Enzymatic Bioprospecting of Fungi Isolated from a Tropical Rainforest in Mexico
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Sampling
2.2. Isolation of the Fungi
2.3. Morphological Identification
2.4. Qualitative Assay in Solid Medium
2.5. Basal Medium for Enzyme Production
2.6. Enzyme Production by Submerged Fermentation
2.6.1. Quantitative Assay Laccase Activity
2.6.2. Quantitative Assay Amylase Activity
2.6.3. Quantitative Assay Xylanase Activity
2.6.4. Quantitative Assay Protease Activity
2.6.5. Quantitative Assay Lipase Activity
2.7. Statistical Analysis
3. Results
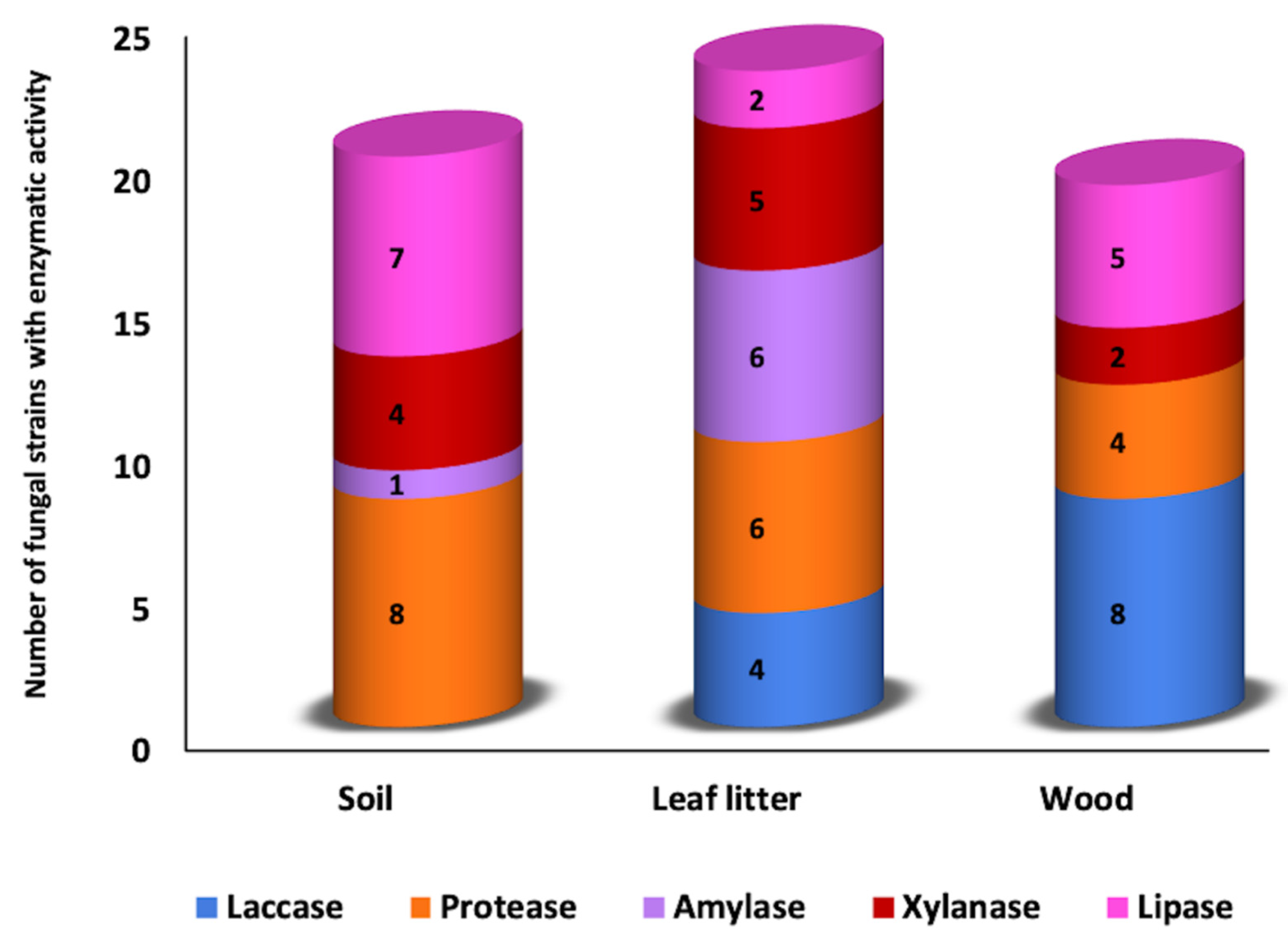
3.1. Qualitative Plate Test
3.2. Enzymatic Activity Quantification
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mittermeier, R.A.; Myers, N.; Tliomsen, J.B.; Olivieri, S. Biodiversity hotspots and major tropical wilderness areas: Approaches to setting conservation priorities. Conserv. Biol. 1998, 12, 516–520. [Google Scholar] [CrossRef]
- Martínez-Meyer, E.; Sosa-Escalante, J.E.; Álvarez, F. El estudio de la biodiversidad en México: A route with a course? Rev. Mex. Biodivers. 2014, 85, S1–S9. [Google Scholar] [CrossRef] [Green Version]
- Guzmán, G. Inventorying the fungi of Mexico. Biodivers. Conserv. 1998, 7, 369–384. [Google Scholar] [CrossRef]
- Godliving Mtui, Y.S. Lignocellulolytic enzymes from tropical fungi: Types, substrates and applications. Sci. Res. Essays 2012, 7, 1544–1555. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Lücking, R. Fungal Diversity Revisited: 2.2 to 3.8 Million Species. Fungal Kingd. 2017, 5, 79–95. [Google Scholar] [CrossRef]
- Wrzosek, M.; Ruszkiewicz-Michalska, M.; Sikora, K.; Damszel, M.; Sierota, Z. The plasticity of fungal interactions. Mycol. Prog. 2017, 16, 101–108. [Google Scholar] [CrossRef] [Green Version]
- Zucconi, L.; Canini, F.; Temporiti, M.E.; Tosi, S. Extracellular enzymes and bioactive compounds from antarctic terrestrial fungi for bioprospecting. Int. J. Environ. Res. Public Health 2020, 17, 6459. [Google Scholar] [CrossRef]
- Hyde, K.D.; Xu, J.; Rapior, S.; Jeewon, R.; Lumyong, S.; Niego, A.G.T.; Abeywickrama, P.D.; Aluthmuhandiram, J.V.S.; Brahamanage, R.S.; Brooks, S.; et al. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 2019, 97, 1–136. [Google Scholar] [CrossRef] [Green Version]
- El-Sawah, A.M.; El-Keblawy, A.; Ali, D.F.I.; Ibrahim, H.M.; El-Sheikh, M.A.; Sharma, A.; Hamoud, Y.A.; Shaghaleh, H.; Brestic, M.; Skalicky, M.; et al. Arbuscular Mycorrhizal Fungi and Plant Growth-Promoting Rhizobacteria Enhance Soil Key Enzymes, Plant Growth, Seed Yield, and Qualitative Attributes of Guar. Agriculture 2021, 11, 194. [Google Scholar] [CrossRef]
- Meyer, V.; Basenko, E.Y.; Benz, J.P.; Braus, G.H.; Caddick, M.X.; Csukai, M.; de Vries, R.P.; Endy, D.; Frisvad, J.C.; Gunde-Cimerman, N.; et al. Growing a circular economy with fungal biotechnology: A white paper. Fungal Biol. Biotechnol. 2020, 7, 5. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.; Yang, D.; Li, R.; Wang, T.; Zhu, Y. Textile Dye Biodecolorization by Manganese Peroxidase: A Review. Molecules 2021, 26, 4403. [Google Scholar] [CrossRef] [PubMed]
- Niyonzima, F.N. Detergent-compatible fungal cellulases. Folia Microbiol. 2021, 66, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Sudeep, K.C.; Upadhyaya, J.; Joshi, D.R.; Lekhak, B.; Chaudhary, D.K.; Pant, B.R.; Bajgai, T.R.; Dhital, R.; Khanal, S.; Koirala, N.; et al. Production, characterization, and industrial application of pectinase enzyme isolated from fungal strains. Fermentation 2020, 6, 59. [Google Scholar] [CrossRef]
- Molina-Gutiérrez, M.; Alcaraz, L.; López, F.A.; Rodríguez-Sánchez, L.; Martínez, M.J.; Prieto, A. Immobilized Forms of the Ophiostoma piceae Lipase for Green Synthesis of Biodiesel. Comparison with Eversa Transform 2.0 and Cal A. J. Fungi 2021, 7, 822. [Google Scholar] [CrossRef]
- Singh, A.K.; Bilal, M.; Iqbal, H.M.N.; Meyer, A.S.; Raj, A. Bioremediation of lignin derivatives and phenolics in wastewater with lignin modifying enzymes: Status, opportunities and challenges. Sci. Total Environ. 2021, 777, 145988. [Google Scholar] [CrossRef]
- Beneyton, T.; Wijaya, I.P.M.; Postros, P.; Najah, M.; Leblond, P.; Couvent, A.; Mayot, E.; Griffiths, A.D.; Drevelle, A. High-throughput screening of filamentous fungi using nanoliter-range droplet-based microfluidics. Sci. Rep. 2016, 6, 27223. [Google Scholar] [CrossRef]
- Tapia-Tussell, R.; Pérez-Brito, D.; Rojas-Herrera, R.; Cortes-Velazquez, A.; Rivera-Muñoz, G.; Solis-Pereira, S. New laccase-producing fungi isolates with biotechnological potential in dye decolorization. African J. Biotechnol. 2011, 10, 10134–10142. [Google Scholar] [CrossRef] [Green Version]
- Chan Cupul, W.; Heredia Abarca, G.P.; Rodríguez Vázquez, R. Aislamiento y evaluación de la actividad enzimática ligninolítica de macromicetos del estado de Veracruz, México. Rev. Int. Contam. Ambient. 2016, 32, 339–351. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Corzo, L.D.; Álvarez-Gutiérrez, P.E.; Meza-Gordillo, R.; Villalobos-Maldonado, J.J.; Enciso-Pinto, S.; Enciso-Sáenz, S. Lignocellulolytic Enzyme Production from Wood Rot Fungi Collected in Chiapas, Mexico, and Their Growth on Lignocellulosic Material. J. Fungi 2021, 7, 450. [Google Scholar] [CrossRef]
- Mueller, G.M.; Bills, G.F.; Foster, M.S. Biodiversity of Fungi: Inventory and Monitoring Methods; Academic, P., Ed.; Elsevier Inc.: Burligton, MA, USA, 2004; ISBN 9780080470269. [Google Scholar]
- Chaparro, D.F.; Rosas, D.C.; Varela, A. Aislamiento y evaluación de la actividad enzimática de hongos descomponedores de madera (Quindío, Colombia). Rev. Iberoam. Micol. 2009, 26, 238–243. [Google Scholar] [CrossRef]
- Ellis, M.B. Dematiaceous Hyphomycetes; Institute, Kew, Commonwealth Mycological: Surrey, UK, 1971. [Google Scholar]
- Phillips, R. Mushrooms & Other Fungi of Great Britain & Europe, 1st ed.; Pan Book: Italy, 1989; ISBN 9780330264419. [Google Scholar]
- Largent, D.L.; Stutnz, D.E. How to Identy Mushrooms to Genus; Eureka, C., Ed.; Mad River Press: Eureka, CA, USA, 1986; ISBN 9780916422004. [Google Scholar]
- Ryvarden, L. Genera of Polypores: Nomenclature and Taxonomy. Syn. Fungorum 1991, 5, 1–363. [Google Scholar]
- Alexopoulos, C.J.; Mims, C.W.; Blackwell, M. Introductory Mycology; John Wiley & Sons: New York, NY, USA, 1996. [Google Scholar]
- Domsch, K.L.; Walter Gams, T.-H.A. Compendium of Soil Fungi, 2nd ed.; Gams, W., Ed.; IHW-Verlag: Eching, Germany, 2007. [Google Scholar]
- Seifert, K.A.; Gams, W. The genera of Hyphomycetes—2011 update. Pers. Mol. Phylogeny Evol. Fungi 2011, 27, 119–129. [Google Scholar] [CrossRef] [Green Version]
- Sunil, S.M.; Renuka, P.S.; Pruthvi, K.; Swetha, M.; Malini, S.; Veena, S.M. Isolation, purification, and characterization of fungal laccase from Pleurotus sp. Enzyme Res. 2011, 2011, 248735. [Google Scholar] [CrossRef] [Green Version]
- Mandic, M.; Djokic, L.; Nikolaivits, E.; Prodanovic, R.; O’Connor, K.; Jeremic, S.; Topakas, E.; Nikodinovic-Runic, J. Identification and Characterization of New Laccase Biocatalysts from Pseudomonas Species Suitable for Degradation of Synthetic Textile Dyes. Catalysts 2019, 9, 629. [Google Scholar] [CrossRef] [Green Version]
- Willerding, A.L.; de Oliveira, L.A.; Moreira, F.W.; Germano, M.G.; Chagas, A.F. Lipase activity among bacteria isolated from amazonian soils. Enzyme Res. 2011, 2011, 720194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopinath, S.C.B.; Anbu, P.; Hilda, A. Extracellular enzymatic activity profiles in fungi isolated from oil-rich environments. Mycoscience 2005, 46, 119–126. [Google Scholar] [CrossRef]
- Abdel-Sater, M.A.; El-Said, A.H.M. Xylan-decomposing fungi and xylanolytic activity in agricultural and industrial wastes. Int. Biodeterior. Biodegrad. 2001, 47, 15–21. [Google Scholar] [CrossRef]
- Kumar, A.; Sharma, V.; Saxena, J.; Yadav, B.; Alam, A.; Prakash, A. Isolation and Screening of Extracellular Protease Enzyme from Bacterial and Fungal Isolates of Soil. Int. J. Sci. Res. Environ. Sci. 2015, 3, 334–340. [Google Scholar] [CrossRef]
- Cruz Ramírez, M.G.; Rivera-Ríos, J.M.; Téllez-Jurado, A.; Maqueda Gálvez, A.P.; Mercado-Flores, Y.; Arana-Cuenca, A. Screening for thermotolerant ligninolytic fungi with laccase, lipase, and protease activity isolated in Mexico. J. Environ. Manage. 2012, 95, S256–S259. [Google Scholar] [CrossRef]
- Sivakumar, R. Isolation, Screening and Optimization of Production Medium for Thermostable Laccase Production from Ganoderma sp. Int. J. Eng. Sci. Technol. 2010, 2, 7133–7141. [Google Scholar]
- Lowry, O.H.; Rosebrogh, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Singh, S.; Singh, S.; Bali, V.; Sharma, L.; Mangla, J. Production of fungal amylases using cheap, readily available agriresidues, for potential application in textile industry. Biomed Res. Int. 2014, 2014, 215748. [Google Scholar] [CrossRef] [Green Version]
- Ravichandra, K.; Yaswanth, V.V.N.; Nikhila, B.; Ahmad, J.; Srinivasa Rao, P.; Uma, A.; Ravindrababu, V.; Prakasham, R.S. Xylanase Production by Isolated Fungal Strain, Aspergillus fumigatus RSP-8 (MTCC 12039): Impact of Agro-industrial Material as Substrate. Sugar Tech. 2016, 18, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Cupp-Enyard, C.; Aldrich, S. Sigma’s non-specific protease activity assay—Casein as a substrate. J. Vis. Exp. 2008, 19, 4–5. [Google Scholar] [CrossRef]
- Winkler, U.K.; Stuckmann, M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J. Bacteriol. 1979, 138, 663–670. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Rathi, P.; Gupta, N.; Bradoo, S. Lipase assays for conventional and molecular screening: An overview. Biotechnol. Appl. Biochem. 2003, 37, 63. [Google Scholar] [CrossRef] [Green Version]
- Bourbonnais, R.; Leech, D.; Paice, M.G. Electrochemical analysis of the interactions of laccase mediators with lignin model compounds. Biochim. Biophys. Acta Gen. Subj. 1998, 1379, 381–390. [Google Scholar] [CrossRef]
- de Salas, F.; Cañadas, R.; Santiago, G.; Virseda-Jerez, A.; Vind, J.; Gentili, P.; Martínez, A.T.; Guallar, V.; Muñoz, I.G.; Camarero, S. Structural and biochemical insights into an engineered high-redox potential laccase overproduced in Aspergillus. Int. J. Biol. Macromol. 2019, 141, 855–866. [Google Scholar] [CrossRef]
- Fraatz, M.A.; Naeve, S.; Hausherr, V.; Zorn, H.; Blank, L.M. A minimal growth medium for the basidiomycete Pleurotus sapidus for metabolic flux analysis. Fungal Biol. Biotechnol. 2014, 1, 9. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Wu, F.; Wei, Z.; Xiao, Y.; Gong, W. Characterization and decolorization ability of a laccase from Panus rudis. Enzyme Microb. Technol. 2006, 39, 92–97. [Google Scholar] [CrossRef]
- Yang, J.; Li, W.; Bun Ng, T.; Deng, X.; Lin, J.; Ye, X. Laccases: Production, expression regulation, and applications in pharmaceutical biodegradation. Front. Microbiol. 2017, 8, 832. [Google Scholar] [CrossRef]
- Cardoso, B.K.; Linde, G.A.; Colauto, N.B.; do Valle, J.S. Panus strigellus laccase decolorizes anthraquinone, azo, and triphenylmethane dyes. Biocatal. Agric. Biotechnol. 2018, 16, 558–563. [Google Scholar] [CrossRef]
- Peralta, R.M.; da Silva, B.P.; Gomes Côrrea, R.C.; Kato, C.G.; Vicente Seixas, F.A.; Bracht, A. Enzymes from Basidiomycetes-Peculiar and Efficient Tools for Biotechnology. In Biotechnology of Microbial Enzymes: Production, Biocatalysis and Industrial Applications; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 119–149. ISBN 9780128037461. [Google Scholar]
- Camacho-Morales, R.L.; Gerardo-Gerardo, J.L.; Guillén Navarro, K.; Sánchez, J.E. Ligninolytic enzyme production by white rot fungi during paraquat (herbicide) degradation. Rev. Argent. Microbiol. 2017, 49, 189–196. [Google Scholar] [CrossRef]
- Rodríguez-Couto, S. Microbial Laccases as Potential Eco-friendly Biocatalysts for the Food Processing Industries. In Microbial Enzyme Technology in Food Applications, 1st ed.; Ray, R.C., Rosell, C.M., Eds.; CRC Press: Boca Ratón, FL, USA, 2017; pp. 255–268. [Google Scholar] [CrossRef]
- Yamanaka, R.; Soares, C.F.; Matheus, D.R.; Machado, K.M.G. Lignolytic enzymes produced by Trametes villosa CCB176 under different culture conditions. Braz. J. Microbiol. 2008, 39, 78–84. [Google Scholar] [CrossRef] [Green Version]
- Ezike, T.C.; Ezugwu, A.L.; Udeh, J.O.; Eze, S.O.O.; Chilaka, F.C. Purification and characterisation of new laccase from Trametes polyzona WRF03. Biotechnol. Reports 2020, 28, e00566. [Google Scholar] [CrossRef]
- Garrido-Bazán, V.; Téllez-Téllez, M.; Herrera-Estrella, A.; Díaz-Godínez, G.; Nava-Galicia, S.; Villalobos-López, M.Á.; Arroyo-Becerra, A.; Bibbins-Martínez, M. Effect of textile dyes on activity and differential regulation of laccase genes from Pleurotus ostreatus grown in submerged fermentation. AMB Express 2016, 6, 93. [Google Scholar] [CrossRef] [Green Version]
- Tinoco, R.; Acevedo, A.; Galindo, E.; Serrano-Carreón, L. Increasing Pleurotus ostreatus laccase production by culture medium optimization and copper/lignin synergistic induction. J. Ind. Microbiol. Biotechnol. 2011, 38, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, M.I.; Tejerina, M.R.; Sawostjanik-Afanasiuk, S.S.; Giorgio, E.M.; Barchuk, M.L.; Zapata, P.D.; Villalba, L.L. Preliminary studies of new strains of Trametes sp. From Argentina for laccase production ability. Braz. J. Microbiol. 2016, 47, 287–297. [Google Scholar] [CrossRef] [Green Version]
- Buddhika, U.V.A.; Savocchia, S.; Steel, C.C. Copper induces transcription of BcLCC2 laccase gene in phytopathogenic fungus, Botrytis cinerea. Mycology 2021, 12, 48. [Google Scholar] [CrossRef] [Green Version]
- Eugenio, M.E.; Carbajo, J.M.; Martín, J.A.; Martín-Sampedro, R.; González, A.E.; Villar, J.C. Synergie effect of inductors on lacease production by Pycnoporus sanguineus. Afinidad 2010, 67, 129–135. [Google Scholar]
- Dhakar, K.; Pandey, A. Laccase production from a temperature and pH tolerant fungal strain of Trametes hirsuta (MTCC 11397). Enzyme Res. 2013, 2013, 869062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zilly, A.; dos Bazanella, G.C.S.; Helm, C.V.; Araújo, C.A.V.; de Souza, C.G.M.; Bracht, A.; Peralta, R.M. Solid-State Bioconversion of Passion Fruit Waste by White-Rot Fungi for Production of Oxidative and Hydrolytic Enzymes. Food Bioprocess Technol. 2012, 5, 1573–1580. [Google Scholar] [CrossRef] [Green Version]
- Kudryavtseva, O.A.; Dunaevsky, Y.E.; Kamzolkina, O.V.; Belozersky, M.A. Fungal proteolytic enzymes: Features of the extracellular proteases of xylotrophic basidiomycetes. Microbiology 2008, 77, 643–653. [Google Scholar] [CrossRef]
- Geisseler, D.; Horwath, W.R. Regulation of extracellular protease activity in soil in response to different sources and concentrations of nitrogen and carbon. Soil Biol. Biochem. 2008, 40, 3040–3048. [Google Scholar] [CrossRef]
- de Souza, P.M.; de Assis Bittencourt, M.L.; Caprara, C.C.; de Freitas, M.; de Almeida, R.P.C.; Silveira, D.; Fonseca, Y.M.; Filho, E.X.F.; Pessoa Junior, A.; Magalhães, P.O. A biotechnology perspective of fungal proteases. Braz. J. Microbiol. 2015, 46, 337–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorneles Inacio, F.; Oliveira Ferreira, R.; Vaz de Araujo, C.A.; Brugnari, T.; Castoldi, R.; Peralta, R.M.; Marques de Souza, C.G. Proteases of wood rot fungi with emphasis on the genus Pleurotus. Biomed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [Green Version]
- Niyonzima, F.N.; More, S.S. Purification and characterization of detergent-compatible protease from Aspergillus terreus gr. 3 Biotech 2015, 5, 61–70. [Google Scholar] [CrossRef] [Green Version]
- Ortega, L.M.; Kikot, G.E.; Astoreca, A.L.; Alconada, T.M. Screening of Fusarium graminearum Isolates for Enzymes Extracellular and Deoxynivalenol Production. J. Mycol. 2013, 2013, 358140. [Google Scholar] [CrossRef] [Green Version]
- León Rodríguez, K.; Higuera, B.L.; Martínez, S.T. Induction of proteases secreted by Fusarium Oxysporum f. sp. Dianthi in the presence of carnation root cell walls. biochemical characterization of a serine protease. J. Plant Pathol. 2017, 99, 609–617. [Google Scholar] [CrossRef]
- Vranova, V.; Rejsek, K.; Formanek, P. Proteolytic activity in soil: A review. Appl. Soil Ecol. 2013, 70, 23–32. [Google Scholar] [CrossRef]
- Vishwanatha, K.S.; Rao, A.G.A.; Singh, S.A. Acid protease production by solid-state fermentation using Aspergillus oryzae MTCC 5341: Optimization of process parameters. J. Ind. Microbiol. Biotechnol. 2010, 37, 129–138. [Google Scholar] [CrossRef]
- Villavicencio, E.V.; Mali, T.; Mattila, H.K.; Lundell, T. Enzyme activity profiles produced on wood and straw by four fungi of different decay strategies. Microorganisms 2020, 8, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Dutt, D.; Tyagi, C.H. Screening of Xylanases from Indigenously Isolated White Rot Fungal Strains for Possible Application in Pulp Biobleaching. Open Accsess Sci. Rep. 2013, 2, 602. [Google Scholar] [CrossRef]
- Tirado-González, D.N.; Jáuregui-Rincón, J.; Tirado-Estrada, G.G.; Martínez-Hernández, P.A.; Guevara-Lara, F.; Miranda-Romero, L.A. Production of cellulases and xylanases by white-rot fungi cultured in corn stover media for ruminant feed applications. Anim. Feed Sci. Technol. 2016, 221, 147–156. [Google Scholar] [CrossRef]
- Martínez-Pacheco, M.M.; Flores-García, A.; Zamudio-Jaramillo, M.A.; Chávez-Parga, M.C.; Alvarez-Navarrete, M. Optimization of production of xylanases with low cellulases in Fusarium solani by means of a solid state fermentation using statistical experimental design. Rev. Argent. Microbiol. 2020, 52, 328–338. [Google Scholar] [CrossRef]
- Maya-Yescas, M.E.; Revah, S.; Le Borgne, S.; Valenzuela, J.; Palacios-González, E.; Terrés-Rojas, E.; Vigueras-Ramírez, G. Growth of Leucoagaricus gongylophorus Möller (Singer) and production of key enzymes in submerged and solid-state cultures with lignocellulosic substrates. Biotechnol. Lett. 2021, 43, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.P.; Singh, H.B.; Prabha, R. Microbial Inoculants in Sustainable Agricultural Productivity; Springer: New York, NY, USA, 2016. [Google Scholar]
- Pessôa, M.G.; Paulino, B.N.; Mano, M.C.R.; Neri-Numa, I.A.; Molina, G.; Pastore, G.M. Fusarium species—a promising tool box for industrial biotechnology. Appl. Microbiol. Biotechnol. 2017, 101, 3493–3511. [Google Scholar] [CrossRef] [PubMed]
- Nwagu, T.N.; Okolo, B.N. Extracellular amylase production of a thermotolerant Fusarium sp: Isolated from Eastern Nigerian soil. Brazilian Arch. Biol. Technol. 2011, 54, 649–658. [Google Scholar] [CrossRef]
- Almeida, M.; Guimaraes, V.; Falkoski, D.L.; Visser, E. Optimization of Endoglucanase and Xylanase Activities from Fusarium verticillioides for Simultaneous Saccharification and Fermentation of Sugarcane Bagasse. Artic. Appl. Biochem. Biotechnol. 2013. [Google Scholar] [CrossRef]
- Uday, U.S.P.; Choudhury, P.; Bandyopadhyay, T.K.; Bhunia, B. Classification, mode of action and production strategy of xylanase and its application for biofuel production from water hyacinth. Int. J. Biol. Macromol. 2016, 82, 1041–1054. [Google Scholar] [CrossRef] [PubMed]
- Peña, C.; Bermúdez, E.; Morales, S.; Farrés, A. Las cutinasas como una herramienta valiosa para la descontaminación de residuos plásticos. Mensaje Bioquímico 2018, 42, 24–35. [Google Scholar]
- Villafana, R.T.; Rampersad, S.N. Diversity, structure, and synteny of the cutinase gene of Colletotrichum species. Ecol. Evol. 2020, 10, 1425–1443. [Google Scholar] [CrossRef] [Green Version]
- Kontkanen, H.; Westerholm-Parvinen, A.; Saloheimo, M.; Bailey, M.; Rättö, M.; Mattila, I.; Mohsina, M.; Kalkkinen, N.; Nakari-Setälä, T.; Buchert, J. Novel Coprinopsis cinerea Polyesterase that hydrolyzes cutin and Suberin. Appl. Environ. Microbiol. 2009, 75, 2148–2157. [Google Scholar] [CrossRef] [Green Version]
- Belge, B.; Llovera, M.; Comabella, E.; Gatius, F.; Guillén, P.; Graell, J.; Lara, I. Characterization of cuticle composition after cold storage of “celeste” and “somerset” sweet cherry fruit. J. Agric. Food Chem. 2014, 62, 8722–8729. [Google Scholar] [CrossRef] [PubMed]
- Baldrian, P. Microbial enzyme-catalyzed processes in soils and their analysis. Plant Soil Environ. 2009, 55, 370–378. [Google Scholar] [CrossRef] [Green Version]
- Adrio, J.L.; Demain, A.L. Microbial enzymes: Tools for biotechnological processes. Biomolecules 2014, 4, 117–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Sampling Site | Source | Fungi | Potency Index (PI) | ||||
|---|---|---|---|---|---|---|---|
| Laccase | Protease | Amylase | Xylanase | Lipase | |||
| Cocona | Wood | Basidiomycete CE10 | 2.3 | ||||
| Cocona | Wood | Marasmius sp. CE25 * | 1.7 | 2.3 | |||
| Cocona | Wood | Basidiomycete CE34 | 3.0 | ||||
| La florida | Wood | Auricularia sp. FE15 | 2.5 | ||||
| La florida | Wood | Neonothopanus sp. FE33 * | 1.2 | 1.6 | |||
| La florida | Wood | Trametes villosa FE35 * | 2.3 | 2.1 | |||
| La florida | Wood | Hexagonia sp. FE40 | 2.0 | ||||
| La florida | Wood | Marasmiellus sp. FE46 | 1.8 | ||||
| La florida | Wood | Rigidoporus sp. FE55 * | 1.9 | 1.8 | |||
| Puyacatengo | Wood | Ascomycete PE37 | 1.8 | ||||
| Puyacatengo | Wood | Marasmius sp. PE38 | 2.2 | ||||
| Puyacatengo | Wood | Panus sp. PE43 | 2.1 | ||||
| Puyacatengo | Wood | Daedalea flavida PE47 * | 2.2 | 2.4 | 2.2 | ||
| Cocona | Leaf litter | Mycelia sterilia CH464 | 1.6 | ||||
| Cocona | Leaf litter | Xylariacea CH240 | 1.4 | ||||
| Cocona | Leaf litter | Mycelia sterilia CH546 | 2.2 | ||||
| Cocona | Leaf litter | Monodictys sp. CH616 | 2.6 | ||||
| Cocona | Leaf litter | Mycelia sterilia CH630 | 2.3 | ||||
| Cocona | Leaf litter | Mycelia sterilia CH631 | 2.1 | ||||
| Cocona | Leaf litter | Aspergillus sp. CH665 | 3.5 | ||||
| Cocona | Leaf litter | Drechslera sp. CH681 | 1.8 | ||||
| Cocona | Leaf litter | Aspergillus sp. CH693 * | 2.5 | 2.3 | |||
| La florida | Leaf litter | Mycelia sterilia FH175 | 2.4 | ||||
| La florida | Leaf litter | Mycelia sterilia FH321 | 2.3 | ||||
| La florida | Leaf litter | Mycelia sterilia FH338 | 2.2 | ||||
| La florida | Leaf litter | Fusarium sp. FH676 | 2.3 | ||||
| La florida | Leaf litter | Fusarium sp. FH702 | 3.2 | ||||
| Puyacatengo | Leaf litter | Fusarium sp. PH20 | 2.2 | ||||
| Puyacatengo | Leaf litter | Fusarium sp. PH79 * | 1.9 | 1.8 | |||
| Puyacatengo | Leaf litter | Xylariaceae PH208 * | 1.9 | 1.9 | |||
| Puyacatengo | Leaf litter | Nodulisporium sp. PH223 | 1.4 | ||||
| Puyacatengo | Leaf litter | Nodulisporium sp. PH225 | 1.8 | ||||
| Puyacatengo | Leaf litter | Mycelia sterilia PH706 | 2.6 | ||||
| Cocona | Soil | Acremonium sp. CS330 | 1.2 | ||||
| Cocona | Soil | Xylariaceae CS708 | 1.4 | ||||
| Cocona | Soil | Aspergillus sp. CS1022 | 2.1 | ||||
| Cocona | Soil | Fusarium sp. CS1053 | 2.2 | ||||
| La florida | Soil | Fusarium sp. FS400 * | 3 | 2.1 | |||
| La florida | Soil | Paecilomyces sp. FS446 | 2.3 | ||||
| La florida | Soil | Cylindrocarpon sp. FS457 | 1.6 | ||||
| La florida | Soil | Paecilomyces sp. FS470 | 2.1 | ||||
| La florida | Soil | Fusarium sp. FS720 * | 2.5 | 1.7 | |||
| La florida | Soil | Graphium sp. FS835 | 2.3 | ||||
| La florida | Soil | Fusarium sp. FS903 | 2.2 | ||||
| La florida | Soil | Mycelia sterilia FS907 | 2.2 | ||||
| La florida | Soil | Mycelia sterilia FS943 | 2.3 | ||||
| Puyacatengo | Soil | Acremonium sp. PS595 | 2.8 | ||||
| Puyacatengo | Soil | Aspergillus sp. PS948 | 2.7 | ||||
| Puyacatengo | Soil | Ascomycete PS1130 | 3.2 | ||||
| Puyacatengo | Soil | Graphium sp. PS1143 | 2.2 | ||||
| Puyacatengo | Soil | Gliomastix sp. PS1298 | 2.4 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peraza-Jiménez, K.; De la Rosa-García, S.; Huijara-Vasconselos, J.J.; Reyes-Estebanez, M.; Gómez-Cornelio, S. Enzymatic Bioprospecting of Fungi Isolated from a Tropical Rainforest in Mexico. J. Fungi 2022, 8, 22. https://doi.org/10.3390/jof8010022
Peraza-Jiménez K, De la Rosa-García S, Huijara-Vasconselos JJ, Reyes-Estebanez M, Gómez-Cornelio S. Enzymatic Bioprospecting of Fungi Isolated from a Tropical Rainforest in Mexico. Journal of Fungi. 2022; 8(1):22. https://doi.org/10.3390/jof8010022
Chicago/Turabian StylePeraza-Jiménez, Karla, Susana De la Rosa-García, José Javier Huijara-Vasconselos, Manuela Reyes-Estebanez, and Sergio Gómez-Cornelio. 2022. "Enzymatic Bioprospecting of Fungi Isolated from a Tropical Rainforest in Mexico" Journal of Fungi 8, no. 1: 22. https://doi.org/10.3390/jof8010022
APA StylePeraza-Jiménez, K., De la Rosa-García, S., Huijara-Vasconselos, J. J., Reyes-Estebanez, M., & Gómez-Cornelio, S. (2022). Enzymatic Bioprospecting of Fungi Isolated from a Tropical Rainforest in Mexico. Journal of Fungi, 8(1), 22. https://doi.org/10.3390/jof8010022







