Complete Genomic Characterization and Identification of Saccharomycopsisphalluae sp. nov., a Novel Pathogen Causes Yellow Rot Disease on Phallus rubrovolvatus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pathogen Isolation and Maintenance
2.2. Pathogenicity Tests
2.3. Morphological, Physiological, and Molecular Characterization
2.4. Whole-Genome Sequencing and Assembly
2.5. Gene Prediction and Genome Annotation
2.6. Orthological, Phylogenetic Tree Construction
3. Results
3.1. Pathogen Isolation, Pathogenicity Tests and Identification
3.2. Features of the S. phalluae Genome
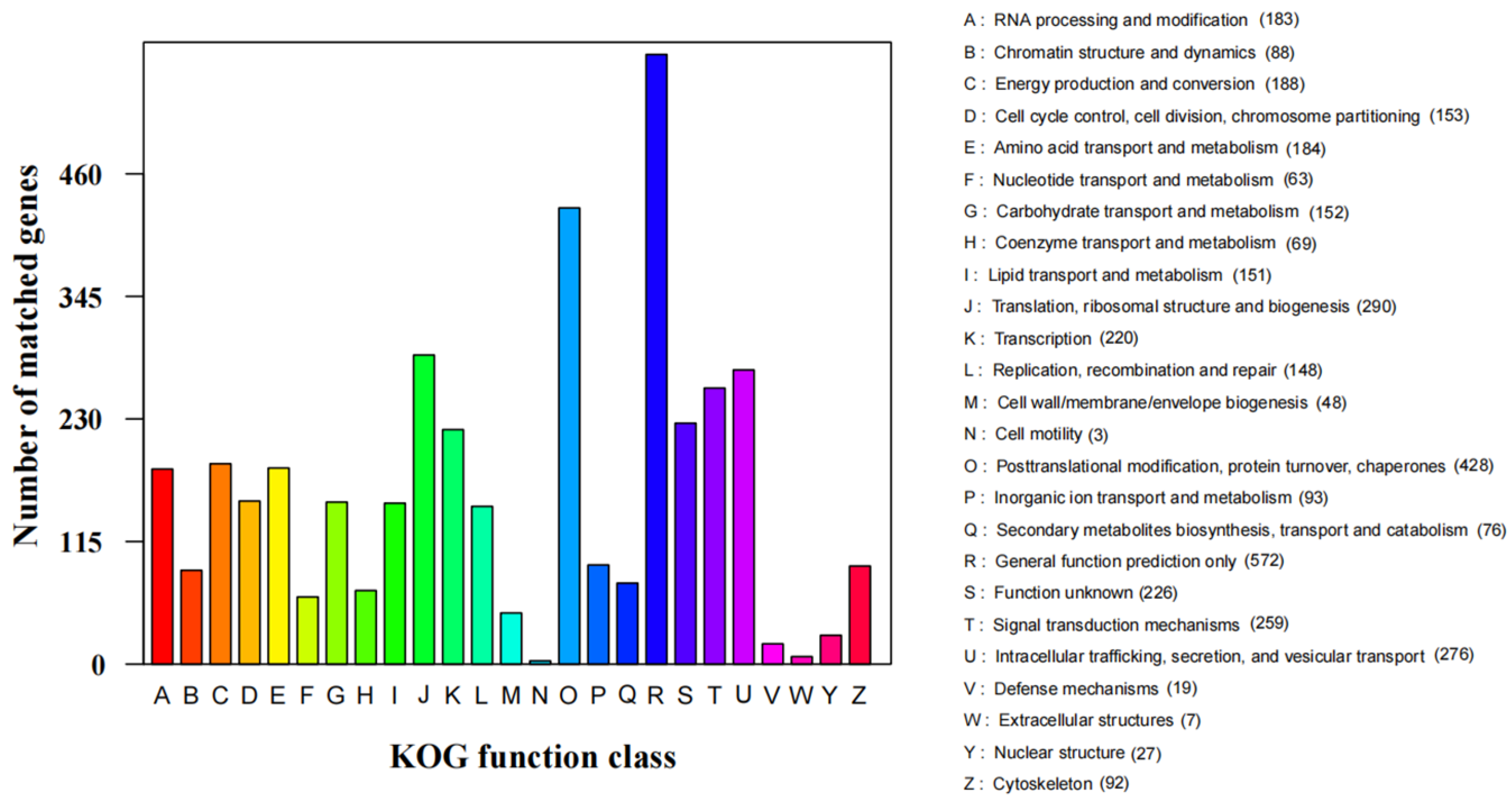
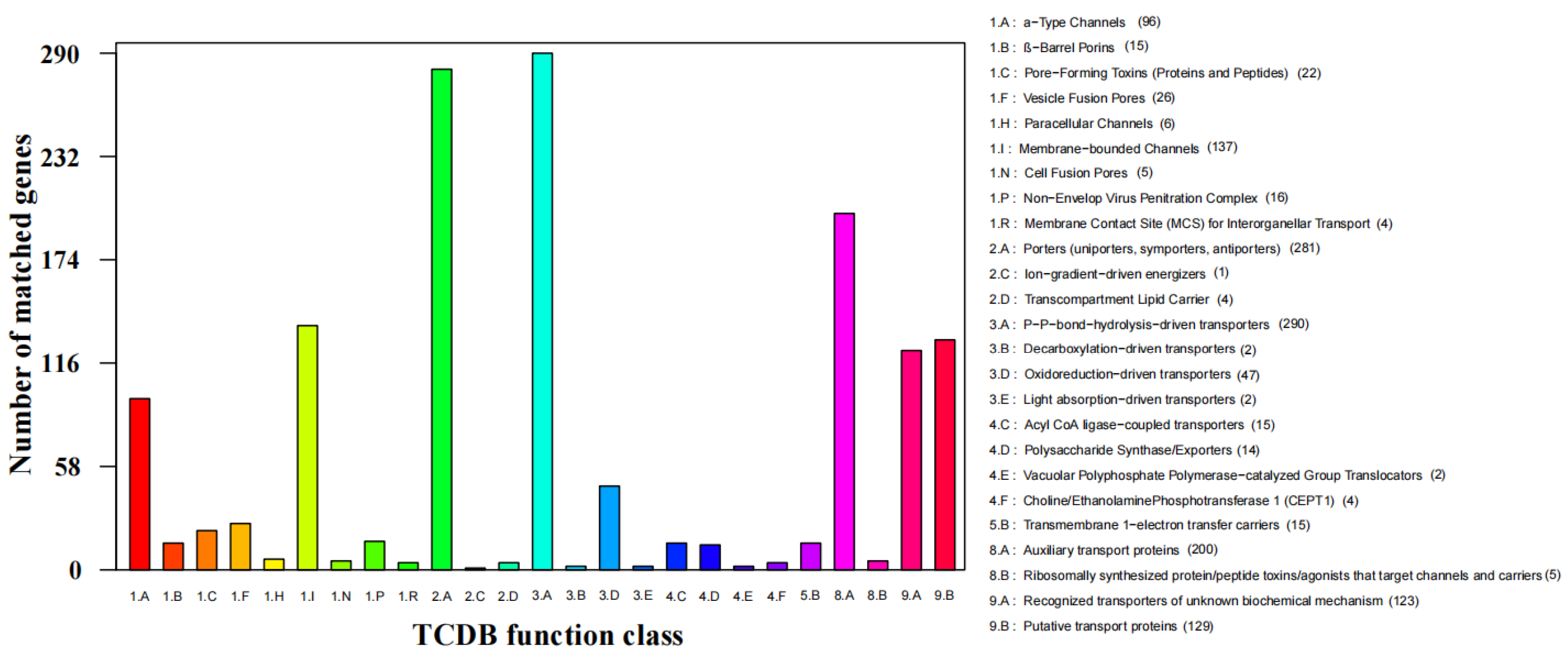
3.3. Functional Annotation of S. phalluae
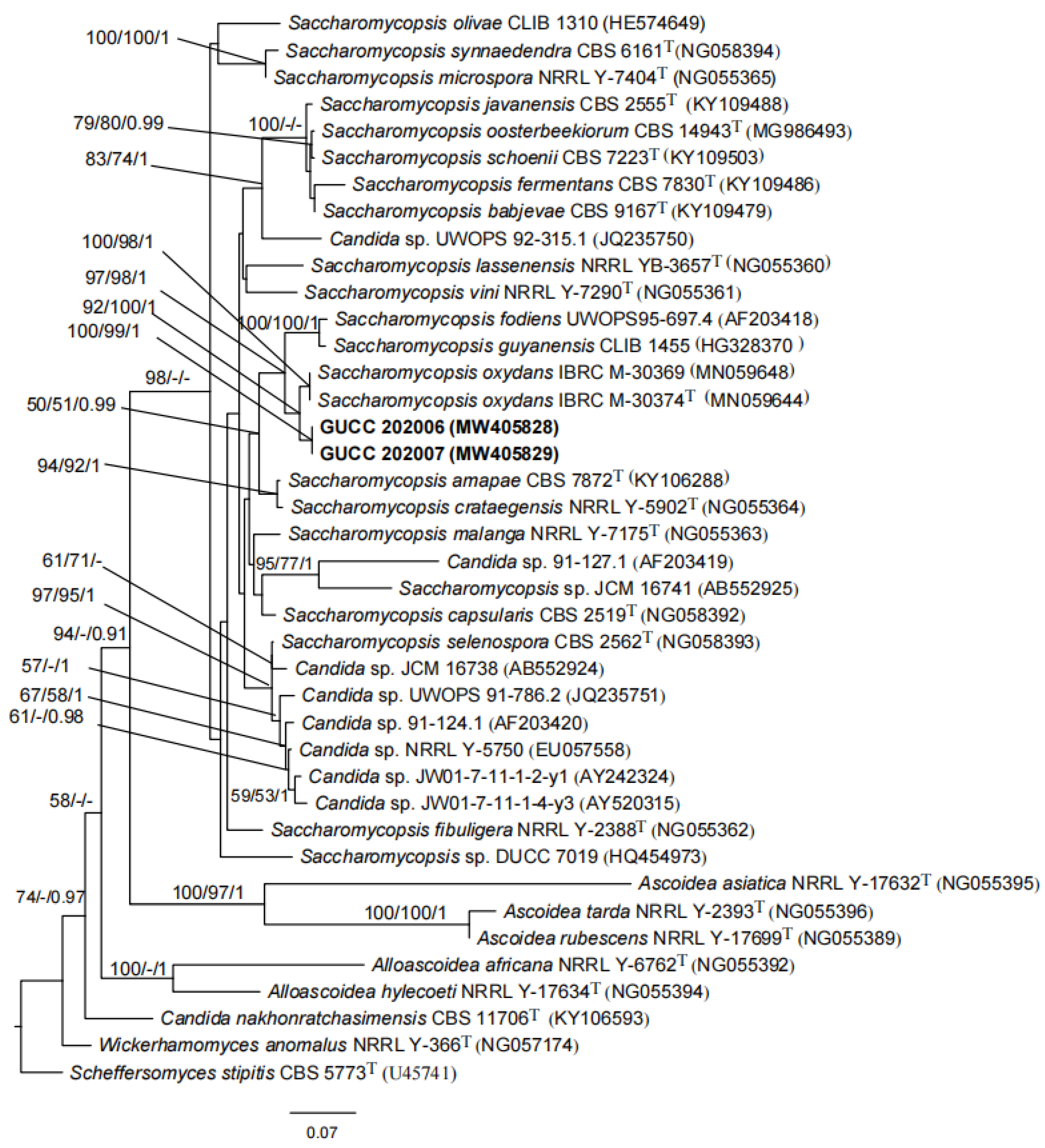
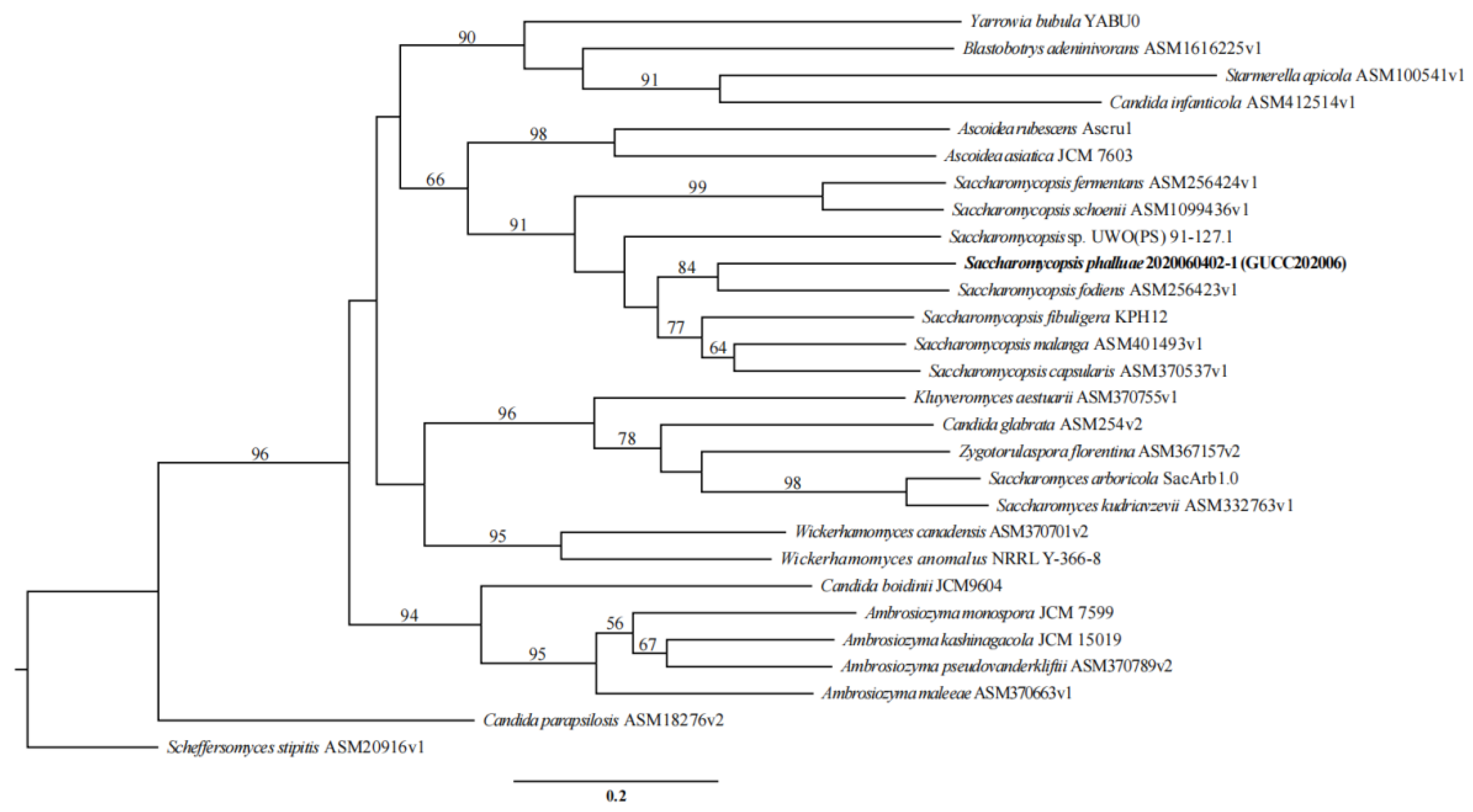
3.4. Phylogenomics Analysis of S. phalluae
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, Y.Y.; Gui, Y.; Lu, G.G.; Yuan, W.S.; Zhu, G.S. Genetic diversity of 18 Dictyophora rubrovolvata germplasm resources from Guizhou. Guizhou Agric. Sci. 2014, 42, 17–20. [Google Scholar]
- Li, Y.; Li, T.H.; Yang, Z.L.; Tu, L.G.E.; Dai, Y.C. Atlas of Chinese Macrofungal Resources; Central Plains Farmers Press: Zhengzhou, China, 2015; p. 1166. [Google Scholar]
- Zhuang, Y.L.; Sun, L.P. Nutritional characteristics of proteins from the volva and pileus in cultivated mushroom Dictyophora rubrovolvata. Int. J. Food Sci. Nutr. 2011, 62, 392–396. [Google Scholar] [CrossRef]
- Sun, L.P.; Bao, C.J.; Chang, W.D.; Zhuang, Y.L. Preparation, characterisation, antioxidant and antiglycation activities of the novel polysaccharides from the pileus of Dictyophora rubrovolvata. Food Sci. Technol.-Braz. 2017, 52, 161–170. [Google Scholar]
- Lu, Y.Y.; Gui, Y.; Chen, Y.Y.; Zeng, G.S.; Zhu, G.S. Preliminary study on the Dictyophora rubrovolvata rot disease. Edible Fungi China 2018, 37, 73–76, 78. [Google Scholar]
- Li, J.Y.; Wu, S.R.; Liu, C.L.; Shang, L.E.; Luo, X.K.; Liu, S.X. Study on fungi infecting in the Dictyophora rubrovolvata rot disease. Edible Fungi China 2021, 40, 109–112. [Google Scholar]
- Chen, X.Y.L.; Zhou, X.H.; Zhao, J.; Tang, X.L.; Pasquali, M.; Migheli, Q.; Berg, G.; Cernava, T. Occurrence of green mold disease on Dictyophora rubrovolvata caused by Trichoderma koningiopsis. J. Plant Pathol. 2021, 103, 981–984. [Google Scholar] [CrossRef]
- Pan, G.C.; Long, H.W.; Wu, D.; Chen, X.; Dai, L.H.; Sun, Y.; Zhou, F.L. Occurrence of bark-rot disease on Dictyophora rubrovolvata and its prevention in Guizhou Province. Edible Fungi China 2015, 34, 72–75. [Google Scholar]
- Schionning, H. Nouveau genre de la famille des Saccharomycetes. Compt. Rend. Trav. Lab Carlsberg 1903, 6, 101–125. [Google Scholar]
- Suh, S.O.; Blackwell, M.; Kurtzman, C.P.; Lachance, M.A. Phylogenetics of Saccharomycetales, the ascomycete yeasts. Mycologia 2006, 98, 1006–1017. [Google Scholar] [CrossRef]
- Jacques, N.; Louis-Mondesir, C.; Coton, M.; Coton, E.; Casaregola, S. Two novel Saccharomycopsis species isolated from black olive brines and a tropical plant. Description of Saccharomycopsis olivae f. a., sp. nov. and Saccharomycopsis guyanensis f. a., sp. nov. Reassignment of Candida amapae to Saccharomycopsis amapae f. a., comb. nov., Candida lassenensis to Saccharomycopsis lassenensis f. a., comb. nov. and Arthroascus babjevae to Saccharomycopsis babjevae f. a., comb. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 2169–2175. [Google Scholar]
- Jin, J.H.; Hanh, N.T.T.; Sanjida, H.; Park, S.H.; Hyewon, O.; Lim, S.; Mok, I.-K.; Li, Y.; Pal, K.; Kim, D. Characteristics of sourdough bread fermented with Pediococcus pentosaceus and Saccharomyces cerevisiae and its bio-preservative effect against Aspergillus flavus. Food Chem. 2021, 345, 128787. [Google Scholar] [CrossRef]
- Iacumin, L.; Manzano, M.; Andyanto, D.; Comi, G. Biocontrol of ochratoxigenic moulds (Aspergillus ochraceus and Penicillium nordicum) by Debaryomyces hansenii and Saccharomycopsis fibuligera during speck production. Food Microbiol. 2017, 62, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.H.; Li, C.T.; Li, Y. First report of Penicillium brevicompactum causing blue mold disease of Grifola frondosa in China. Plant Dis. 2017, 101, 1549. [Google Scholar] [CrossRef]
- Kurtzman, C.P. Two new members of the Saccharomycopsis clade: Saccharomycopsis microspora comb. nov. and Candida lassenensis, sp. nov. Mycotaxon 1999, 71, 241–250. [Google Scholar]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. The Yeasts, a Taxonomic Study, 5th ed.; Elsevier Science: Amsterdam, The Netherlands, 2011; pp. 87–110. [Google Scholar]
- Hajihosseinali, M.; Nasr, S.; Amoozegar, M.A.; Yurkov, A. Saccharomycopsis oxydans sp. nov., a new non-fermentative member in the genus Saccharomycopsis isolated from a traditional dairy product of Iran. Int. J. Syst. Evol. Microbiol. 2020, 70, 1059–1063. [Google Scholar] [CrossRef]
- Barnett, J.A.; Payne, R.W.; Yarrow, D. Yeasts: Characteristics and Identification, 3rd ed.; Cambridge University Press: Cambridge, UK, 2000; pp. 8–16. [Google Scholar]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press Inc.: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- O’Donnell, K. Fusarium and Its Near Relatives. The Fungal Holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematic; Reynolds, D.R., Taylor, J.W., Eds.; CAB International: Wallingford, UK, 1993; pp. 225–233. [Google Scholar]
- Kurtzman, C.P.; Robnett, C.J. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in 5′ end of the large subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 1997, 35, 1216–1223. [Google Scholar] [CrossRef] [Green Version]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef]
- Lachance, M.A.; Rosa, C.A.; Carvajal, E.J.; Freitas, L.F.D.; Bowles, J.M. Saccharomycopsis fodiens sp., nov., a rare predacious yeast from three distant localities. Int. J. Syst. Evol. Microbiol. 2012, 62, 2793–2798. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Gateway Computing Environments Workshop (GCE); IEEE Computer Society: Washington, DC, USA, 2010. [Google Scholar]
- Berlin, K.; Koren, S.; Chin, C.S.; Drake, J.; Landolin, J.M.; Phillippy, A.M. Assembling large genomes with single-molecule sequencing and locality sensitive hashing. Nat. Biotechnol. 2015, 33, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Phillippy, A.M. One chromosome, one contig: Complete microbial genomes from long-read sequencing and assembly. Curr. Opin. Microbiol. 2015, 23, 110–120. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.H.; Deng, W.Q.; Yan, W.J.; Li, T.H. Whole genome sequence of an edible and potential medicinal fungus, Cordyceps guangdongensis. G3 Genes Genom. Genet. 2018, 8, 1863–1870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanke, M.; Keller, O.; Gunduz, I.; Hayes, A.; Waack, S.; Morgenstern, B. AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006, 34, W435–W439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, S.; Bridges, S.; Magbanua, Z.V.; Peterson, D.G. Empirical comparison of ab initio repeat finding programs. Nucleic Acids Res. 2008, 36, 2284–2294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [Green Version]
- Li, W.Z.; Jaroszewski, L.; Godzik, A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 2002, 18, 77–82. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Mistry, J.; Schuster-Böckler, B.; Griffiths-Jones, S.; Hollich, V.; Lassmann, T.; Moxon, S.; Marshall, M.; Khanna, A.; Durbin, R.; et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006, 34, D247–D251. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence data bank and its new supplement TrEMBL. Nucleic Acids Res. 1996, 25, 31–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saier, M.H., Jr.; Reddy, V.S.; Tamang, D.G.; Västermark, Å. The transporter classification database. Nucleic Acids Res. 2014, 42, D251–D258. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.; Pant, R.; Raghunath, A.; Irvine, A.G.; Pedro, H.; Hammond-Kosack, K.E. The Pathogen-Host Interactions database (PHI-base): Additions and future developments. Nucleic Acids Res. 2015, 43, D645–D655. [Google Scholar] [CrossRef] [Green Version]
- Coleine, C.; Masonjones, S.; Sterflinger, K.; Onofri, S.; Selbmann, L.; Stajich, J.E. Peculiar genomic traits in the stress-adapted cryptoendolithic Antarctic fungus Friedmanniomyces endolithicus. Fungal Biol. UK 2020, 124, 458–467. [Google Scholar] [CrossRef]
- Nelson, D.R. The cytochrome P450 homepage. Hum. Genom. 2009, 4, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Gong, Y.; Cai, Y.; Liu, W.; Zhou, Y.; Xiao, Y.; Xu, Z.; Liu, Y.; Lei, X.; Wang, G.; et al. Genome Sequence of the Edible Cultivated Mushroom Lentinula edodes (Shiitake) Reveals Insights into Lignocellulose Degradation. PLoS ONE 2016, 11, e0160336. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. Clustal omega for making accurate alignments of many protein sequences. Protein Sci. 2017, 27, 135–145. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Von Arx, J.A.; Rodríguez de Miranda, L.; Smith, M.T.; Yarrow, D. The genera of yeasts and the yeast-like fungi. Stud. Mycol. 1977, 14, 9–10. [Google Scholar]
- Bashyal, B.M.; Rawat, K.; Sharma, S.; Kulshreshtha, D.; Krishnan, S.G.; Singh, A.K.; Dubey, H.; Solanke, A.U.; Sharma, T.R.; Aggarwal, R. Whole Genome Sequencing of Fusarium fujikuroi Provides Insight into the Role of Secretory Proteins and Cell Wall Degrading Enzymes in Causing Bakanae Disease of Rice. Front. Plant Sci. 2017, 8, 2013. [Google Scholar] [CrossRef]
- Xu, R.; Liu, X.; Peng, B.; Liu, P.; Li, Z.; Dai, Y.; Xiao, S. Genomic Features of Cladobotryum dendroides, Which Causes Cobweb Disease in Edible Mushrooms, and Identification of Genes Related to Pathogenicity and Mycoparasitism. Pathogens 2020, 9, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Q.-S.; Xie, X.-L.; Liang, G.; Gong, F.; Wang, Y.; Wei, X.-Q.; Wang, Q.; Ji, Z.-L.; Chen, Q.-X. The GH18 family of chitinases: Their domain architectures, functions and evolutions. Glycobiology 2011, 22, 23–34. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, A.; Ghosh, S.; Sahoo, D.; Jha, G. Fungal effectors, the double edge sword of phytopathogens. Curr. Genet. 2021, 67, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Sossah, F.L.; Sun, L.; Fu, Y.P.; Li, Y. Genome analysis of Hypomyces perniciosus, the causal agent of wet bubble disease of button mushroom (Agaricus bisporus). Genes 2019, 10, 417. [Google Scholar] [CrossRef] [Green Version]
- Sossah, F.L.; Liu, Z.H.; Yang, C.T.; Okorley, B.A.; Sun, L.; Fu, Y.P.; Li, Y. Genome sequencing of Cladobotryum protrusum provides insights into the evolution and pathogenic mechanisms of the cobweb disease pathogen on cultivated mushroom. Genes 2019, 10, 124. [Google Scholar] [CrossRef] [Green Version]
- Yin, Z.Y.; Liu, H.Q.; Li, Z.P.; Ke, X.W.; Dou, D.L.; Gao, X.N.; Song, N.; Dai, Q.; Wu, Y.; Xu, J.; et al. Genome sequence of Valsa canker pathogens uncovers a potential adaptation of colonization of woody bark. New Phytol. 2015, 208, 1202–1216. [Google Scholar] [CrossRef]






| Characteristic | 1 | 2 a | 3 b | 4 c | 5 d |
|---|---|---|---|---|---|
| D-glucose fermentation | − | W/− | W/V | − | − |
| Maximum growth temperature | 35 °C | ND | 33 °C | 35 °C | 28 °C |
| Growth in 5% NaCl | − | ND | − | ND | + |
| Growth in 10% NaCl | − | − | ND | + | − |
| Potassium nitrate | + | − | − | + | − |
| Scaffold Characteristic | |
|---|---|
| Total counts of contigs sequences | 8 |
| Total length of contigs sequences (bp) | 14,148,124 |
| Contigs N50 (bp) | 1,822,654 |
| Contigs N90 (bp) | 1,540,684 |
| Largest scaffold length (bp) | 2,335,166 |
| Genome coverage | 600× |
| GC content (%) | 43.55 |
| Genome Characteristic | |
| Gene number | 5966 |
| Average gene length (bp) | 1480 |
| Gene density (Unit:gene_number/1000 bp) | 0.4 |
| Exon number per Gene | 1.14 |
| Exon average length (bp) | 1298 |
| Genome GC percent (%) | 43.55 |
| Exon GC percent (%) | 46.56 |
| Database Used for Gene/Protein Annotation | Number of Genes |
|---|---|
| nr | 4735 |
| GO | 3921 |
| KEGG | 1968 |
| KOG | 4015 |
| TCDB | 1461 |
| PHI | 1779 |
| CAZy | 220 |
| P450 | 264 |
| Classification | Number |
|---|---|
| Carbohydrate-binding module (CBMs) | 4 |
| Carbohydrate esterase (CEs) | 15 |
| Glycoside hydrolase (GHs) | 85 |
| Glycosyl transferase (GTs) | 92 |
| Polysaccharide lyase (PLs) | 3 |
| Auxiliary activities (AAs) | 21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, X.; Peng, K.; Li, C.; Zhao, Z.; Zeng, X.; Tian, F.; Li, Y. Complete Genomic Characterization and Identification of Saccharomycopsisphalluae sp. nov., a Novel Pathogen Causes Yellow Rot Disease on Phallus rubrovolvatus. J. Fungi 2021, 7, 707. https://doi.org/10.3390/jof7090707
Yuan X, Peng K, Li C, Zhao Z, Zeng X, Tian F, Li Y. Complete Genomic Characterization and Identification of Saccharomycopsisphalluae sp. nov., a Novel Pathogen Causes Yellow Rot Disease on Phallus rubrovolvatus. Journal of Fungi. 2021; 7(9):707. https://doi.org/10.3390/jof7090707
Chicago/Turabian StyleYuan, Xiaoxiao, Keqin Peng, Changtian Li, Zhibo Zhao, Xiangyu Zeng, Fenghua Tian, and Yu Li. 2021. "Complete Genomic Characterization and Identification of Saccharomycopsisphalluae sp. nov., a Novel Pathogen Causes Yellow Rot Disease on Phallus rubrovolvatus" Journal of Fungi 7, no. 9: 707. https://doi.org/10.3390/jof7090707
APA StyleYuan, X., Peng, K., Li, C., Zhao, Z., Zeng, X., Tian, F., & Li, Y. (2021). Complete Genomic Characterization and Identification of Saccharomycopsisphalluae sp. nov., a Novel Pathogen Causes Yellow Rot Disease on Phallus rubrovolvatus. Journal of Fungi, 7(9), 707. https://doi.org/10.3390/jof7090707






