Genomic and Metabolomic Analyses of the Marine Fungus Emericellopsis cladophorae: Insights into Saltwater Adaptability Mechanisms and Its Biosynthetic Potential
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture Conditions and DNA Extraction
2.2. Genome Sequencing, Assembly, and Prediction
2.3. Genome Annotation and Functional Analysis
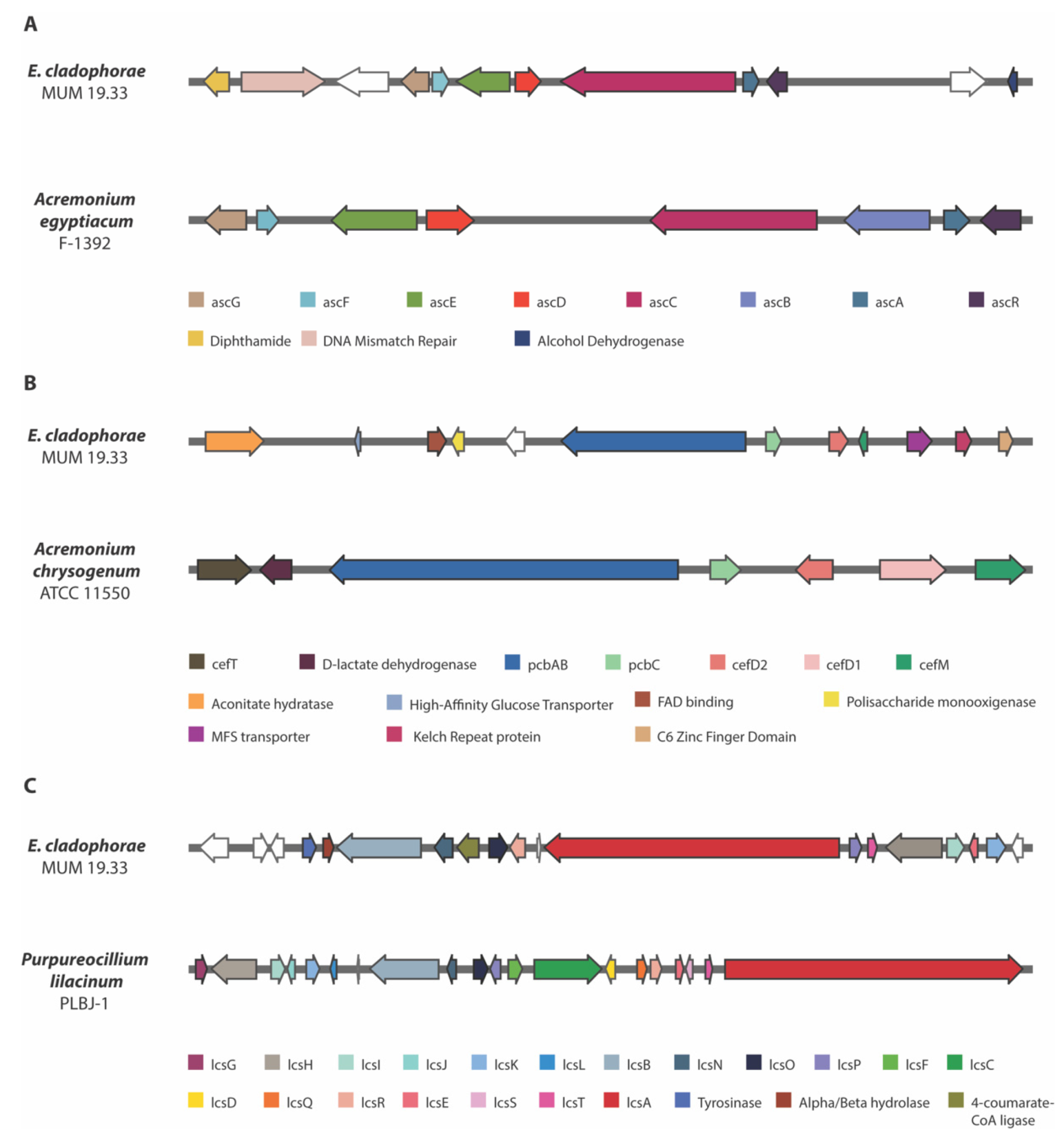
2.4. Comparative Analyses with E. atlantica
2.5. Small-Scale Fermentation and Extraction of Metabolites
2.6. LC-MS Data Analysis, Processing, and Visualization
3. Results and Discussion
3.1. Sequencing, Assembly Data and Genomic Characteristics
3.2. Repetitive Sequences and of tRNAs
3.3. Gene Annotation
3.4. Carbohydrate-Active Enzymes (CAZymes)
3.5. Transporter Proteins
3.6. Biosynthetic Gene Clusters
3.7. High-Osmolarity Glycerol (HOG) Pathway
3.8. Comparison of Genome Features between E. cladophorae MUM 19.33 and E. atlantica TS7
3.9. Metabolome Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kingma, F.V.B.T. Beschreibung einiger neuer Pilzarten aus dem Centraalbureau voor Schimmelcultures, Baarn (Nederland). Antonie Van Leeuwenhoek 1939, 6, 263–290. [Google Scholar] [CrossRef]
- Zuccaro, A.; Summerbell, R.C.; Gams, W.; Schoers, H.J.; Mitchell, J.I. A new Acremonium species associated with Fucus spp., and its affinity with a phylogenetically distinct marine Emericellopsis clade. Stud. Mycol. 2004, 50, 283–297. [Google Scholar]
- Konovalova, O.; Logacheva, M. Mitochondrial genome of two marine fungal species. Mitochondrial DNA Part A 2016, 27, 4280–4281. [Google Scholar] [CrossRef] [PubMed]
- Mohammadian, E.; Arzanlou, M.; Babai-Ahari, A. Two new hyphomycete species from petroleum-contaminated soils for mycobiota of Iran. Mycol. Iran. 2016, 3, 135–140. [Google Scholar]
- Tubaki, K. Aquatic sediment as a habitat of Emericellopsis, with a description of an undescribed species of Cephalosporium. Mycologia 1973, 65, 938–941. [Google Scholar] [CrossRef]
- Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of Soil Fungi, 2nd ed.; IHW Verlag: Eching, Germany, 2007; 384p. [Google Scholar]
- Gonçalves, M.F.M.; Vicente, T.F.; Esteves, A.C.; Alves, A. Novel halotolerant species of Emericellopsis and Parasarocladium associated with macroalgae in an estuarine environment. Mycologia 2020, 112, 154–171. [Google Scholar] [CrossRef]
- Kuvarina, A.E.; Gavryushina, I.A.; Kulko, A.B.; Ivanov, I.A.; Rogozhin, E.A.; Georgieva, M.L.; Sadykova, V.S. The Emericellipsins A–E from an alkalophilic fungus Emericellopsis alkalina show potent activity against multidrug-resistant pathogenic fungi. J. Fungi 2021, 7, 153. [Google Scholar] [CrossRef]
- Agrawal, S.; Saha, S. The genus Simplicillium and Emericellopsis: A review of phytochemistry and pharmacology. Biotechnology and Applied Biochemistry. Appl. Biochem. Biotechnol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Flissi, A.; Ricart, E.; Campart, C.; Chevalier, M.; Dufresne, Y.; Michalik, J.; Jacques, P.; Flahaut, C.; Lisacek, F.; Leclère, V.; et al. Norine: Update of the nonribosomal peptide resource. Nucleic Acids Res. 2020, 48, D465–D469. [Google Scholar] [PubMed]
- Jaworski, A.; Brückner, H. New sequences and new fungal producers of peptaibol antibiotics antiamoebins. J. Pept. Sci. 2000, 6, 149–167. [Google Scholar] [CrossRef]
- Berg, A.; Ritzau, M.; Ihn, W.; Schlegel, B.; Fleck, W.F.; Heinze, S.; Gräfe, U. Isolation and structure of bergofungin, a new antifungal peptaibol from Emericellopsis donezkii HKI 0059. J. Antibiot. 1996, 49, 817–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gessmann, R.; Axford, D.; Brückner, H.; Berg, A.; Petratos, K. A natural, single-residue substitution yields a less active peptaibiotic: The structure of bergofungin A at atomic resolution. Acta Crystallogr. F. Struct. Biol. Commun. 2017, 73, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Argoudelis, A.D.; Johnson, L.E. Emerimicins II, III and IV, antibiotics produced by Emericellopsis microspora in media supplemented with trans-4-n-propyl-L-proline. J. Antibiot. 1974, 27, 274–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inostroza, A.; Lara, L.; Paz, C.; Perez, A.; Galleguillos, F.; Hernandez, V.; Becerra, J.; González-Rocha, G.; Silva, M. Antibiotic activity of Emerimicin IV isolated from Emericellopsis minima from Talcahuano Bay, Chile. Nat. Prod. Res. 2018, 32, 1361–1364. [Google Scholar] [CrossRef]
- Ishiyama, D.; Satou, T.; Senda, H.; Fujimaki, T.; Honda, R.; Kanazawa, S. Heptaibin, a novel antifungal peptaibol antibiotic from Emericellopsis sp. BAUA8289. J. Antibiot. 2000, 53, 728–732. [Google Scholar] [CrossRef] [PubMed]
- Rinehart Jr, K.L.; Gaudioso, L.A.; Moore, M.L.; Pandey, R.C.; Cook Jr, J.C.; Barber, M.; Donald, S.R.; Bordoli, R.S.; Tyler, A.N.; Green, B.N. Structures of eleven zervamicin and two emerimicin peptide antibiotics studied by fast atom bombardment mass spectrometry. J. Am. Chem. Soc. 1981, 103, 6517–6520. [Google Scholar] [CrossRef]
- Gonçalves, M.F.M.; Paço, A.; Escada, L.F.; Albuquerque, M.S.F.; Pinto, C.A.; Saraiva, J.A.; Duarte, A.S.; Rocha-Santos, T.A.P.; Esteves, A.C.; Alves, A. Unveiling biological activities of marine fungi: The effect of sea salt. Appl. Sci. 2021, 11, 6008. [Google Scholar] [CrossRef]
- Vargas-Gastélum, L.; Riquelme, M. The mycobiota of the deep sea: What omics can offer. Life 2020, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Oppong-Danquah, E.; Passaretti, C.; Chianese, O.; Blümel, M.; Tasdemir, D. Mining the metabolome and the agricultural and pharmaceutical potential of sea foam-derived fungi. Mar. Drugs 2020, 18, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, L.E.; Marner, M.; Labes, A.; Tasdemir, D. Rapid metabolome and bioactivity profiling of fungi associated with the leaf and rhizosphere of the Baltic seagrass Zostera marina. Mar. Drugs 2019, 17, 419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagestad, O.C.; Hou, L.; Andersen, J.H.; Hansen, E.H.; Altermark, B.; Li, C.; Kuhnert, E.; Cox, R.J.; Crous, P.W.; Spatafora, J.W.; et al. Genomic characterization of three marine fungi, including Emericellopsis atlantica sp. nov. with signatures of a generalist lifestyle and marine biomass degradation. IMA Fungus 2021, 12, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Pitcher, D.G.; Saunders, N.A.; Owen, R.J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 1989, 8, 151–156. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanke, M.; Steinkamp, R.; Waack, S.; Morgenstern, B. AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res. 2004, 32, W309–W312. [Google Scholar] [CrossRef] [Green Version]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2018. Institute for Systems Biology. 2015. Available online: https://www.repeatmasker.org/ (accessed on 5 February 2021).
- Gelfand, Y.; Rodriguez, A.; Benson, G. TRDB—The tandem repeats database. Nucleic Acids Res. 2007, 35, D80–D87. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012, 40, W445–W451. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Saier Jr, M.H.; Reddy, V.S.; Tsu, B.V.; Ahmed, M.S.; Li, C.; Moreno-Hagelsieb, G. The transporter classification database (TCDB): Recent advances. Nucleic Acids Res. 2016, 44, D372–D379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, A.M.; Nascimento, A.S.; Polikarpov, I. Structural diversity of carbohydrate esterases. Biotechnol. Res. Innov. 2017, 1, 35–51. [Google Scholar] [CrossRef]
- Hehemann, J.H.; Truong, L.V.; Unfried, F.; Welsch, N.; Kabisch, J.; Heiden, S.E.; Junker, S.; Becher, D.; Thürmer, A.; Daniel, R.; et al. Aquatic adaptation of a laterally acquired pectin degradation pathway in marine gammaproteobacteria. Environ. Microbiol. 2017, 19, 2320–2333. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, J.K.; Hettle, A.G.; Vickers, C.; Boraston, A.B. Biochemical reconstruction of a metabolic pathway from a marine bacterium reveals its mechanism of pectin depolymerization. Appl. Environ. Microbiol. 2019, 85, e02114–e02118. [Google Scholar] [CrossRef] [Green Version]
- Ruocco, N.; Costantini, S.; Guariniello, S.; Costantini, M. Polysaccharides from the marine environment with pharmacological, cosmeceutical and nutraceutical potential. Molecules 2016, 21, 551. [Google Scholar] [CrossRef]
- dos Santos, S.C.; Teixeira, M.C.; Dias, P.J.; Sá-Correia, I. MFS transporter required for multidrug/multixenobiotic (MD/MX) resistance in the model yeast: Understanding their physiological function through post-genomic approaches. Front. Physiol. 2014, 5, 180. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Chen, J.; Xu, H.; Li, D. Role of a major facilitator superfamily transporter in adaptation capacity of Penicillium funiculosum under extreme acidic stress. Fungal Genet. Biol. 2014, 69, 75–83. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.H.; Tsai, H.C.; Yu, P.L.; Chung, K.R. A major facilitator superfamily transporter-mediated resistance to oxidative stress and fungicides requires Yap1, Skn7, and MAP kinases in the citrus fungal pathogen Alternaria alternata. PLoS ONE 2017, 12, e0169103. [Google Scholar] [CrossRef] [Green Version]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kogej, T.; Stein, M.; Volkmann, M.; Gorbushina, A.A.; Galinski, E.A.; Gunde-Cimerman, N. Osmotic adaptation of the halophilic fungus Hortaea werneckii: Role of osmolytes and melanization. Microbiology 2007, 153, 4261–4273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anwar, T.; Samudrala, G. Bioinformatics analysis and functional prediction of transmembrane proteins in entamoeba histolytica. Genes 2018, 9, 499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabir, F.; Loureiro-Dias, M.C.; Prista, C. Comparative analysis of sequences, polymorphisms and topology of yeasts aquaporins and aquaglyceroporins. FEMS Yeast Res. 2016, 16, fow025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmadpour, D.; Geijer, C.; Tamás, M.J.; Lindkvist-Petersson, K.; Hohmann, S. Yeast reveals unexpected roles and regulatory features of aquaporins and aquaglyceroporins. BBA Gen. Subj. 2014, 1840, 1482–1491. [Google Scholar] [CrossRef] [PubMed]
- Araki, Y.; Awakawa, T.; Matsuzaki, M.; Cho, R.; Matsuda, Y.; Hoshino, S.; Shinohara, Y.; Yamamoto, M.; Kido, Y.; Inaoka, D.K.; et al. Complete biosynthetic pathways of ascofuranone and ascochlorin in Acremonium egyptiacum. Proc. Natl. Acad. Sci. USA 2019, 116, 8269–8274. [Google Scholar] [CrossRef] [Green Version]
- Seephonkai, P.; Isaka, M.; Kittakoop, P.; Boonudomlap, U.; Thebtaranonth, Y. A novel ascochlorin glycoside from the insect pathogenic fungus Verticillium hemipterigenum BCC 2370. J. Antibiot. 2004, 57, 10–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Zhu, B. Study on genetic engineering of Acremonium chrysogenum, the cephalosporin C producer. Synth. Syst. Biotechnol. 2016, 1, 143–149. [Google Scholar] [CrossRef] [Green Version]
- Sarookhani, M.R.; Moazzami, N. Isolation of Acremonium species producing cephalosporine C (CPC) from forest soil in Gilan province, Iran. Afr. J. Biotechnol. 2007, 22, 2506–2510. [Google Scholar] [CrossRef]
- Ullán, R.; Liu, G.; Casqueiro, J.; Gutiérrez, S.; Banuelos, O.; Martín, J.F. The cefT gene of Acremonium chrysogenum C10 encodes a putative multidrug efflux pump protein that significantly increases cephalosporin C production. Mol. Genet. Genom. 2002, 267, 673–683. [Google Scholar] [CrossRef]
- Teijeira, F.; Ullán, R.V.; Guerra, S.M.; García-Estrada, C.; Vaca, I.; Martín, J.F. The transporter CefM involved in translocation of biosynthetic intermediates is essential for cephalosporin production. Biochem. J. 2009, 418, 113–124. [Google Scholar] [CrossRef] [Green Version]
- Arai, T.; Mikami, Y.; Fukushima, K.; Utsumi, T.; Yazawa, K. A new antibiotic, leucinostatin, derived from Penicillium lilacinum. J. Antibiot. 1973, 26, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Liu, Z.; Lin, R.; Li, E.; Mao, Z.; Ling, J.; Yang, Y.; Yin, W.B.; Xie, B. Biosynthesis of antibiotic leucinostatins in bio-control fungus Purpureocillium lilacinum and their inhibition on Phytophthora revealed by genome mining. PLoS Pathog. 2016, 12, e1005685. [Google Scholar] [CrossRef] [Green Version]
- Strobel, G.A.; Torczynski, R.; Bollon, A. Acremonium sp.—A leucinostatin A producing endophyte of European yew (Taxus baccata). Plant Sci. 1997, 128, 97–108. [Google Scholar] [CrossRef]
- Scharf, D.H.; Brakhage, A.A. Engineering fungal secondary metabolism: A roadmap to novel compounds. J. Biotechnol. 2013, 163, 179–183. [Google Scholar] [CrossRef]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: A literature review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, C.Y.; Chen, S.A.; Hsueh, Y.P. The high osmolarity glycerol (HOG) pathway functions in osmosensing, trap morphogenesis and conidiation of the nematode-trapping fungus Arthrobotrys oligospora. J. Fungi 2020, 6, 191. [Google Scholar] [CrossRef]
- Ren, W.; Liu, N.; Yang, Y.; Yang, Q.; Chen, C.; Gao, Q. The sensor proteins BcSho1 and BcSln1 are involved in, though not essential to, vegetative differentiation, pathogenicity and osmotic stress tolerance in Botrytis cinerea. Front. Microbiol. 2019, 10, 328. [Google Scholar] [CrossRef]
- de Assis, L.J.; Silva, L.P.; Liu, L.; Schmitt, K.; Valerius, O.; Braus, G.H.; Ries, L.N.A.; Goldman, G.H. The high osmolarity glycerol mitogen-activated protein kinase regulates glucose catabolite repression in filamentous fungi. PLoS Genetics 2020, 16, 1008996. [Google Scholar] [CrossRef] [PubMed]
- Gladfelter, A.S.; James, T.Y.; Amend, A.S. Marine fungi. Curr. Biol. 2019, 29, R191–R195. [Google Scholar] [CrossRef]
- Román, E.; Correia, I.; Prieto, D.; Alonso, R.; Pla, J. The HOG MAPK pathway in Candida albicans: More than an osmosensing pathway. Int. Microbiol. 2020, 2, 23–29. [Google Scholar] [CrossRef]
- Miskei, M.; Karányi, Z.; Pócsi, I. Annotation of stress–response proteins in the aspergilli. Fungal Genet. Biol. 2009, 46, S105–S120. [Google Scholar] [CrossRef] [Green Version]
- Huberman, L.B.; Coradetti, S.T.; Glass, N.L. Network of nutrient-sensing pathways and a conserved kinase cascade integrate osmolarity and carbon sensing in Neurospora crassa. Proc. Natl. Acad. Sci. USA 2017, 114, E8665–E8674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takayama, T.; Yamamoto, K.; Saito, H.; Tatebayashi, K. Interaction between the transmembrane domains of Sho1 and Opy2 enhances the signaling efficiency of the Hog1 MAP kinase cascade in Saccharomyces cerevisiae. PLoS ONE 2019, 14, e0211380. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, A.I.; Coutinho, A.J.; Costa Lima, S.A.; Reis, S. Marine polysaccharides in pharmaceutical applications: Fucoidan and chitosan as key players in the drug delivery match field. Mar. Drugs 2019, 17, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bremer, E.; Krämer, R. Responses of microorganisms to osmotic stress. Annu. Rev. Microbiol. 2019, 73, 313–334. [Google Scholar] [CrossRef] [PubMed]
- Bidochka, M.J.; Low, N.H.; Khachatourians, G.G. Carbohydrate storage in the entomopathogenic fungus Beauveria bassiana. Appl. Environ. Microbiol. 1990, 56, 3186–3190. [Google Scholar] [CrossRef] [Green Version]
- Cairns, T.C.; Nai, C.; Meyer, V. How a fungus shapes biotechnology: 100 years of Aspergillus niger research. Fungal Biol. Biotechnol. 2018, 5, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef]
- Meir, Z.; Osherov, N. Vitamin biosynthesis as an antifungal target. J. Fungi 2018, 4, 72. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.Z.; Yan, X.; Tang, X.X.; Lin, J.G.; Qiu, Y.K. New bis-alkenoic acid derivatives from a marine-derived fungus Fusarium solani H915. Mar. Drugs 2018, 16, 483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Averianova, L.A.; Balabanova, L.A.; Son, O.M.; Podvolotskaya, A.B.; Tekutyeva, L.A. Production of vitamin B2 (riboflavin) by microorganisms: An overview. Front. Bioeng. Biotechnol. 2020, 8, 570828. [Google Scholar] [CrossRef]
- Abbas, C.A.; Sibirny, A.A. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol. Mol. Biol. Rev. 2011, 75, 321–360. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, A.M.D.M.L. Study of Bioactive Compounds of Marine-Derived Fungi. Master’s Thesis, Instituto de Ciências Biomédicas Abel Salazar–University of Porto, Porto, Portugal, 2012. [Google Scholar]
- Hernández-Chávez, G.; Martinez, A.; Gosset, G. Metabolic engineering strategies for caffeic acid production in Escherichia coli. Electron. J. Biotechnol. 2019, 38, 19–26. [Google Scholar] [CrossRef]
- Kumara, P.M.; Soujanya, K.N.; Ravikanth, G.; Vasudeva, R.; Ganeshaiah, K.N.; Shaanker, R.U. Rohitukine, a chromone alkaloid and a precursor of flavopiridol, is produced by endophytic fungi isolated from Dysoxylum binectariferum Hook. f and Amoora rohituka (Roxb). Wight & Arn. Phytomedicine 2014, 21, 541–546. [Google Scholar] [PubMed]
- Wu, X.; Zhang, G.; Hu, M.; Pan, J.; Li, A.; Zhang, Y. Molecular characteristics of gallocatechin gallate affecting protein glycation. Food Hydrocoll. 2020, 105, 105782. [Google Scholar] [CrossRef]
- Liu, S.; Hu, W.; Wang, Z.; Chen, T. Production of riboflavin and related cofactors by biotechnological processes. Microb. Cell Factories 2020, 19, 1–16. [Google Scholar] [CrossRef]
- Dietl, A.M.; Meir, Z.; Shadkchan, Y.; Osherov, N.; Haas, H. Riboflavin and pantothenic acid biosynthesis are crucial for iron homeostasis and virulence in the pathogenic mold Aspergillus fumigatus. Virulence 2018, 9, 1036–1049. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.D.; Wang, P.; Zhang, J.; Wang, W.; Yao, L.P.; Gu, C.B.; Efferth, T.; Fu, Y.J. 2′ O-galloylhyperin attenuates LPS-induced acute lung injury via up-regulation antioxidation and inhibition of inflammatory responses in vivo. Chem. Biol. Interact. 2019, 304, 20–27. [Google Scholar] [CrossRef]
- Kongkiatpaiboon, S.; Vongsak, B.; Machana, S.; Weerakul, T.; Pattarapanich, C. Simultaneous HPLC quantitative analysis of mangostin derivatives in Tetragonula pagdeni propolis extracts. J. King Saud. Univ. Sci. 2016, 28, 131–135. [Google Scholar] [CrossRef] [Green Version]
- Masui, H.; Kondo, T.; Kojima, M. An antifungal compound, 9, 12, 13-trihydroxy-(E)-10-octadecenoic acid, from Colocasia antiquorum inoculated with Ceratocystis fimbriata. Phytochemistry 1989, 28, 2613–2615. [Google Scholar] [CrossRef]
- Sun, M.Y.; Ye, Y.; Xiao, L.; Rahman, K.; Xia, W.; Zhang, H. Daidzein: A review of pharmacological effects. Afr. J. Tradit. Complement. Altern. Med. 2016, 13, 117–132. [Google Scholar] [CrossRef] [Green Version]
- Howlett, D.R.; George, A.R.; Owen, D.E.; Ward, R.V.; Markwell, R.E. Common structural features determine the effectiveness of carvedilol, daunomycin and rolitetracycline as inhibitors of Alzheimer β-amyloid fibril formation. Biochem. J. 1999, 343, 419–423. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Kaihatsu, K.; Nishino, K.; Ogawa, M.; Kato, N.; Yamaguchi, A. Antibacterial and antifungal activities of new acylated derivatives of epigallocatechin gallate. Front. Microbiol. 2012, 3, 53. [Google Scholar] [CrossRef] [Green Version]
- Panaccione, D.G. Origins and significance of ergot alkaloid diversity in fungi. FEMS Microbiol. Lett. 2005, 251, 9–17. [Google Scholar] [CrossRef] [Green Version]
- Ledesma-Amaro, R.; Buey, R.M.; Revuelta, J.L. Increased production of inosine and guanosine by means of metabolic engineering of the purine pathway in Ashbya gossypii. Microb. Cell Factories 2015, 14, 58. [Google Scholar] [CrossRef] [Green Version]
- Ramu, A.K.; Ali, D.; Alarifi, S.; Abuthakir, M.H.S.; Abdul, B.A.A. Reserpine inhibits DNA repair, cell proliferation, invasion and induces apoptosis in oral carcinogenesis via modulation of TGF-β signaling. Life Sci. 2021, 264, 118730. [Google Scholar] [CrossRef]
- Qin, Z.; Yan, Q.; Yang, S.; Jiang, Z. Modulating the function of a β-1, 3-glucanosyltransferase to that of an endo-β-1, 3-glucanase by structure-based protein engineering. Appl. Microbiol. Biotechnol. 2016, 100, 1765–1776. [Google Scholar] [CrossRef]
- Zonaras, V.; Tyrpenou, A.; Alexis, M.; Koupparis, M. Determination of sulfadiazine, trimethoprim, and N4-acetyl-sulfadiazine in fish muscle plus skin by Liquid Chromatography–Mass Spectrometry. Withdrawal-time calculation after in-feed administration in gilthead sea bream (Sparus aurata L.) fed two different diets. J. Vet. Pharmacol. Ther. 2016, 39, 504–513. [Google Scholar]
- Tsuchikado, R.; Kami, S.; Takahashi, S.; Nishida, H. Novobiocin inhibits membrane synthesis and vacuole formation of Enterococcus faecalis protoplasts. Microb. Cell 2020, 7, 300. [Google Scholar] [CrossRef]
- Kawaguti, H.Y.; Sato, H.H. Palatinose production by free and Ca-alginate gel immobilized cells of Erwinia sp. Biochem. Eng. J. 2007, 36, 202–208. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, M.; Dong, Z.; Zhao, D.; An, L.; Zhu, H.; Xia, Q.; Zhao, P. Synthesis, secretion, and antifungal mechanism of a phosphatidylethanolamine-binding protein from the silk gland of the silkworm Bombyx mori. Int. J. Biol. Macromol. 2020, 149, 1000–1007. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Narukami, T.; Nameki, M.; Ozawa, T.; Kamimura, Y.; Hoshino, T.; Takaya, N. Heme-biosynthetic porphobilinogen deaminase protects Aspergillus nidulans from nitrosative stress. Appl. Environ. Microbiol. 2012, 78, 103–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, J.; Xie, Y.; Yu, H.; Guo, Y.; Cheng, Y.; Qian, H.; Yao, W. Synergistic antifungal mechanism of thymol and salicylic acid on Fusarium solani. LWT 2021, 140, 110787. [Google Scholar] [CrossRef]
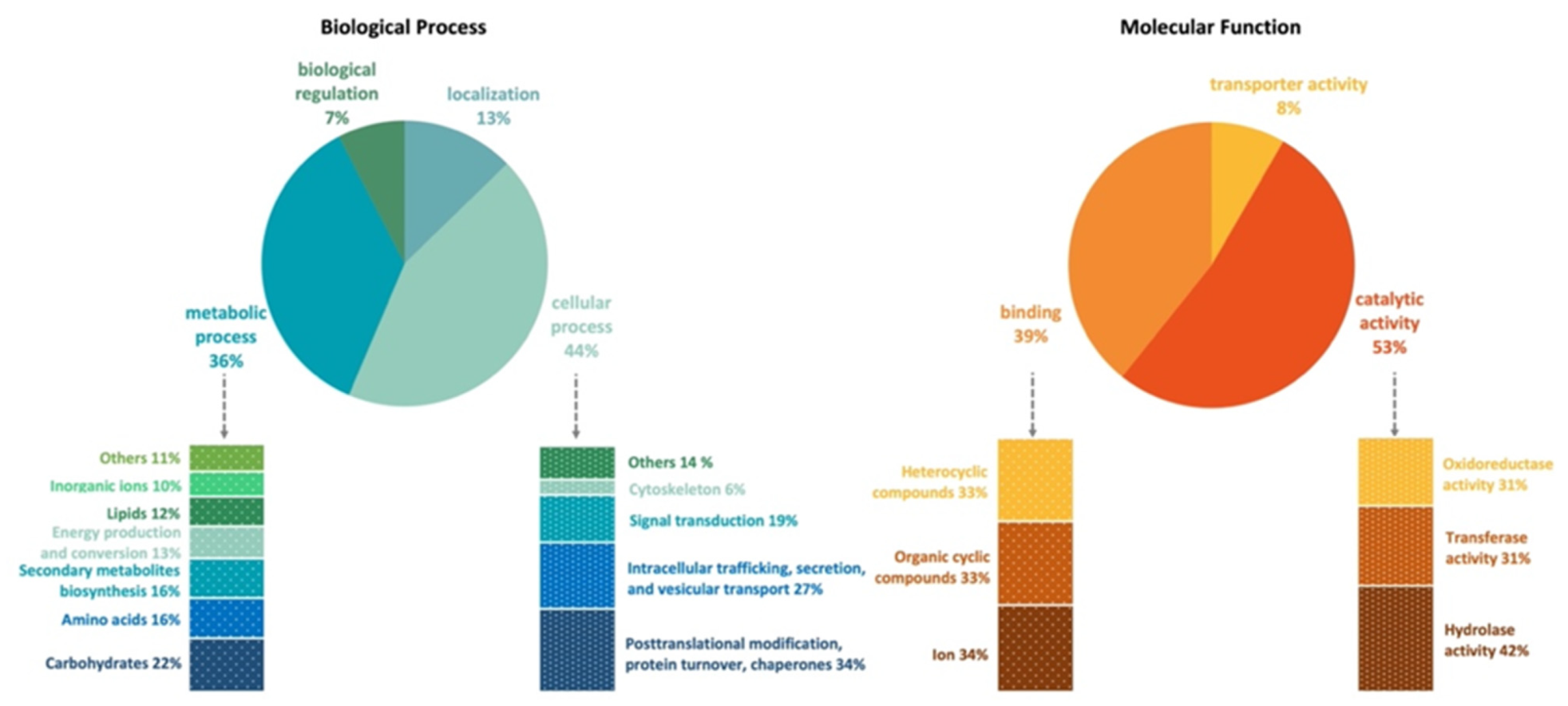



| General Features | |
|---|---|
| Genome assembled | 26.9 Mb |
| Number of contigs (>500 bp) | 300 |
| Largest contig length | 1,489,480 bp |
| N50 | 315,653 bp |
| N75 | 183,754 bp |
| GC content | 54.34% |
| Number of predicted genes | 8572 |
| Total length of predicted genes | 13,253,623 bp |
| Average length of predicted genes | 1546 bp |
| Total length of predicted genes/Genome assembled | 49.2% |
| Average of exons per gene | 3 |
| Average of introns per gene | 2 |
| Type | Number | Total Length (bp) | Percentage in Genome (%) | |
|---|---|---|---|---|
| Interspersed repeat | SINEs | 0 | 0 | 0.0000 |
| LINEs | 3 | 196 | 0.0007 | |
| LTRs | 128 | 48,958 | 0.1817 | |
| DNA transposons | 22 | 1362 | 0.0051 | |
| Rolling-circles | 1 | 37 | 0.0001 | |
| Unclassified | 0 | 0 | 0.0000 | |
| Small RNA | 62 | 9505 | 0.0353 | |
| Satellites | 5 | 698 | 0.0026 | |
| Simple repeats | 4656 | 183,030 | 0.6794 | |
| Low complexity | 381 | 17,309 | 0.0643 | |
| Total | 5258 | 261,095 | 0.9692 | |
| Tandem repeat tRNAs | 2365 122 | 232,036 10,432 | 0.8614 0.0387 |
| Transporter Class | Number of Genes (n) |
|---|---|
| Channels and pores (TC 1) | 461 |
| Electrochemical potential-driven transporters (TC 2) | 570 |
| Primary active transporters (TC 3) | 405 |
| Group translocators (TC 4) | 77 |
| Transmembrane electron carriers (TC 5) | 30 |
| Accessory factors involved in transport (TC 8) | 270 |
| Incompletely characterized transport systems (TC 9) | 384 |
| Total | 2197 |
| E. cladophorae MUM 19.33 | E. atlantica TS7 | ||
|---|---|---|---|
| Genome assembled | 26.9 Mb | 27.3 Mb | |
| Coverage | 130 | 225.6 | |
| GC content | 54.34% | 54.2% | |
| Number of genes | 8572 | 9964 | |
| Average genes length | 1546 bp | 1832 bp | |
| Genes encoding CAZymes | 407 | 396 | |
| AA | 72 | 53 | |
| CBM | 8 | 40 | |
| CE | 28 | 21 | |
| GH | 200 | 217 | |
| GT | 83 | 93 | |
| PL | 16 | 17 | |
| BGCs | 37 | 35 | |
| NRPS | 10 | 8 | |
| NRPS-like | 7 | 6 | |
| PKs | 6 | 6 | |
| NRPS-PKs | 5 | 3 | |
| NRPS-like-PKs | 1 | 0 | |
| NRPS-PKs-hybrid | 0 | 1 | |
| Terpenes | 7 | 9 | |
| Indole | 0 | 1 | |
| Phosphonate | 1 | 1 |
| Putative Metabolite | m/z | Rt | Adduct | Molecular Formula | Class | Function |
|---|---|---|---|---|---|---|
| (-)-Gallocatechin 3-gallate | 169.0130 | 6.87 | [M−H-C15H12O6]− | C22H18O11 | Benzopyrans | Antioxidant activity and inhibitory ability on α-amylase and α-glucosidase related to diabetes mellitus [79] |
| (-)-Riboflavin | 375.1300 | 8.16 | [M−H]− | C17H20N4O6 | Vitamin | Known as vitamin B2 and is the central source of all important flavins [80]. It may be an attractive target for antifungal therapy [81] |
| 2’-O-Galloylhyperin | 307.0484 | 10.47 | [M−2H]− | C28H24O16 | Carboxilic Acid | Antioxidant and anti-inflammatory [82] |
| 3-Isomangostin | 427.1782 | 4.91 | [M + OH]− | C24H26O6 | Xanthone | Derivative of mangostin that has antioxidant, anti-inflammatory, anticancer and anti-microbial activities [83] |
| 3,4-dihydroxycinnamic acid | 264.0863 | 17.01 | [M−H]− | C13H15NO5 | Carboxylic Acid | Antioxidant, anti-cancer, anti-viral and anti- inflammatory [77] |
| 9,12,13-Trihydroxyoctadec-10-enoic acid | 329.2324 | 23.40 | [M−H]− | C18H34O5 | Carboxilic Acid | Antifungal [84] |
| Citric Acid | 191.0185 | 1.56 | [M−H]− | C6H8O7 | Carboxylic Acid | Antioxidant, preservative, acidulant and pH- regulator [70] |
| Daidzein | 253.0498 | 14.82 | [M−H]− | C15H10O4 | Flavonoids | Anticancer, anti-inflammatory, protective effects against osteoporosis, diabetes, and cardiovascular diseases [85] |
| Daunomycinone | 379.0825 | 1.05 | [M−H-H2O]− | C21H18O8 | Naphthacene | Antibiotic with anti-cancer activity [86] |
| (-)-Epigallocatechin | 611.1352 | 2.55 | [2M−H]− | C15H14O7 | Benzopyrans | Antiviral, antimicrobial, antitoxin and anticancer [87] |
| Ergocryptine | 558.0951 | 16.35 | [M−H]− | C32H41N5O5 | Alkaloids | Cause ergot in cereal grains and fescue toxicoses in animals [88] |
| Flavopiridol | 382.0995 | 3.92 | [M−H]− | C21H20ClNO5 | Piperidines | Treatment of chronic lymphocytic leukemia [78] |
| Guanosine | 282.0838 | 2.27 | [M−H]− | C10H13N5O5 | Nucleosides | Antioxidant, neuroprotective, cardiotonic and immuno-modulatory properties [89] |
| Hymeglusin | 647.3769 | 21.57 | [2M−H]− | C18H28O5 | Lactones | Fungal beta-lactone antibiotic with anti-fungal activity [73] |
| Isoreserpin | 607.2677 | 2.33 | [M−H]− | C33H40N2O9 | Alkaloids | Anticancer [90] |
| Laminaritetraose | 701.1903 | 1.10 | [M + Cl]− | C24H42O21 | Carbohydrates | Obtained from hydrolysis of laminarin, which is a carbohydrate food reserve [91] |
| N4-Acetylsulfadiazine | 291.0537 | 15.90 | [M−H]− | C12H12N4O3S | Sulfonamide | Marine xenobiotic which is the main constituent of sulfadiazine (antibiotic) [92] |
| NovobiocinA | 611.2305 | 4.49 | [M−H]− | C31H36N2O11 | Glycoside | Antibacterial [93] |
| Palatinose | 341.1078 | 1.05 | [M−H]− | C12H22O11 | Carbohydrates | Obtained from the enzymatic conversion of sucrose, used in food industries as a sugar substitute [94] |
| Pantothenic acid | 18.1024 | 3.84 | [M−H]− | C9H17NO5 | Vitamin | Known as vitamin B5 and is essential for fatty acid and carbohydrate metabolism. It may be an attractive target for antifungal therapy [81] |
| Phosphatidylethanolamine | 612.3720 | 7.33 | [M−H]− | C32H56NO8P | Glycerophospholipids | Antifungal [95] |
| Porphobilinogen | 225.0870 | 3.72 | [M−H]− | C10H14N2O4 | Pirrole | Involved in the heme biosynthetic pathway and protection from nitrosative stress [96] |
| Salicylic acid | 137.0237 | 4.12 | [M−H]− | C7H6O3 | Carboxilic Acid | Antifungal [97] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves, M.F.M.; Hilário, S.; Van de Peer, Y.; Esteves, A.C.; Alves, A. Genomic and Metabolomic Analyses of the Marine Fungus Emericellopsis cladophorae: Insights into Saltwater Adaptability Mechanisms and Its Biosynthetic Potential. J. Fungi 2022, 8, 31. https://doi.org/10.3390/jof8010031
Gonçalves MFM, Hilário S, Van de Peer Y, Esteves AC, Alves A. Genomic and Metabolomic Analyses of the Marine Fungus Emericellopsis cladophorae: Insights into Saltwater Adaptability Mechanisms and Its Biosynthetic Potential. Journal of Fungi. 2022; 8(1):31. https://doi.org/10.3390/jof8010031
Chicago/Turabian StyleGonçalves, Micael F. M., Sandra Hilário, Yves Van de Peer, Ana C. Esteves, and Artur Alves. 2022. "Genomic and Metabolomic Analyses of the Marine Fungus Emericellopsis cladophorae: Insights into Saltwater Adaptability Mechanisms and Its Biosynthetic Potential" Journal of Fungi 8, no. 1: 31. https://doi.org/10.3390/jof8010031
APA StyleGonçalves, M. F. M., Hilário, S., Van de Peer, Y., Esteves, A. C., & Alves, A. (2022). Genomic and Metabolomic Analyses of the Marine Fungus Emericellopsis cladophorae: Insights into Saltwater Adaptability Mechanisms and Its Biosynthetic Potential. Journal of Fungi, 8(1), 31. https://doi.org/10.3390/jof8010031










