Noninvasive Combined Diagnosis and Monitoring of Aspergillus and Pseudomonas Infections: Proof of Concept
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture, Serology and MALDI-TOF
2.2. Liquid Chromatography and Mass Spectrometry Assay for Biomarker Monitoring
2.3. Biomarker Standardization and Quantitation in Liquids and Tissues
2.4. Animal Experiments
3. Results
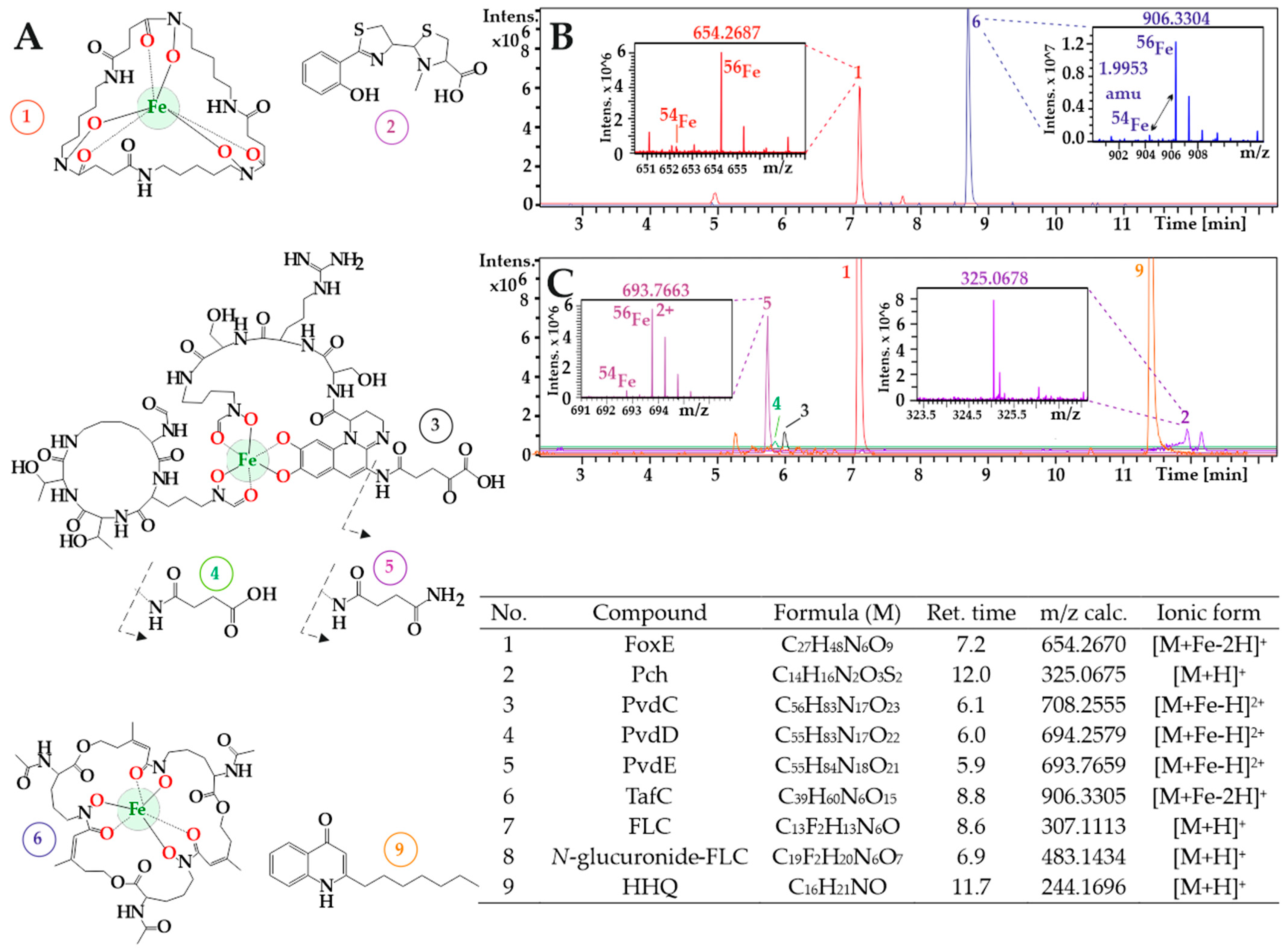
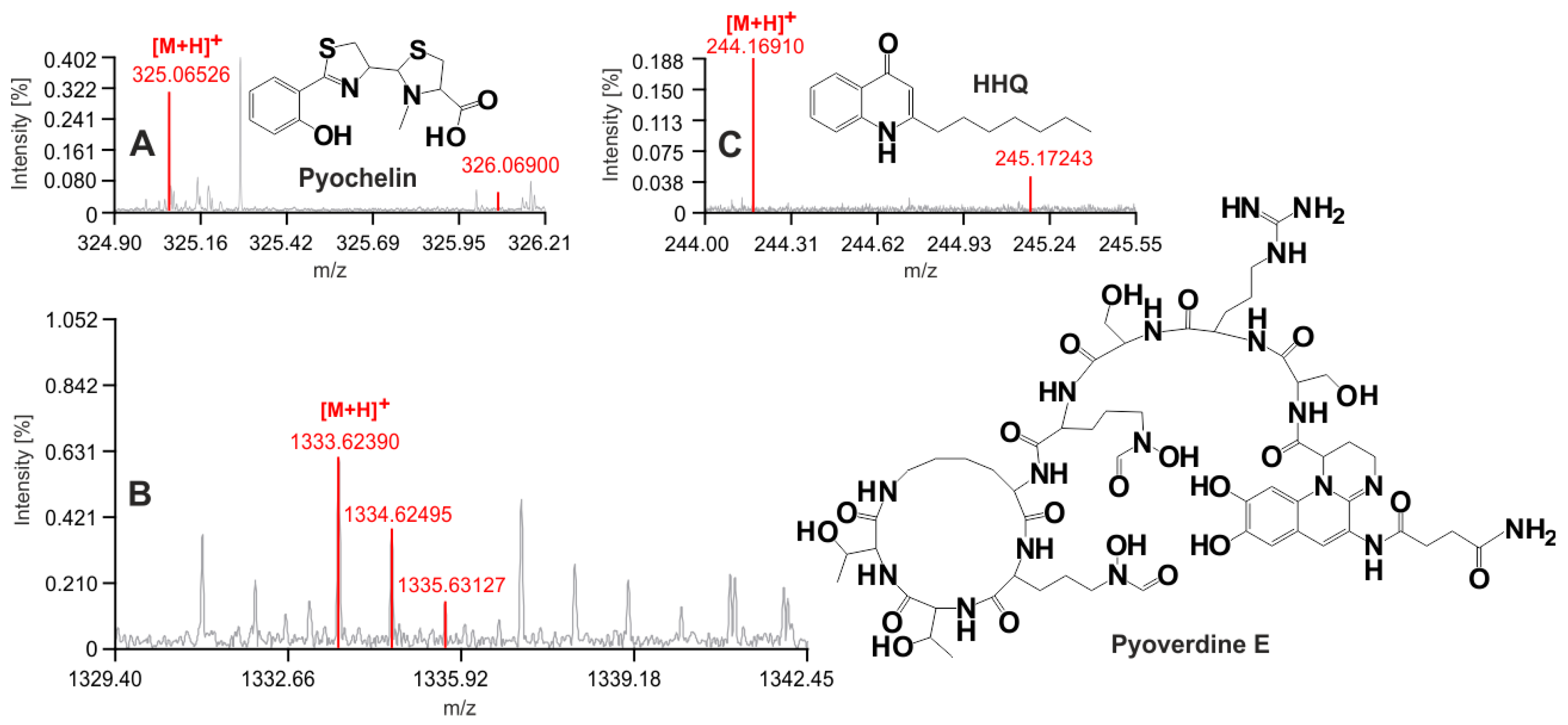
3.1. A. fumigatus and P. aeruginosa in Co-Infection
3.2. New Urine Biomarkers of Pseudomonas aeruginosa
3.3. Animal Infection Models
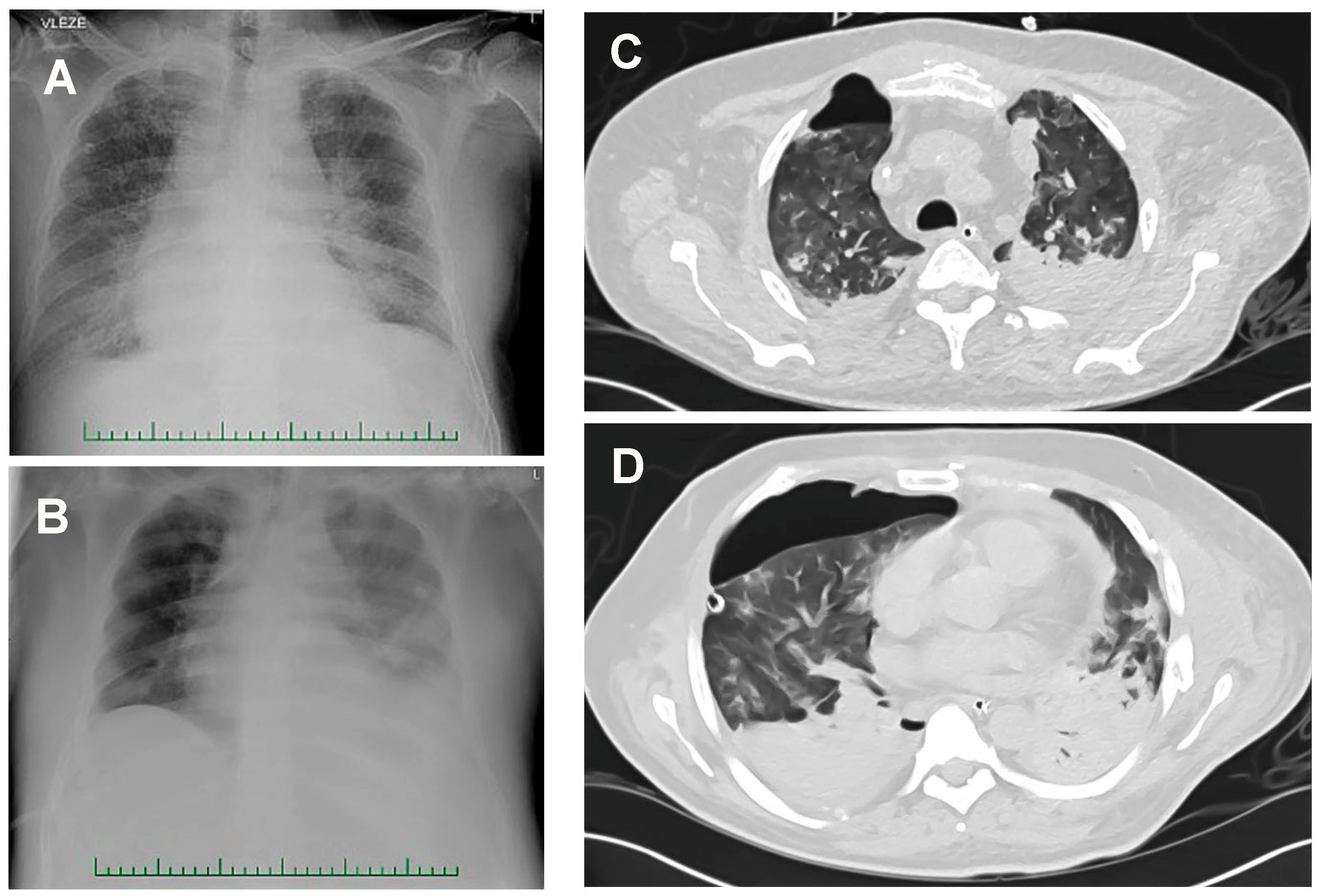
3.4. Pseudomonas aeruginosa in COVID-19-Associated Pneumonia
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Janbon, G.; Quintin, J.; Lanternier, F.; d’Enfert, C. Studying fungal pathogens of humans and fungal infections: Fungal diversity and diversity of approaches. Genes Immun. 2019, 20, 403–414. [Google Scholar] [CrossRef]
- Köhler, J.R.; Casadevall, A.; Perfect, J. The spectrum of fungi that infects humans. Cold Spring Harb. Perspect. Med. 2014, 5, a019273. [Google Scholar] [CrossRef] [Green Version]
- Salmanton-García, J.; Sprute, R.; Stemler, J.; Bartoletti, M.; Dupont, D.; Valerio, M.; Garcia-Vidal, C.; Falces-Romero, I.; Machado, M.; de la Villa, S.; et al. COVID-19–associated pulmonary aspergillosis, March–August 2020. Emerg. Infect. Dis. 2021, 27, 1077. [Google Scholar] [CrossRef]
- Verweij, P.E.; Rijnders, B.J.A.; Bruggemann, R.J.M.; Azoulay, E.; Bassetti, M.; Blot, S.; Calandra, T.; Clancy, C.J.; Cornely, O.A.; Chiller, T.; et al. Review of influenza-associated pulmonary aspergillosis in ICU patients and proposal for a case definition: An expert opinion. Intensive Care Med. 2020, 46, 1524–1535. [Google Scholar] [CrossRef]
- Shortridge, D.; Gales, A.C.; Streit, J.M.; Huband, M.D.; Tsakris, A.; Jones, R.N. Geographic and Temporal Patterns of Antimicrobial Resistance in Pseudomonas aeruginosa Over 20 Years From the SENTRY Antimicrobial Surveillance Program, 1997–2016. Open Forum Infect. Dis. 2019, 6, S63–S68. [Google Scholar] [CrossRef]
- Curran, C.S.; Bolig, T.; Torabi-Parizi, P. Mechanisms and targeted therapies for Pseudomonas aeruginosa lung infection. Am. J. Respir. Crit. Care Med. 2018, 197, 708–727. [Google Scholar] [CrossRef] [PubMed]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Sass, G.; Nazik, H.; Penner, J.; Shah, H.; Ansari, S.R.; Clemons, K.V.; Groleau, M.-C.; Dietl, A.-M.; Visca, P.; Haas, H.; et al. Aspergillus-Pseudomonas interaction, relevant to competition in airways. Med. Mycol. 2019, 57, S228–S232. [Google Scholar] [CrossRef] [PubMed]
- Novák, J.; Škríba, A.; Havlíček, V. Cyclobranch 2: Molecular formula annotations applied to imzML data sets in bimodal fusion and LC-MS data files. Anal. Chem. 2020, 92, 6844–6849. [Google Scholar] [CrossRef]
- Škríba, A.; Pluháček, T.; Palyzová, A.; Nový, Z.; Lemr, K.; Hajdúch, M.; Petřík, M.; Havlíček, V. Early and non-invasive diagnosis of aspergillosis revealed by infection kinetics monitored in a rat model. Front. Microbiol. 2018, 9, 2356. [Google Scholar] [CrossRef]
- Petrik, M.; Umlaufova, E.; Raclavsky, V.; Palyzova, A.; Havlicek, V.; Pfister, J.; Mair, C.; Novy, Z.; Popper, M.; Hajduch, M.; et al. 68Ga-labelled desferrioxamine-B for bacterial infection imaging. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 372–382. [Google Scholar] [CrossRef]
- Raymond, K.N.; Dertz, E.A.; Kim, S.S. Enterobactin: An archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 2003, 100, 3584–3588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazik, H.; Sass, G.; Ansari, S.R.; Ertekin, R.; Haas, H.; Deziel, E.; Stevens, D.A. Novel intermicrobial molecular interaction: Pseudomonas aeruginosa Quinolone Signal (PQS) modulates Aspergillus fumigatus response to iron. Microbiology 2020, 166, 44–55. [Google Scholar] [CrossRef]
- Petřík, M.; Umlaufová, E.; Raclavský, V.; Palyzová, A.; Havlíček, V.; Haas, H.; Nový, Z.; Doležal, D.; Hajduch, M.; Decristoforo, C. Imaging of Pseudomonas aeruginosa infection with Ga-68 labelled pyoverdine for positron emission tomography. Sci. Rep. 2018, 8, 15698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthaiou, E.I.; Sass, G.; Stevens, D.A.; Hsu, J.L. Iron: An essential nutrient for Aspergillus fumigatus and a fulcrum for pathogenesis. Curr. Opin. Infect. Dis. 2018, 31, 506–511. [Google Scholar] [CrossRef] [PubMed]
- Sass, G.; Nazik, H.; Chatterjee, P.; Stevens, D.A. Under nonlimiting iron conditions pyocyanin is a major antifungal molecule, and differences between prototypic Pseudomonas aeruginosa strains. Med. Mycol. 2021, 59, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Moura-Alves, P.; Puyskens, A.; Stinn, A.; Klemm, M.; Guhlich-Bornhof, U.; Dorhoi, A.; Furkert, J.; Kreuchwig, A.; Protze, J.; Lozza, L.; et al. Host monitoring of quorum sensing during Pseudomonas aeruginosa infection. Science 2019, 366, eaaw1629. [Google Scholar] [CrossRef]
- Allegretta, G.; Maurer, C.K.; Eberhard, J.; Maura, D.; Hartmann, R.W.; Rahme, L.; Empting, M. In-depth profiling of mvfr-regulated small molecules in Pseudomonas aeruginosa after quorum sensing inhibitor treatment. Front. Microbiol. 2017, 8, 924. [Google Scholar] [CrossRef]
- Bassetti, M.; Azoulay, E.; Kullberg, B.J.; Ruhnke, M.; Shoham, S.; Vazquez, J.; Giacobbe, D.R.; Calandra, T. EORTC/MSGERC definitions of invasive fungal diseases: Summary of activities of the intensive care unit working group. Clin. Infect. Dis. 2021, 72, 121–127. [Google Scholar] [CrossRef]
- An, J.; McDowell, A.; Kim, Y.-K.; Kim, T.-B. Extracellular vesicle-derived microbiome obtained from exhaled breath condensate in patients with asthma. Ann. Allergy Asthma Immunol. 2021, 126, 729–731. [Google Scholar] [CrossRef]
- Škríba, A.; Patil, R.H.; Hubáček, P.; Dobiáš, R.; Palyzová, A.; Marešová, H.; Pluháček, T.; Havlíček, V. Rhizoferrin glycosylation in Rhizopus microsporus. J. Fungi 2020, 6, 89. [Google Scholar] [CrossRef] [PubMed]
- Luptáková, D.; Pluháček, T.; Petřík, M.; Novák, J.; Palyzová, A.; Sokolová, L.; Škríba, A.; Šedivá, B.; Lemr, K.; Havlíček, V. Non-invasive and invasive diagnoses of aspergillosis in a rat model by mass spectrometry. Sci. Rep. 2017, 7, 16523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Coopersmith, C.M.; et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
| Sample | Concentration (ng/mL) | |||||
|---|---|---|---|---|---|---|
| Specimen | PvdE | PvdD | PvdC | Pch | HHQ | |
| Patient 1 | condensate | 85.3 ± 3.4 | DET | 12.0 ± 0.8 | 135.1 ± 4.2 | 12.9 ± 1.2 |
| Patient 2 | urine | 23.7 ± 0.1 | - | 82.7 ± 0.3 | 90.8 ± 1.1 | 1.4 ± 0.5 |
| Patient 3 | ETA | - | - | - | 19.5 ± 0.9 & | 1.4 ± 0.1 & |
| Patient 4 | ETA | - | - | - | 24.2 ± 1.1 & | 0.2 ± 0.01 & |
| Patient 5 | ETA | - | - | - | 252.4 ± 2.8 & | 1.0 ± 0.04 & |
| Rat 1 | urine | 14,334 ± 352 | 102.6 ± 3.7 | 390.7 ± 13.1 | 1233.9 ± 26.3 | - |
| serum | 53.0 ± 3.0 | 3.4 ± 0.2 | - | - | - | |
| Rat 2 | urine | 12,366 ± 843 | 74.3 ± 4.1 | 261.4 ± 11.9 | 1349.1 ± 15.7 | - |
| serum | 281.8 ± 6.2 | 14.7 ± 0.7 | 9.8 ± 0.3 | DET | - | |
| Blank | inoculum ** | DET | - | - | 74 ± 2 * | - |
| Content (μg/g) | ||||||
| Rat 2 | lung tissue | 3.9 ± 0.3 | - | - | - | - |
| Sample | Concentration (ng/mL) | ||||
|---|---|---|---|---|---|
| Specimen | PvdE | Pch | Fc | TafC | |
| Patient 1 | condensate | 85.3 ± 3.4 | 135.1 ± 4.2 | - | - |
| urine | - | - | - | 23 ± 1 | |
| Rat 3 | urine | 234 497 ± 13 961 | 69 460 ± 23 125 | 5 761 ± 191 | 1854 ± 49 |
| Rat 4 | urine | 88 513 ± 2 553 | 20 711 ± 3474 | - | - |
| Rat 5 | urine | - | - | 21 138 ± 2935 | 1 818 ± 124 |
| Rat 6 | urine | 179 916 ± 16 104 | 66 965 ± 14 432 | DET | 564 ± 133 |
| Rat 7 | urine | 108 313 ± 8 036 | 20 246 ± 2 609 | DET | 378 ± 14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dobiáš, R.; Škríba, A.; Pluháček, T.; Petřík, M.; Palyzová, A.; Káňová, M.; Čubová, E.; Houšť, J.; Novák, J.; Stevens, D.A.; et al. Noninvasive Combined Diagnosis and Monitoring of Aspergillus and Pseudomonas Infections: Proof of Concept. J. Fungi 2021, 7, 730. https://doi.org/10.3390/jof7090730
Dobiáš R, Škríba A, Pluháček T, Petřík M, Palyzová A, Káňová M, Čubová E, Houšť J, Novák J, Stevens DA, et al. Noninvasive Combined Diagnosis and Monitoring of Aspergillus and Pseudomonas Infections: Proof of Concept. Journal of Fungi. 2021; 7(9):730. https://doi.org/10.3390/jof7090730
Chicago/Turabian StyleDobiáš, Radim, Anton Škríba, Tomáš Pluháček, Miloš Petřík, Andrea Palyzová, Marcela Káňová, Eva Čubová, Jiří Houšť, Jiří Novák, David A. Stevens, and et al. 2021. "Noninvasive Combined Diagnosis and Monitoring of Aspergillus and Pseudomonas Infections: Proof of Concept" Journal of Fungi 7, no. 9: 730. https://doi.org/10.3390/jof7090730
APA StyleDobiáš, R., Škríba, A., Pluháček, T., Petřík, M., Palyzová, A., Káňová, M., Čubová, E., Houšť, J., Novák, J., Stevens, D. A., Mitulovič, G., Krejčí, E., Hubáček, P., & Havlíček, V. (2021). Noninvasive Combined Diagnosis and Monitoring of Aspergillus and Pseudomonas Infections: Proof of Concept. Journal of Fungi, 7(9), 730. https://doi.org/10.3390/jof7090730






