2.12. Statistical Analyses and Softwares
3. Results
3.1. Penicillium Chrysogenum Strain A3 DSM105774
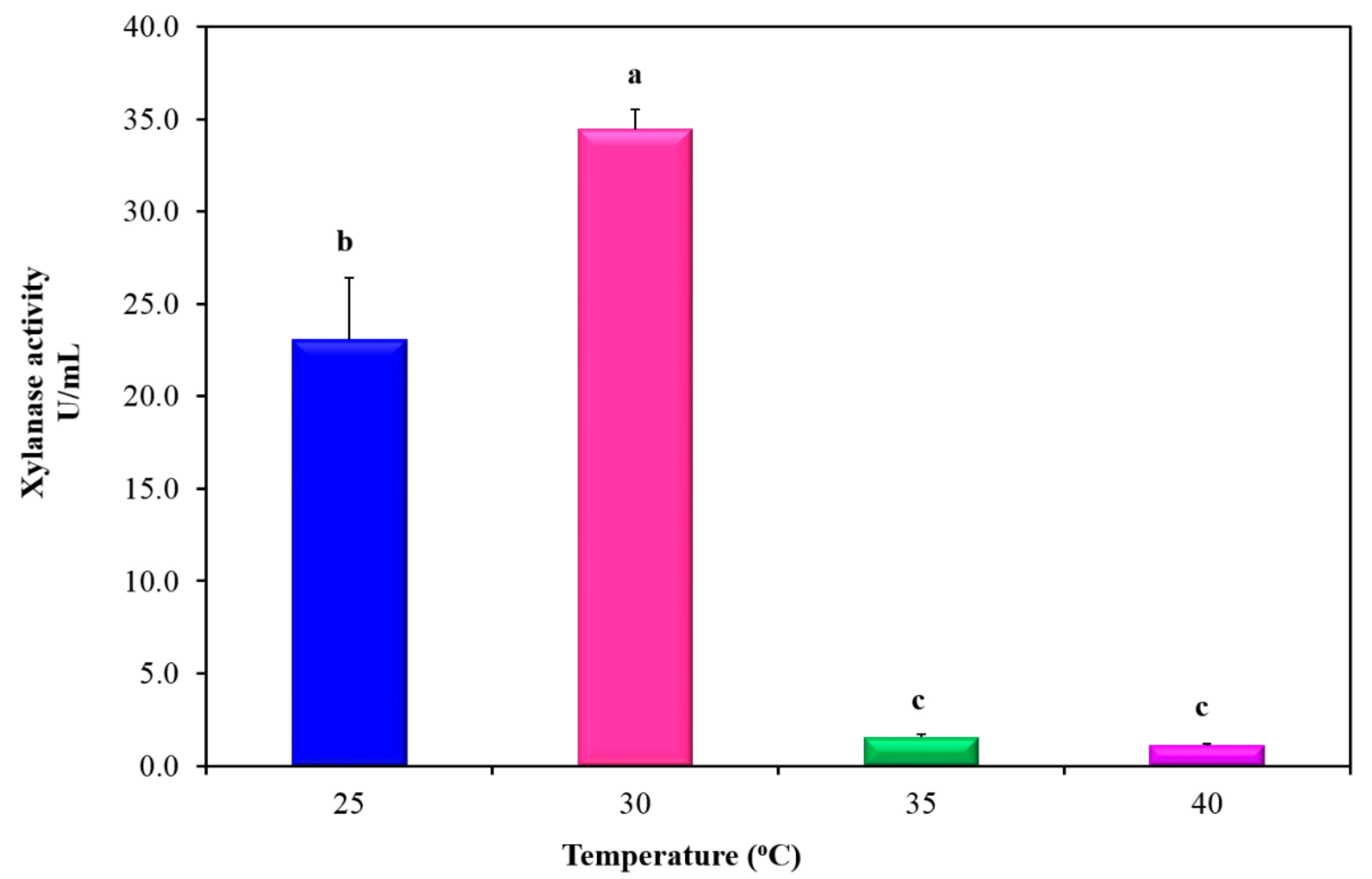
3.2. Key Factors Directing Xylanase Production
3.3. Screening of Key Factors Influencing Xylanase Production Using PBD
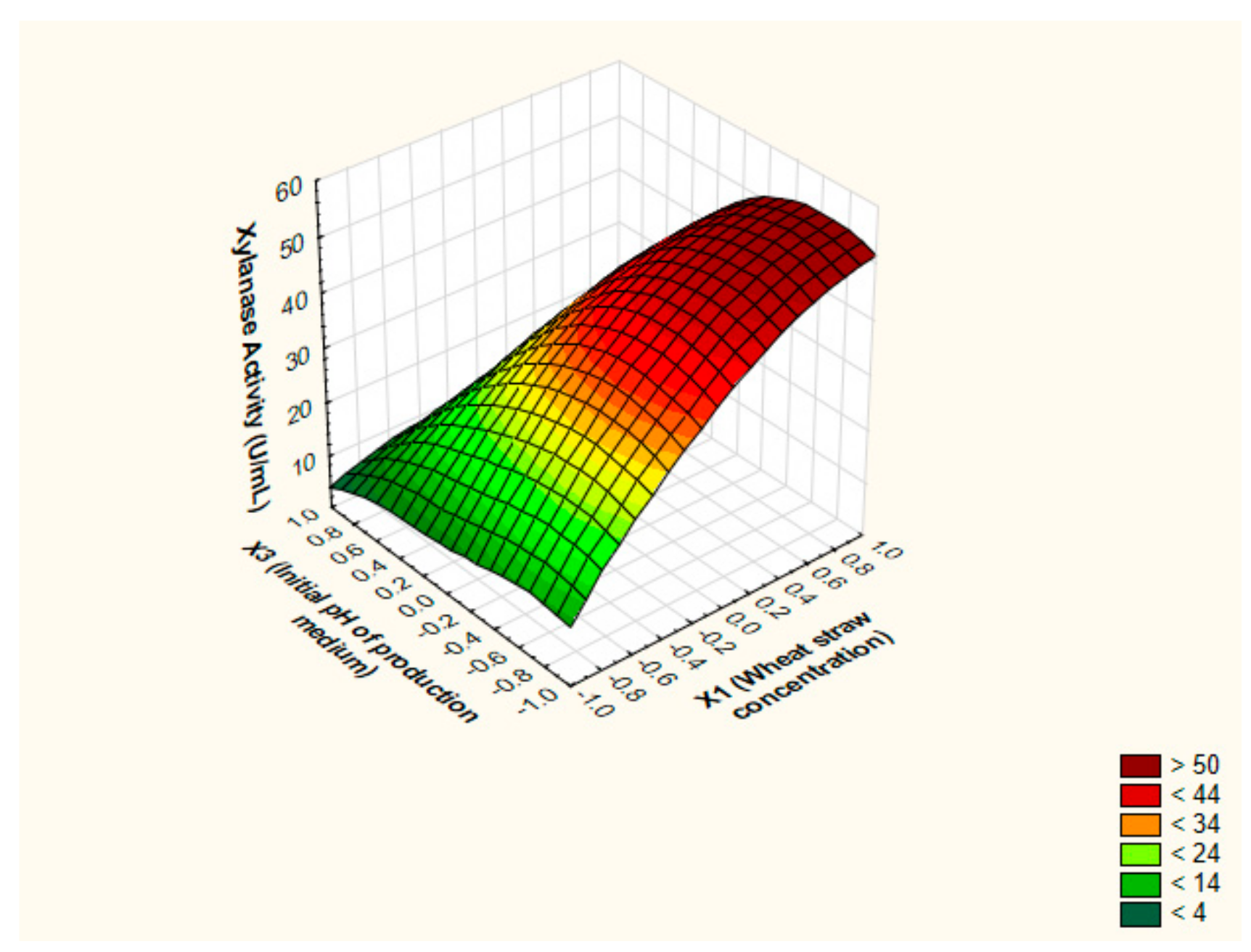
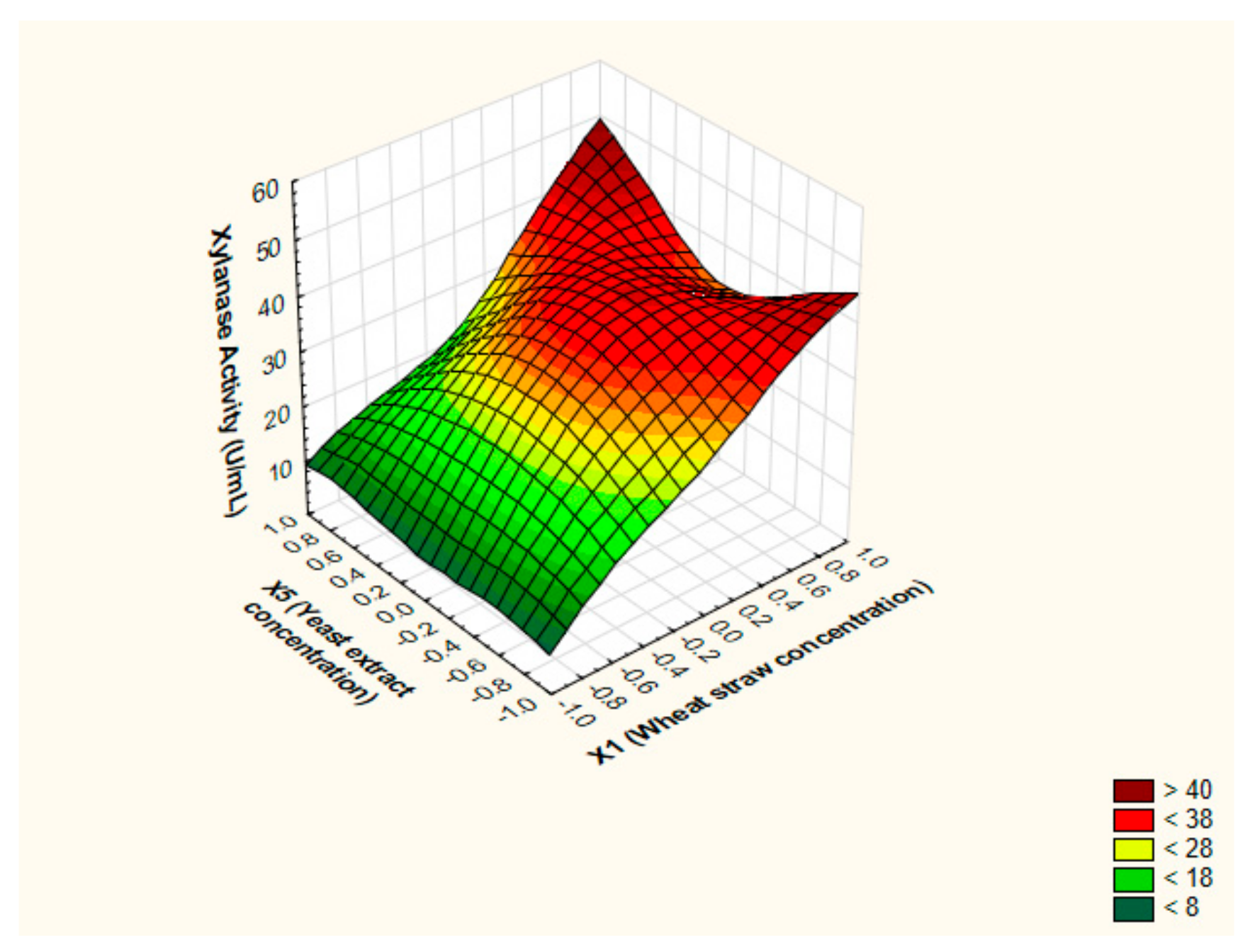
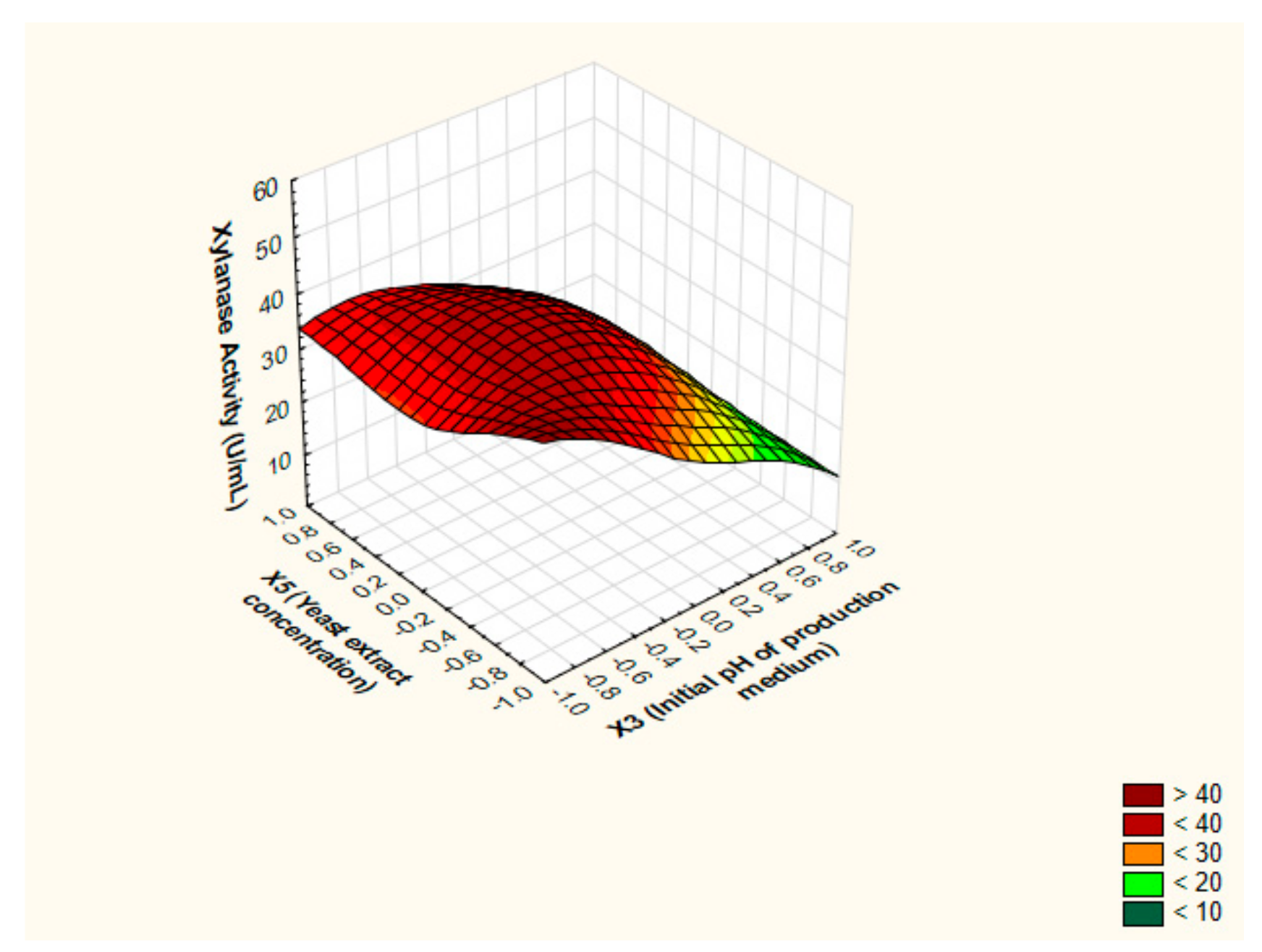
3.4. Pinpointing the Optimal Values of the Key Factors Switching the Production of Xylanase by BBD
3.5. Canonical and Ridge Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Anwar, Z.; Gulfraz, M.; Irshad, M. Agro-industrial lignocellulosic biomass a key to unlock the future bio-energy: A brief review. J. Radiat. Res. Appl. Sc. 2014, 7, 163–173. [Google Scholar] [CrossRef]
- Gubitz, G.M.; Mansfield, S.D.; Bohm, D.; Saddler, J.N. Effect of endoglucanases and hemicelluloses in magnetic and floatation deinking of xerographic and laser printed papers. J. Biotechnol. 1998, 65, 209–215. [Google Scholar] [CrossRef]
- McKendry, P. Energy production from biomass (part 1): Overview of biomass. Bioresour. Technol. 2002, 83, 37–46. [Google Scholar] [CrossRef]
- Yasin, M.; Bhutto, A.W.; Bazmi, A.A.; Karim, S. Efficient utilization of rice-wheat straw to produce value added composite products. Int. J. Chem. Env. Eng. 2010, 1, 136–143. [Google Scholar]
- Adapa, P.K.; Tabil, L.G.; Schoenau, G.J.; Canam, T.; Dumonceaux, T. Quantitative analysis of lignocellulosic components of non-treated and steam exploded barley, canola, oat and wheat straw using fourier transform infrared spectroscopy. J. Agric. Sci. Technol. 2011, B1, 177–188. [Google Scholar]
- WASDE. World Agricultural Supply and Demand Estimates, 2010; World Agricultural Outlook Board; United States Department of Agriculture: North Bend, WA, USA, 2010. [Google Scholar]
- Drankham, K.; Carter, J.; Madl, R.; Klopfenstein, C.; Padula, F.; Lu, Y.; Warren, T.; Schmitz, N.; Takemoto, D. Antitumor activity of wheats with high orthophenolic content. Nutr. Cancer 2003, 47, 188–194. [Google Scholar] [CrossRef]
- Mojsov, K. Application of solid state fermentation for cellulase enzyme production using Trichoderma viride. Perspect. Innov. Econ. Bus. 2010, 5, 108–110. [Google Scholar] [CrossRef][Green Version]
- Murray, T.D.; Bruehl, G.W. Composition of wheat straw infested with Cephalosporium gramineum and implications for its decomposition in soil. Phytopathology 1983, 73, 1046–1048. [Google Scholar] [CrossRef]
- Goodall, C. Biochar: Ten Technologies to Save the Planet; Greystone Books: Vancouver, BC, Canada, 2010. [Google Scholar]
- CWC and Domtar Inc. Wheat Straw as a Fiber Source: Recycling Technology Assistance Partnership (RTAP); A Program of the Clean Washington Center; Clean Washington Center: Seattle, WA, USA, 1997. [Google Scholar]
- Dupont, L.; Bounanda, J.; Domonceau, J.; Aplincourt, M. Metal ions binding onto a lignocellulosic substrate extracted from wheat bran: A NICA-donnon approach. J. Colloid Interface Sci. 2003, 263, 35–41. [Google Scholar] [CrossRef]
- Al-Kindi, S.; Abed, R.M. Effect of Biostimulation using sewage sludge, soybean meal, and wheat straw on oil degradation and bacterial community composition in a contaminated desert soil. Front. Microbiol. 2016, 7, 240. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Agrawal, R.; Singhania, R.R.; Satlewal, A.; Mathur, A.; Tuli, D.; Adsul, M. Untreated wheat straw: Potential source for diverse cellulolytic enzyme secretion by Penicillium janthinellum EMS-UV-8 mutant. Biores. Tech. 2015, 196, 518–524. [Google Scholar] [CrossRef]
- Gao, H.; Chu, X.; Wang, Y.; Zhou, F.; Zhao, K.; Mu, Z.; Liu, Q. Media optimization for laccase production by Trichoderma harzianum ZF-2 using response surface methodology. J. Microbiol. Biotechnol. 2013, 23, 1757–1764. [Google Scholar] [CrossRef]
- Balkan, B.; Ertan, F. Production of alpha amylase from Penicillium chrysogenum under solid-state fermentation by using some agricultural by-products. Food Technol. Biotechnol. 2007, 45, 439–442. [Google Scholar]
- Yang, S.Q.; Yan, Q.J.; Jiang, Z.Q.; Li, L.T.; Tian, H.M.; Wang, Y.Z. High-level of xylanase production by the thermophilic Paecilomyces themophila J18 on wheat straw in solid-state fermentation. Biores. Technol. 2006, 97, 1794–1800. [Google Scholar] [CrossRef] [PubMed]
- Michelin, M.; Maria de Lourdes, T.M.; Ruzene, D.S.; Silva, D.P.; Vicente, A.A.; Jorge, J.A.; Terenzi, H.F.; Teixeira, J.A. Xylanase and β-xylosidase Production by Aspergillus ochraceus: New perspectives for the application of wheat straw autohydrolysis liquor. Appl. Biochem. Biotechnol. 2012, 166, 336–347. [Google Scholar] [CrossRef]
- Azzouz, Z.; Bettache, A.; Boucherba, N.; Prieto, A.; Martinez, M.J.; Benallaoua, S.; de Eugenio, L.I. Optimization of β-1,4-endoxylanase production by an Aspergillus niger strain growing on wheat straw and application in xylooligosaccharides production. Molecules 2021, 26, 2527. [Google Scholar] [CrossRef] [PubMed]
- Ninawe, S.; Kuhad, R.C. Bleaching of wheat straw-rich soda pulp with xylanase from a thermoalkalophilic Streptomyces cyaneus SN32. Biores. Technol. 2006, 97, 2291–2295. [Google Scholar] [CrossRef] [PubMed]
- Ali, U.F.; El-Dein, H.S.S. Production and partial purification of cellulase complex by Aspergillus niger and A. nidulans grown on water hyacinth blend. J. Appl. Sci. Res. 2008, 4, 875–891. [Google Scholar]
- Sonia, K.G.; Chadha, B.S.; Saini, H.S. Sorghum straw for xylanase hyper-production by Thermomyces lianuginosus (D2W3) under solid-state fermentation. Biores. Technol. 2005, 96, 1561–1569. [Google Scholar] [CrossRef]
- Matrawy, A.A.; Khalil, A.I.; Marey, H.S.; Embaby, A.M. Biovalorization of the raw agro-industrial waste rice husk through directed production of xylanase by Thermomyces lanuginosus strain A3-1 DSM 105773: A statistical sequential model. Biomass Conv. Bioref. 2020. [Google Scholar] [CrossRef]
- Rajoka, M.I.; Huma, T.; Khalid, A.M.; Latif, F. Kinetics of enhanced substrate consumption and endo-β-xylanase production by a mutant derivative of Humicola lanuginosa in solid-state fermentation. World J. Microbiol. Biotechnol. 2005, 21, 869–876. [Google Scholar] [CrossRef]
- Sarkar, N.; Aikat, K. Cellulose and xylanase production from rice straw by a locally isolated fungus Aspergillus fumigatus NITDGPKA3 under solid state fermentation-statistical optimization by response surface methodology. J. Technol. Innov. Renew. Energy 2012, 1, 54–62. [Google Scholar]
- Okafor, U.A.; Emezue, T.N.; Okochi, V.I.; Onyegeme-Okerenta, B.M.; NwodoChinedu, S. Xylanase production by Penicillium chrysogenum (PCL501) fermented on cellulosic wastes. Afr. J. Biochem. Res. 2007, 1, 48–53. [Google Scholar]
- Yang, S.; Wang, Y.; Jiang, Z.; Hua, C. Crystallization and preliminary X-ray analysis of a 1,3–1,4-β-glucanase from Paecilomyces thermophile. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008, 64, 754–756. [Google Scholar] [CrossRef]
- Biswas, R.; Sahai, V.; Mishra, S.; Bisaria, V.S. Bioprocess strategies for enhanced production of xylanase by Melanocarpus albomyces IITD3A on agro-residual extract. J. Biosci. Bioeng. 2010, 110, 702–708. [Google Scholar] [CrossRef]
- Goyal, M.; Kalra, E.L.; Sarren, V.K.; Soni, G. Xylanase production with xylan rich lignocellulosic wastes by a local soil isolate of Trichoderma viride. Braz. J. Microbiol. 2008, 39, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Fengjie, C.; Zhiqiang, L.; Yingying, X.; Hui, Z. Improvement of xylanase production by Penicillium oxalicum ZH-30 using response surface methodology. Enzym. Microb. Technol. 2007, 40, 1381–1388. [Google Scholar]
- Zahari, N.I.; Shah, U.K.M.; Asa’ari, A.Z.M.; Mohamad, R. Selection of potential fungi for production of cellulase-poor xylanase from rice straw. Bioresources 2016, 11, 1162–1175. [Google Scholar] [CrossRef][Green Version]
- Brown, J.A.; Collin, S.A.; Wood, T.M. Development of a medium for high cellulase, xylanase and fl-glucosidase production by a mutant strain (NTG III/6) of the cellulolytic fungus Penicillium pinophilum. Enzym. Microbl. Technol. 1987, 9, 355–360. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, R.; Zhang, Y. Production of lignocellulolytic enzymes by Trametes gallica and detection of polysaccharide hydrolase and laccase activities in polyacrylamide gels. J. Basic Microbiol. 2004, 44, 220–231. [Google Scholar] [CrossRef]
- Oliveira, L.; Porto, A.; Tambourgi, E. Production of Xylanase and Protease by Penicillium janthinellum CRC 87M-115 from Different Agricultural Wastes. Bioresour. Technol. 2006, 97, 862–867. [Google Scholar] [CrossRef]
- Sunkar, B.; Kannoju, B.; Bhukya, B. Optimized production of xylanase by Penicillium purpurogenum and ultrasound impact on enzyme kinetics for the production of monomeric sugars from pretreated corn cobs. Front. Microbiol. 2020, 11, 772. [Google Scholar] [CrossRef]
- Camassola, M.; Dillon, A.J.J. Production of cellulases and hemicellulases by Penicillium echinulatum grown on pretreated sugar cane bagasse and wheat bran in solid-state fermentation. Appl. Microbiol. 2007, 103, 2196–2204. [Google Scholar] [CrossRef] [PubMed]
- Querido, A.; Coelho, J.; Araújo, E.; Chaves-Alves, V. Partial purification and characterization of xylanase produced by Penicillium expansum. Braz. Arch. Biol. Technol. 2006, 49, 475–480. [Google Scholar] [CrossRef]
- Bakri, Y.; Jacques, P.; Thonart, P. Xylanase production by Penicillium canescens 10-10c in Solid-state fermentation. Appl. Biochem. Biotechnol. 2003, 105–108, 737–748. [Google Scholar] [CrossRef]
- Fusawat, P.; Rakariyatham, N. Potential of cellulase and xylanase production by fungal strains using corn husks as substrate. Asia Pac. J. Sci. Technol. 2014, 19, 229–234. [Google Scholar]
- Adeleke, M.A.; Akatah, H.A.; Hassan, A.O.; Adebimpe, W.O. Microbial load and multiple drug resistance of pathogenic bacteria isolated from feaces and body surfaces of cockroaches in an urban area of southwestern Nigeria. J. Microbiol. Biotechnol. Food Sci. 2012, 1, 1448. [Google Scholar]
- Smit, E.; Leeflang, P.; Glandorf, B.; Van Elsas, J.D.; Wernars, K. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 1999, 65, 2614–2621. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Plackett, R.L.; Burman, J.P. The design of optimum multifactorial experiments. Biometrika 1946, 33, 305–325. [Google Scholar] [CrossRef]
- Box, G.E.; Behnken, D.W. Some new three level designs for the study of quantitative variables. Technometrics 1960, 2, 455–475. [Google Scholar] [CrossRef]
- Myers, R.H. Response Surface Methodology; Edwards Brothers: Ann Arbor, MI, USA, 1976. [Google Scholar]
- Draper, N.R. Ridge analysis of response surfaces. Technometrics 1963, 5, 469–479. [Google Scholar]
- Khucharoenphaisan, K.; Tokuyama, S.; Kitpreechavanich, V. Statistical optimization of activity and stability of β-xylanase produced by newly isolated Thermomyces lanuginosus THKU-49 using central composite design. Afr. J. Biotechnol. 2008, 7, 3599–3602. [Google Scholar]
- Embaby, A.M.; Masoud, A.A.; Marey, H.S.; Shaban, N.Z.; Ghonaim, T.M. Raw agro-industrial orange peel waste as a low cost effective inducer for alkaline polygalacturonase production from Bacillus licheniformis SHG10. SpringerPlus 2014, 3, 327. [Google Scholar] [CrossRef] [PubMed]
- Anan, A.; Ghanem, K.M.; Embaby, A.M.; Hussein, A.; El-Naggar, M.Y. Statistically optimized ceftriaxone sodium biotransformation through Achromobacter xylosoxidans strain Cef6: Unusual insight for bioremediation. J. Basic Microbiol. 2018, 58, 120–130. [Google Scholar] [CrossRef]
- Embaby, A.M.; Hussein, M.N.; Hussein, A. Monascus orange and red pigments production by Monascus purpureus ATCC16436 through co-solid state fermentation of corn cob and glycerol: An eco-friendly environmental low cost approach. PLoS ONE 2018, 13, e0207755. [Google Scholar] [CrossRef] [PubMed]
- Embaby, A.M.; Melika, R.R.; Hussein, A.; El-Kamel, A.; Marey, H.S. A Novel non-cumbersome approach towards biosynthesis of pectic-oligosaccharides by non-aflatoxigenic Aspergillus sp. Section Flavi Strain EGY1 DSM 101520 through citrus pectin fermentation. PLoS ONE 2016, 11, e0167981. [Google Scholar] [CrossRef]
- Embaby, A.M.; Melika, R.R.; Hussein, A.; El-Kamel, A.H.; Marey, H.S. Biosynthesis of chitosan-oligosaccharides (COS) by non-aflatoxigenic Aspergillus sp. strain EGY1 DSM 101520: A robust biotechnological approach. Process. Biochem. 2018, 64, 16–30. [Google Scholar] [CrossRef]
- Embaby, A.M.; Marey, H.S.; Hussein, A. A statistical-mathematical model to optimize chicken feather waste bioconversion via Bacillus licheniformis SHG10: A low cost effective and ecologically safe approach. J. Bioprocess Biotech. 2015, 5, 1. [Google Scholar]
- Terrone, C.; Freitas, C.; Fanchini Terrasan, C.R.; Almeida, A.L.; Carmona, E. Agroindustrial biomass for xylanase production by Penicillium chrysogenum: Purification, biochemical properties, and hydrolysis of hemicelluloses. Electron. J. Biotechnol. 2018, 33, 39–45. [Google Scholar] [CrossRef]
- Krishna, C. Solid-state fermentation systems—An overview. Crit. Rev. Biotechnol. 2005, 25, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.P.; Ghosh, S. Optimization of xylanase production by Penicillium citrinum xym2 and application in saccharification of agro-residues. Biocatal. Agric. Biotechnol. 2014, 3, 188–196. [Google Scholar] [CrossRef]
- Knob, A.; Beitel, S.M.; Fortkamp, D.; Terrasan, C.R.F.; Almeida, A.F.D. Production, purification, and characterization of a major Penicillium glabrum xylanase using brewer’s spent grain as substrate. BioMed Res. Int. 2013, 2013, 728–735. [Google Scholar] [CrossRef]
- Mogk, A.; Mayer, M.P.; Deuerling, E. Mechanisms of protein folding: Molecular chaperones and their application in biotechnology. Chembiochem 2002, 3, 807–814. [Google Scholar] [CrossRef]
- Kumar, A.; Gautam, A.; Dutt, D. Co-Cultivation of Penicillium sp. AKB-24 and Aspergillus nidulans AKB-25 as a cost-effective method to produce cellulases for the hydrolysis of pearl millet stover. Fermentation 2016, 2, 12. [Google Scholar] [CrossRef]
- Bagewadi, Z.K.; Mulla, S.I.; Shouche, Y.; Ninnekar, H.Z. Xylanase production from Penicillium citrinum isolate HZN13 using response surface methodology and characterization of immobilized xylanase on glutaraldehyde-activated calcium-alginate beads. 3 Biotech 2016, 6, 164. [Google Scholar] [CrossRef]
- Cui, F.; Zhao, L. Optimization of xylanase production from Penicillium sp. WX-Z1 by a two-step statistical strategy: Plackett-Burman and Box-Behnken experimental design. Int. J. Mol. Sci. 2012, 13, 10630–10646. [Google Scholar] [CrossRef]
- Sridevi, A.; Golla, N.; Suvarnalatha, P. Production of xylanase by Penicillium sp. and its biobleaching efficiency in paper and pulp industry. Int. J. Pharm. Sci. Res. 2019, 10, 1307–1311. [Google Scholar]
- Khalil., A.I.; Krakowiak, A.; Russel, S. Production of extracellular cellulase and xylanase by the ligninolytic white-rot fungus Trametes versicolor grown on agricultural wastes. Ann. Agric. Sci. 2002, 47, 161–173. [Google Scholar]

| (a) | |||||||
| Independent Factors | Y Xylanase Activity (U/mL) | ||||||
| Trial # | X1 | X2 | X3 | X4 | X5 | Exp. a | Pred. b |
| 1 | 1(1.0) | −1(0.065) | 1(7.0) | −1(4.0) | −1(0.025) | 14.43 | 14.39 |
| 2 | 1(1.0) | 1(0.25) | −1(5.0) | 1(8.0) | −1(0.025) | 15.00 | 15.54 |
| 3 | −1(0.25) | 1(0.25) | 1(7.0) | −1(4.0) | 1(0.15) | 6.54 | 5.92 |
| 4 | 1(1.0) | −1(0.065) | 1(7.0) | 1(8.0) | −1(0.025) | 13.47 | 13.98 |
| 5 | 1(1.0) | 1(0.25) | −1(5.0) | 1(8.0) | 1(0.15) | 14.32 | 13.04 |
| 6 | 1(1.0) | 1(0.25) | 1(7.0) | −1(4.0) | 1(0.15) | 9.63 | 11.98 |
| 7 | −1(0.25) | 1(0.25) | 1(7.0) | 1(8.0) | −1(0.025) | 9.39 | 7.99 |
| 8 | −1(0.25) | −1(0.065) | 1(7.0) | 1(8.0) | 1(0.15) | 6.21 | 5.42 |
| 9 | −1(0.25) | −1(0.065) | −1(5.0) | 1(8.0) | 1(0.15) | 4.47 | 6.89 |
| 10 | 1(1.0) | −1(0.065) | −1(5.0) | −1(4.0) | 1(0.15) | 15.45 | 13.38 |
| 11 | −1(0.25) | 1(0.25) | −1(5.0) | −1(4.0) | −1(0.025) | 9.48 | 9.89 |
| 12 | −1(0.25) | −1(0.065) | −1(5.0) | −1(4.0) | −1(0.025) | 9.84 | 9.81 |
| (b) | |||||||
| Independent Factor | Estimate Symbol | Estimate | t-Value | p-Value | Confidence Level (%) | ||
| Intercept | B° | 10.68575 | 34.2592 | 7.64 × 10−18 | 100.00 * | ||
| X1 | B1 | 3.030083 | 9.714641 | 1.39 × 10−8 | 99.99 * | ||
| X2 | B2 | 0.040083 | 0.12851 | 0.899171 | |||
| X3 | B3 | −0.74008 | −2.37275 | 0.029001 | 97.10 * | ||
| X4 | B4 | −0.20925 | −0.67087 | 0.510822 | |||
| X5 | B5 | −1.24925 | −4.00518 | 0.000830 | 99.91 * | ||
| (a) | |||||
| Trial # | X1 | X3 | X5 | Y6 Xylanase Activity (U/mL) | |
| Exp. a | Pred. b | ||||
| 1 | −1(0.1) | −1(4.5) | 0(0.056) | 10.77 | 8.45 |
| 2 | 1(1.9) | −1(4.5) | 0(0.056) | 51.42 | 56.52 |
| 3 | −1(0.1) | 1(9.5) | 0(0.056) | 3.84 | −1.26 |
| 4 | 1(1.9) | 1(9.5) | 0(0.056) | 12.03 | 14.35 |
| 5 | −1(0.1) | 0(7.0) | −1(0.012) | 7.26 | 13.32 |
| 6 | 1(1.9) | 0(7.0) | −1(0.012) | 45.48 | 44.13 |
| 7 | −1(0.1) | 0(7.0) | 1(0.1) | 9.45 | 10.80 |
| 8 | 1(1.9) | 0(7.0) | 1(0.1) | 49.74 | 43.68 |
| 9 | 0(1.0) | −1(4.5) | −1(0.012) | 43.50 | 39.75 |
| 10 | 0(1.0) | 1(9.5) | −1(0.012) | 11.13 | 10.17 |
| 11 | 0(1.0) | −1(4.5) | 1(0.1) | 33.66 | 34.62 |
| 12 | 0(1.0) | 1(9.5) | 1(0.1) | 8.58 | 12.33 |
| 13 | 0(1.0) | 0(7.0) | 0(0.56) | 37.47 | 38.45 |
| 14 | 0(1.0) | 0(7.0) | 0(0.56) | 38.82 | 38.45 |
| 15 | 0(1.0) | 0(7.0) | 0(0.56) | 39.06 | 38.45 |
| (b) | |||||
| Independent Factor | Estimate Symbol | Estimate | t-Value | p-Value | Confidence Level (%) |
| Intercept | B° | 38.45 | 22.75182 | 9.1 × 10−6 | 100.00 * |
| X1 | B1 | 15.92 | 15.38201 | 1.51 × 10−12 | 99.99 * |
| X3 | B2 | −12.97 | −12.5339 | 6.28 × 10−11 | 99.999 * |
| X5 | B3 | −0.74 | −0.71746 | 0.481384 | - |
| X1.X1 | B11 | −7.59 | −4.97925 | 7.21 × 10−5 | 99.99 * |
| X3.X3 | B22 | −11.35 | −7.45082 | 3.44 × 10−7 | 99.999 * |
| X5.X5 | B33 | −2.88 | −1.89225 | 0.073025 | - |
| X1.X3 | B12 | −8.11 | −5.5447 | 1.99 × 10−5 | 99.99 * |
| X1.X5 | B13 | 0.52 | 0.35359 | 0.727346 | - |
| X3.X5 | B23 | 1.82 | 1.245251 | 0.227433 | - |
| Distance (d) | Independent Factor * | Xylanase Activity (U/mL) | ||
|---|---|---|---|---|
| X1 | X3 | X5 | ||
| 0.0 | 0.000 | 0.000 | 0.000 | 38.46 |
| 0.1 | 0.078 | −0.062 | −0.004 | 40.44 |
| 0.3 | 0.237 | −0.184 | −1.093 | 44.16 |
| 0.5 | 0.397 | −0.184 | −0.030 | 47.46 |
| 0.7 | 0.559 | −0.184 | −0.049 | 50.37 |
| 0.9 | 0.722 | −0.533 | −0.072 | 52.86 |
| 1.1 | 0.884 | −0.647 | −0.101 | 54.99 |
| 1.3 ** | 1.046 | −0.759 | −0.137 | 56.7 |
| 1.5 | 1.208 | −0.871 | −0.179 | 58.05 |
| 1.7 | 1.368 | −0.982 | −0.230 | 58.98 |
| 1.9 | 1.527 | −1.093 | −0.290 | 59.55 |
| 0.0 | 0.000 | 0.000 | 0.000 | 38.46 |
| 0.1 | 0.078 | −0.062 | −0.004 | 40.44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matrawy, A.A.; Khalil, A.I.; Marey, H.S.; Embaby, A.M. Use of Wheat Straw for Value-Added Product Xylanase by Penicillium chrysogenum Strain A3 DSM105774. J. Fungi 2021, 7, 696. https://doi.org/10.3390/jof7090696
Matrawy AA, Khalil AI, Marey HS, Embaby AM. Use of Wheat Straw for Value-Added Product Xylanase by Penicillium chrysogenum Strain A3 DSM105774. Journal of Fungi. 2021; 7(9):696. https://doi.org/10.3390/jof7090696
Chicago/Turabian StyleMatrawy, Amira A., Ahmed I. Khalil, Heba S. Marey, and Amira M. Embaby. 2021. "Use of Wheat Straw for Value-Added Product Xylanase by Penicillium chrysogenum Strain A3 DSM105774" Journal of Fungi 7, no. 9: 696. https://doi.org/10.3390/jof7090696
APA StyleMatrawy, A. A., Khalil, A. I., Marey, H. S., & Embaby, A. M. (2021). Use of Wheat Straw for Value-Added Product Xylanase by Penicillium chrysogenum Strain A3 DSM105774. Journal of Fungi, 7(9), 696. https://doi.org/10.3390/jof7090696






