Predication of the Effector Proteins Secreted by Fusarium sacchari Using Genomic Analysis and Heterogenous Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Prediction of Fungal CSEPs
2.3. Structural Characters of the CSEPs
2.4. Plasmid Construction and Preparation
2.5. Transient Expression of Target CSEPs in N. benthamiana
2.6. Verification of the Secretory Function of the Signal Peptide
2.7. qRT-PCR Validation of Target Genes
3. Results
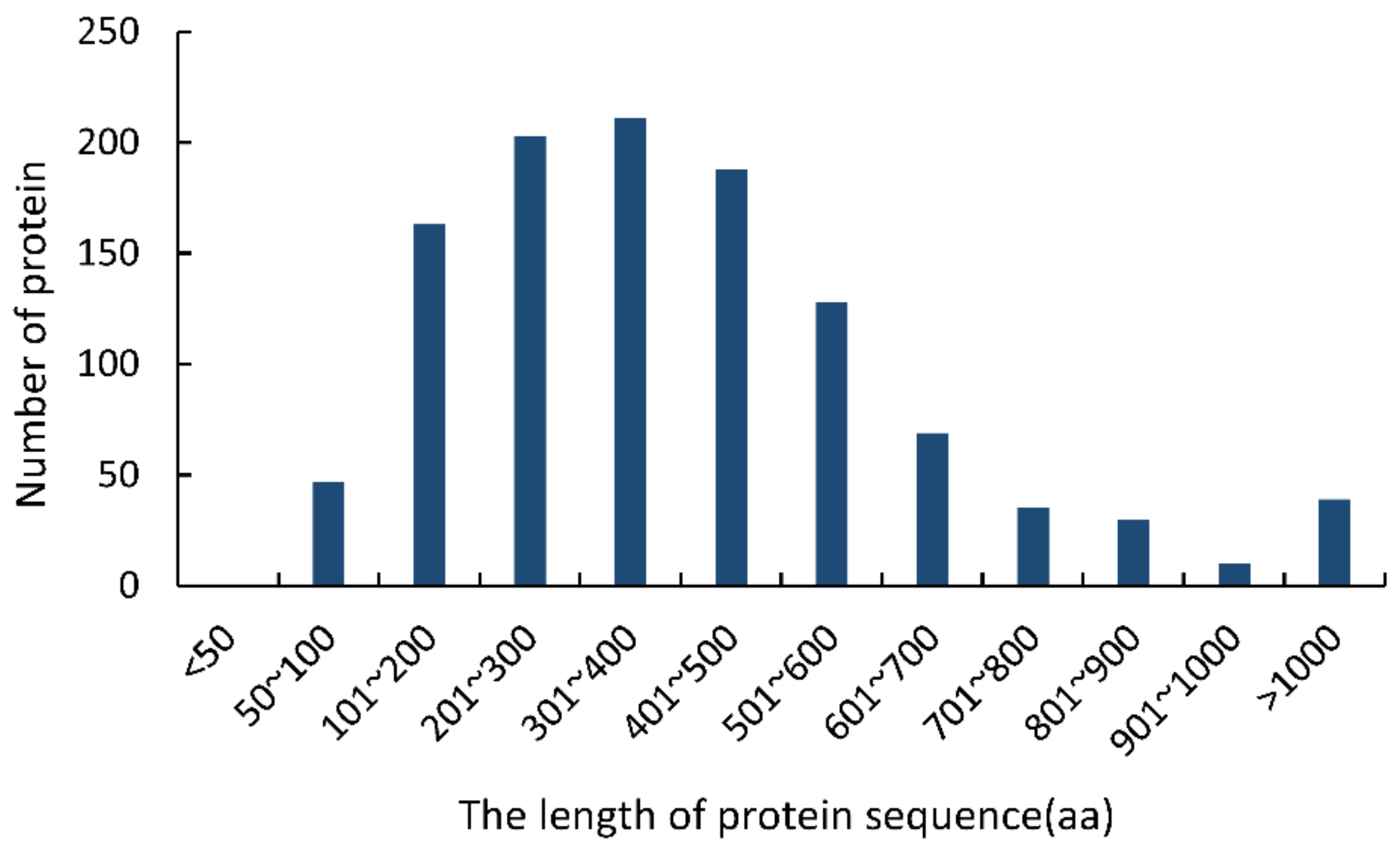
3.1. Comprehensive CSEP Prediction
3.2. The CSEPs of F. sacchari Had Typical Structural Characteristics
3.3. Certain CSEPs Induced PCD or Suppressed BAX-Triggered PCD in N. benthamiana
3.4. Validation of the Signal Peptides of Candidate Effector Proteins
3.5. CSEP Expression Profiles in F. sacchari at Different Stages of Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, A.; Chandra, A. Identification of New Leuconostoc Species Responsible for Post-harvest Sucrose Losses in Sugarcane. Sugar Tech 2018, 20, 492–496. [Google Scholar] [CrossRef]
- Li, Y.-R.; Song, X.-P.; Wu, J.-M.; Li, C.-N.; Liang, Q.; Liu, X.-H.; Wang, W.-Z.; Tan, H.-W.; Yang, L.-T. Sugar Industry and Improved Sugarcane Farming Technologies in China. Sugar Tech 2016, 18, 603–611. [Google Scholar] [CrossRef]
- Lin, Z.; Xu, S.; Que, Y.; Wang, J.; Comstock, J.C.; Wei, J.; Mccord, P.H.; Chen, B.; Chen, R.; Zhang, M. Species-Specific Detection and Identification of Fusarium Species Complex, the Causal Agent of Sugarcane Pokkah Boeng in China. PLoS ONE 2014, 9, e104195. [Google Scholar] [CrossRef]
- Yao, Z.; Zou, C.; Peng, N.; Zhu, Y.; Bao, Y.; Zhou, Q.; Wu, Q.; Chen, B.; Zhang, M. Virome Identification and Characterization of Fusarium sacchari and F. andiyazi: Causative Agents of Pokkah Boeng Disease in Sugarcane. Front. Microbiol. 2020, 11, 240. [Google Scholar] [CrossRef] [Green Version]
- Viswanathan, R.; Balaji, C.G.; Selvakumar, R.; Malathi, P.; Sundar, A.R.; Prasanth, C.N.; Chhabra, M.L.; Parameswari, B. Epidemiology of Fusarium Diseases in Sugarcane: A New Discovery of Same Fusarium sacchari Causing Two Distinct Diseases, Wilt and Pokkah Boeng. Sugar Tech 2017, 19, 638–646. [Google Scholar] [CrossRef]
- Viswanathan, R. Fusarium diseases affecting sugarcane production in India. Indian Phytopathol. 2020, 73, 415–424. [Google Scholar] [CrossRef]
- Meng, J.R.; Huang, H.J.; Li, Y.X.; Li, Y.J.; Li, J.Q.; Chen, B.S. First Report of Fusarium sacchari Causing Sugarcane Pokkah Boeng in China. Plant Dis. 2020, 104, 1553. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling Mechanisms in Pattern-Triggered Immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, J.; Minren, H.; Li’An, X. LysM Domains and Its Roles in Plant-Fungus Interactions. Chin. Bull. Bot. 2014, 49, 221. [Google Scholar] [CrossRef]
- Liang, W.; Tong, M.; Li, X. SUSA2 is an F-box protein required for autoimmunity mediated by paired NLRs SOC3-CHS1 and SOC3-TN2. Nat. Commun. 2020, 11, 5190. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, aaf6395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- HH, F. Host-parasite interactions in flax rust-its genetics and other implications. Phytopathology 1955, 45, 680–685. [Google Scholar]
- Stergiopoulos, I.; de Wit, P.J.G.M. Fungal Effector Proteins. Annu. Rev. Phytopathol. 2009, 47, 233–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocafort, M.; Fudal, I.; Mesarich, C.H. Apoplastic effector proteins of plant-associated fungi and oomycetes. Curr. Opin. Plant Biol. 2020, 56, 9–19. [Google Scholar] [CrossRef]
- Vleeshouwers, V.G.A.A.; Oliver, R.P. Effectors as Tools in Disease Resistance Breeding Against Biotrophic, Hemibiotrophic, and Necrotrophic Plant Pathogens. Mol. Plant-Microbe Interact. 2014, 27, 196–206. [Google Scholar] [CrossRef] [Green Version]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef] [Green Version]
- Houterman, P.M.; Cornelissen, B.J.C.; Rep, M. Suppression of Plant Resistance Gene-Based Immunity by a Fungal Effector. PLOS Pathog. 2008, 4, e1000061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhse, S.; Djamei, A. Effectors of plant-colonizing fungi and beyond. PLoS Pathog. 2018, 14, e1006992. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, A.; Ghosh, S.; Sahoo, D.; Jha, G. Fungal effectors, the double edge sword of phytopathogens. Curr. Genet. 2021, 67, 27–40. [Google Scholar] [CrossRef]
- Van de Wouw, A.P.; Idnurm, A. Biotechnological potential of engineering pathogen effector proteins for use in plant disease management. Biotechnol. Adv. 2019, 37, 107387. [Google Scholar] [CrossRef]
- van Kan, J.A.; van den Ackerveken, G.F.; de Wit, P.J. Cloning and Characterization of cDNA of Avirulence Geneavr9 of the Fungal PathogenCladosporium fulvum, Causal Agent of Tomato Leaf Mold. Mol. Plant-Microbe Interact. 1991, 4, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.A.; Bertazzoni, S.; Turo, C.J.; Syme, R.A.; Hane, J.K. Bioinformatic prediction of plant–pathogenicity effector proteins of fungi. Curr. Opin. Microbiol. 2018, 46, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Dodds, P.N.; Gardiner, D.M.; Manners, J.M.; Singh, K.B.; Taylor, J.M. Advances and Challenges in Computational Prediction of Effectors from Plant Pathogenic Fungi. PLoS Pathog. 2015, 11, e1004806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.-T.; Jeon, J.; Choi, J.; Cheong, K.; Song, H.; Choi, G.; Kang, S.; Lee, Y.-H. Kingdom-Wide Analysis of Fungal Small Secreted Proteins (SSPs) Reveals their Potential Role in Host Association. Front. Plant Sci. 2016, 7, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Q.; Wang, H.; Xu, B.; Zhu, S.; Hu, L.; Huang, M. Discovery of a novel small secreted protein family with conserved N-terminal IGY motif in Dikarya fungi. BMC Genom. 2014, 15, 1151. [Google Scholar] [CrossRef] [Green Version]
- Birch, P.R.; Boevink, P.C.; Gilroy, E.M.; Hein, I.; Pritchard, L.; Whisson, S.C. Oomycete RXLR effectors: Delivery, functional redundancy and durable disease resistance. Curr. Opin. Plant Biol. 2008, 11, 373–379. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, J.; Qi, Y.; Li, J.; Amin, R.; Yang, W.; Liu, D. Predicating the Effector Proteins Secreted by Puccinia triticina Through Transcriptomic Analysis and Multiple Prediction Approaches. Front. Microbiol. 2020, 11, 2295. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H. Predicting Secretory Proteins with SignalP. Methods Mol. Biol. 2017, 1611, 73. [Google Scholar]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting Subcellular Localization of Proteins Based on their N-terminal Amino Acid Sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [Green Version]
- Emanuelsson, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Eisenhaber, B.; Schneider, G.; Wildpaner, M.; Eisenhaber, F. A Sensitive Predictor for Potential GPI Lipid Modification Sites in Fungal Protein Sequences and its Application to Genome-wide Studies for Aspergillus nidulans, Candida albicans Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe. J. Mol. Biol. 2004, 337, 243–253. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, H.-G.; Zhang, X.-H.; Wang, T.-T.; Wei, W.-L.; Wang, Y.-X.; Chen, J.; Zhou, Y.-B.; Chen, M.; Ma, Y.-Z.; Xu, Z.-S.; et al. Genome-Wide Identification, Evolution, and Expression of GDSL-Type Esterase/Lipase Gene Family in Soybean. Front. Plant Sci. 2020, 11, 726. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xu, L.; Jia, Q.; Pan, R.; Oelmüller, R.; Zhang, W.; Wu, C. Arms race: Diverse effector proteins with conserved motifs. Plant Signal. Behav. 2019, 14, e1557008. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Kanneganti, T.-D.; Huitema, E.; Kamoun, S. In planta Expression of Oomycete and Fungal Genes. Methods Mol. Biol. 2007, 354, 35–43. [Google Scholar] [CrossRef]
- Ma, L.; Lukasik, E.; Gawehns, F.; Takken, F.L.W. The Use of Agroinfiltration for Transient Expression of Plant Resistance and Fungal Effector Proteins in Nicotiana benthamiana Leaves. Methods Mol. Biol. 2012, 835, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Nelson, R.S.; Sherwood, J.L. Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. BioTechniques 1994, 16, 664–668. [Google Scholar] [PubMed]
- Jacobs, K.A.; Collins-Racie, L.A.; Colbert, M.; Duckett, M.; Golden-Fleet, M.; Kelleher, K.; Kriz, R.; LaVallie, E.R.; Merberg, D.; Spaulding, V.; et al. A genetic selection for isolating cDNAs encoding secreted proteins. Gene 1997, 198, 289–296. [Google Scholar] [CrossRef]
- Huang, Z.; Li, H.; Zhou, Y.; Bao, Y.; Zhang, M.; Yao, W. Identification and functional analysis of Nep1-like proteins of Fusarium sacchari, the pathogen of sugarcane pokkah boeng disease. Acta Phytopathol. Sin. 2022, 1–13. [Google Scholar] [CrossRef]
- Lin, Z.; Wang, J.; Bao, Y.; Guo, Q.; Powell, C.A.; Xu, S.; Chen, B.; Zhang, M. Deciphering the transcriptomic response of Fusarium verticillioides in relation to nitrogen availability and the development of sugarcane pokkah boeng disease. Sci. Rep. 2016, 6, 29692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Oh, S.-K.; Young, C.; Lee, M.; Oliva, R.; Bozkurt, T.O.; Cano, L.M.; Win, J.; Bos, J.I.; Liu, H.-Y.; van Damme, M.; et al. In Planta Expression Screens of Phytophthora infestans RXLR Effectors Reveal Diverse Phenotypes, Including Activation of the Solanum bulbocastanum Disease Resistance Protein Rpi-blb2. Plant Cell 2009, 21, 2928–2947. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Huang, Z.; Zhou, Y.; Bao, Y.; Yao, Z.; Zhang, M.; Yao, W. Identification and Functional Analysis of Carbohydrate Binding Module Effector Gene Fs11724 of Fusarium sacchari. Chin. J. Trop. Crop. 2022, pp. 1–11. Available online: https://kns.cnki.net/kcms/detail/46.1019.S.20210729.0925.002.html (accessed on 29 July 2021).
- Mentges, M.; Glasenapp, A.; Boenisch, M.; Malz, S.; Henrissat, B.; Frandsen, R.J.; Güldener, U.; Münsterkötter, M.; Bormann, J.; Lebrun, M.H.; et al. Infection cushions of Fusarium graminearum are fungal arsenals for wheat infection. Mol. Plant Pathol. 2020, 21, 1070–1087. [Google Scholar] [CrossRef]
- Wang, X.; Zhai, T.; Zhang, X.; Tang, C.; Zhuang, R.; Zhao, H.; Xu, Q.; Cheng, Y.; Wang, J.; Duplessis, S.; et al. Two stripe rust effectors impair wheat resistance by suppressing import of host Fe–S protein into chloroplasts. Plant Physiol. 2021, 187, 2530–2543. [Google Scholar] [CrossRef]
- Sánchez-Vallet, A.; Tian, H.; Rodriguez-Moreno, L.; Valkenburg, D.-J.; Saleem-Batcha, R.; Wawra, S.; Kombrink, A.; Verhage, L.; de Jonge, R.; van Esse, H.P.; et al. A secreted LysM effector protects fungal hyphae through chitin-dependent homodimer polymerization. PLoS Pathog. 2020, 16, e1008652. [Google Scholar] [CrossRef]
- Navarrete, F.; Grujic, N.; Stirnberg, A.; Aleksza, D.; Gallei, M.; Adi, H.; Bindics, J.; Trujillo, M.; Djamei, A. The Pleiades cluster of fungal effector genes inhibit host defenses. PLoS Pathog. 2021, 17, e1009641. [Google Scholar] [CrossRef]
- Park, C.-H.; Chen, S.; Shirsekar, G.; Zhou, B.; Khang, C.H.; Songkumarn, P.; Afzal, A.J.; Ning, Y.; Wang, R.; Bellizzi, M.; et al. The Magnaporthe oryzae Effector AvrPiz-t Targets the RING E3 Ubiquitin Ligase APIP6 to Suppress Pathogen-Associated Molecular Pattern–Triggered Immunity in Rice. Plant Cell 2012, 24, 4748–4762. [Google Scholar] [CrossRef] [Green Version]
- Yin, Z.; Wang, N.; Duan, W.; Pi, L.; Shen, D.; Dou, D. Phytophthora capsici CBM1-containing protein CBP3 is an apoplastic effector with plant immunity-inducing activity. Mol. Plant Pathol. 2021, 22, 1358–1369. [Google Scholar] [CrossRef] [PubMed]
- von Heijne, G. Protein transport—Life and death of a signal peptide. Nature 1998, 396, 111–113. [Google Scholar] [CrossRef]
- LiBin, Y.; ShuQin, X.; ChunSheng, X. Prediction and analysis of candidate effectors from the genome of Setosphaeria turcica. J. Shenyang Agric. Univ. 2017, 48, 15–20. [Google Scholar]
- Han, C. Prediction for candidate effector proteins from Phytophthora cinnamomi genome. J. Nanjing For. Univ. Nat. Sci. Ed. 2015, 39, 69–74. [Google Scholar]
- Lu, S.; Edwards, M.C. Genome-Wide Analysis of Small Secreted Cysteine-Rich Proteins Identifies Candidate Effector Proteins Potentially Involved in Fusarium graminearum−Wheat Interactions. Phytopathology 2016, 106, 166–176. [Google Scholar] [CrossRef] [Green Version]
- Gui, Y.-J.; Chen, J.-Y.; Zhang, D.-D.; Li, N.-Y.; Li, T.-G.; Zhang, W.-Q.; Wang, X.-Y.; Short, D.P.G.; Li, L.; Guo, W.; et al. Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1. Environ. Microbiol. 2017, 19, 1914–1932. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Wang, Z.; Li, J.; Wang, Y.; Yuan, J.; Zhan, J.; Wang, P.; Lin, Y.; Li, F.; Ge, X. Verticillium dahliae secreted protein Vd424Y is required for full virulence, targets the nucleus of plant cells, and induces cell death. Mol. Plant Pathol. 2021, 22, 1109–1120. [Google Scholar] [CrossRef]
- Mouyna, I.; Hartl, L.; Latgé, J.-P. β-1,3-glucan modifying enzymes in Aspergillus fumigatus. Front. Microbiol. 2013, 4, 81. [Google Scholar] [CrossRef] [Green Version]
- Wessels, J.G.H. Fruiting in the Higher Fungi. Adv. Microb. Physiol. 1993, 34, 147–202. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Watanabe, H.; Nagai, M.; Nakade, K.; Takahashi, M.; Sato, T. Lentinula edodes tlg1 Encodes a Thaumatin-Like Protein That Is Involved in Lentinan Degradation and Fruiting Body Senescence. Plant Physiol. 2006, 141, 793–801. [Google Scholar] [CrossRef] [Green Version]
- Xiang, M.; ZhiYuan, Y.; JiaJun, N.; LiLi, H. Functional degradation domain and potential degradation pathway of the CAP superfamily protein VmPR1c from Valsa mali. Mycosystema 2019, 38, 1470–1479. [Google Scholar]
- Schneiter, R.; Di Pietro, A. The CAP protein superfamily: Function in sterol export and fungal virulence. Biomol. Concepts 2013, 4, 519–525. [Google Scholar] [CrossRef]
- Buist, G.; Steen, A.; Kok, J.; Kuipers, O.P. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 2008, 68, 838–847. [Google Scholar] [CrossRef] [Green Version]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef] [Green Version]
- Bolton, M.D.; van Esse, H.P.; Vossen, J.H.; de Jonge, R.; Stergiopoulos, I.; Stulemeijer, I.J.E.; Berg, G.C.M.V.D.; Borrás-Hidalgo, O.; Dekker, H.L.; de Koster, C.G.; et al. The novelCladosporium fulvumlysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 2008, 69, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Mentlak, T.A.; Kombrink, A.; Shinya, T.; Ryder, L.S.; Otomo, I.; Saitoh, H.; Terauchi, R.; Nishizawa, Y.; Shibuya, N.; Thomma, B.P.H.J.; et al. Effector-Mediated Suppression of Chitin-Triggered Immunity by Magnaporthe oryzae Is Necessary for Rice Blast Disease. Plant Cell 2012, 24, 322–335. [Google Scholar] [CrossRef] [Green Version]
- Marshall, R.; Kombrink, A.; Motteram, J.; Loza-Reyes, E.; Lucas, J.; Hammond-Kosack, K.E.; Thomma, B.P.H.J.; Rudd, J.J. Analysis of Two in Planta Expressed LysM Effector Homologs from the Fungus Mycosphaerella graminicola Reveals Novel Functional Properties and Varying Contributions to Virulence on Wheat. Plant Physiol. 2011, 156, 756–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Percudani, R.; Montanini, B.; Ottonello, S. The anti-HIV cyanovirin-N domain is evolutionarily conserved and occurs as a protein module in eukaryotes. Proteins Struct. Funct. Bioinform. 2005, 60, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.; Shen, C.; Fu, Y.; Xie, J.; Jiang, D.; Li, G.; Cheng, J. Comparative genomic and transcriptional analyses of the carbohydrate-active enzymes and secretomes of phytopathogenic fungi reveal their significant roles during infection and development. Sci. Rep. 2015, 5, 15565. [Google Scholar] [CrossRef] [PubMed]
- Laugé, R.; Joosten, M.H.A.J.; Ackerveken, G.F.J.M.V.D.; Broek, H.W.J.V.D.; De Wit, P.J.G.M. The In Planta-Produced Extracellular Proteins ECP1 and ECP2 of Cladosporium fulvum are Virulence Factors. Mol. Plant-Microbe Interact. 1997, 10, 725–734. [Google Scholar] [CrossRef] [Green Version]
- Stergiopoulos, I.; Kourmpetis, Y.A.I.; Slot, J.C.; Bakker, F.T.; De Wit, P.J.G.M.; Rokas, A. In Silico Characterization and Molecular Evolutionary Analysis of a Novel Superfamily of Fungal Effector Proteins. Mol. Biol. Evol. 2012, 29, 3371–3384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Kock, M.J.D.; Iskandar, H.M.; Brandwagt, B.F.; Laugé, R.; De Wit, P.J.G.M.; Lindhout, P. Recognition of Cladosporium fulvum Ecp2 elicitor by non-host Nicotiana spp. is mediated by a single dominant gene that is not homologous to known Cf-genes. Mol. Plant Pathol. 2004, 5, 397–408. [Google Scholar] [CrossRef]
- Fellbrich, G.; Romanski, A.; Varet, A.; Blume, B.; Brunner, F.; Engelhardt, S.; Felix, G.; Kemmerling, B.; Krzymowska, M.; Nürnberger, T. NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J. 2002, 32, 375–390. [Google Scholar] [CrossRef]
- Gijzen, M.; Nürnberger, T. Nep1-like proteins from plant pathogens: Recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 2006, 67, 1800–1807. [Google Scholar] [CrossRef] [PubMed]
- Pazzagli, L.; Cappugi, G.; Manao, G.; Camici, G.; Santini, A.; Scala, A. Purification, Characterization, and Amino Acid Sequence of Cerato-platanin, a New Phytotoxic Protein from Ceratocystis fimbriata f. sp. platani. J. Biol. Chem. 1999, 274, 24959–24964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Zhang, Y.; Li, B.; Yang, X.; Dong, Y.; Qiu, D. A Verticillium dahliae Pectate Lyase Induces Plant Immune Responses and Contributes to Virulence. Front. Plant Sci. 2018, 9, 1271. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Li, Z.; Lin, B.; Liao, J.; Zhuo, K. A Meloidogyne graminicola Pectate Lyase Is Involved in Virulence and Activation of Host Defense Responses. Front. Plant Sci. 2021, 12, 651627. [Google Scholar] [CrossRef]
- Oliva, R.; Win, J.; Raffaele, S.; Boutemy, L.; Bozkurt, T.O.; Chaparro-Garcia, A.; Segretin, M.E.; Stam, R.; Schornack, S.; Cano, L.M.; et al. Recent developments in effector biology of filamentous plant pathogens. Cell. Microbiol. 2010, 12, 1015. [Google Scholar] [CrossRef]
- Godfrey, D.; Böhlenius, H.; Pedersen, C.; Zhang, Z.; Emmersen, J.; Thordal-Christensen, H. Powdery mildew fungal effector candidates share N-terminal Y/F/WxC-motif. BMC Genom. 2010, 11, 317. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, C.; van Themaat, E.V.L.; McGuffin, L.J.; Abbott, J.C.; Burgis, T.A.; Barton, G.; Bindschedler, L.V.; Lu, X.; Maekawa, T.; Weßling, R.; et al. Structure and evolution of barley powdery mildew effector candidates. BMC Genom. 2012, 13, 694. [Google Scholar] [CrossRef] [Green Version]
- Duplessis, S.; Cuomo, C.A.; Lin, Y.-C.; Aerts, A.; Tisserant, E.; Veneault-Fourrey, C.; Joly, D.; Hacquard, S.; Amselem, J.; Cantarel, B.L.; et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA 2011, 108, 9166–9171. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, K.; Saitoh, H.; Fujisawa, S.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Tosa, Y.; Chuma, I.; Takano, Y.; Win, J.; et al. Association Genetics Reveals Three Novel Avirulence Genes from the Rice Blast Fungal Pathogen Magnaporthe oryzae. Plant Cell 2009, 21, 1573–1591. [Google Scholar] [CrossRef] [Green Version]
- Gout, L.; Fudal, I.; Kuhn, M.L.; Blaise, F.; Eckert, M.; Cattolico, L.; Balesdent, M.H.; Rouxel, T. Lost in the middle of nowhere: The AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol. Microbiol. 2006, 60, 67–80. [Google Scholar] [CrossRef]
- Ma, L.-J.; Van Der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.-J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Manning, V.A.; Hamilton, S.M.; Karplus, P.; Ciuffetti, L.M. The Arg-Gly-Asp–Containing, Solvent-Exposed Loop of Ptr ToxA Is Required for Internalization. Mol. Plant-Microbe Interact. 2008, 21, 315–325. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Tang, C.; Wang, X.; Sun, S.; Zhao, J.; Kang, Z.; Wang, X. An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. Nat. Commun. 2019, 10, 5571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, G.; Yang, Y.; Li, T.; Lu, W.; Du, Y.; Qiang, X.; Wen, Q.; Shan, W. A Phytophthora capsici RXLR Effector Targets and Inhibits a Plant PPIase to Suppress Endoplasmic Reticulum-Mediated Immunity. Mol. Plant 2018, 11, 1067–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Huai, B.; Lu, Y.; Cai, K.; Guo, J.; Zhu, X.; Kang, Z.; Guo, J. A stripe rust effector Pst18363 targets and stabilises TaNUDX23 that promotes stripe rust disease. New Phytol. 2019, 225, 880–895. [Google Scholar] [CrossRef]
- Chen, C.; Chen, Y.; Jian, H.; Yang, D.; Dai, Y.; Pan, L.; Shi, F.; Yang, S.; Liu, Q. Large-Scale Identification and Characterization of Heterodera avenae Putative Effectors Suppressing or Inducing Cell Death in Nicotiana benthamiana. Front. Plant Sci. 2018, 8, 2062. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Han, C.; Ferreira, A.O.; Yu, X.; Ye, W.; Tripathy, S.; Kale, S.D.; Gu, B.; Sheng, Y.; Sui, Y.; et al. Transcriptional Programming and Functional Interactions within the Phytophthora sojae RXLR Effector Repertoire. Plant Cell 2011, 23, 2064–2086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhang, K.; Fang, A.; Han, Y.; Yang, J.; Xue, M.; Bao, J.; Hu, D.; Zhou, B.; Sun, X.; et al. Specific adaptation of Ustilaginoidea virens in occupying host florets revealed by comparative and functional genomics. Nat. Commun. 2014, 5, 3849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Songkumarn, P.; Venu, R.C.; Gowda, M.; Bellizzi, M.; Hu, J.; Liu, W.; Ebbole, D.; Meyers, B.; Mitchell, T.; et al. Identification and Characterization of in planta–Expressed Secreted Effector Proteins from Magnaporthe oryzae That Induce Cell Death in Rice. Mol. Plant-Microbe Interact. 2013, 26, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Niu, X.; Yang, G.; Lin, H.; Liu, Y.; Li, P.; Zheng, A. A Novel, Small Cysteine-Rich Effector, RsSCR10 in Rhizoctonia solani Is Sufficient to Trigger Plant Cell Death. Front. Microbiol. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Kang, H.; Chen, X.; Kemppainen, M.; Pardo, A.G.; Veneault-Fourrey, C.; Kohler, A.; Martin, F.M. The small secreted effector protein MiSSP7.6 of Laccaria bicolor is required for the establishment of ectomycorrhizal symbiosis. Environ. Microbiol. 2020, 22, 1435–1446. [Google Scholar] [CrossRef]
- Wang, D.; Tian, L.; Zhang, D.; Song, J.; Song, S.; Yin, C.; Zhou, L.; Liu, Y.; Wang, B.; Kong, Z.; et al. Functional analyses of small secreted cysteine-rich proteins identified candidate effectors in Verticillium dahliae. Mol. Plant Pathol. 2020, 21, 667–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukada, F.; Rössel, N.; Münch, K.; Glatter, T.; Kahmann, R. A small Ustilago maydis effector acts as a novel adhesin for hyphal aggregation in plant tumors. New Phytol. 2021, 231, 416–431. [Google Scholar] [CrossRef]
- Tian, H.; MacKenzie, C.I.; Rodriguez-Moreno, L.; Berg, G.C.M.V.D.; Chen, H.; Rudd, J.J.; Mesters, J.R.; Thomma, B.P.H.J. Three LysM effectors of Zymoseptoria tritici collectively disarm chitin-triggered plant immunity. Mol. Plant Pathol. 2021, 22, 683–693. [Google Scholar] [CrossRef]
- Liu, D.; Yao, Z.; Lai, X.; Yao, X.; Zhang, M.; Zhou, C.; Chen, B. Microscopic observation on the infection of sugarcane leaves by pokkah boeng pathogen Fusarium verticillioides. Sugar Crop. China 2019, 41, 41–45. [Google Scholar] [CrossRef]






| Description | Number | |
|---|---|---|
| Superfamily | alpha_CA superfamily | 2 |
| CAP superfamily | 3 | |
| SGNH_hydrolase superfamily | 7 | |
| FkpA superfamily | 1 | |
| cupin_like superfamily | 2 | |
| ML superfamily | 1 | |
| ZnMc superfamily | 3 | |
| DUF1961 superfamily | 1 | |
| LysM superfamily | 2 | |
| Cupredoxin superfamily | 2 | |
| PRK11907 superfamily | 1 | |
| CE4_SF superfamily | 2 | |
| Glyco_hydro_12 superfamily | 1 | |
| SurE superfamily | 1 | |
| GAT_1 superfamily | 2 | |
| CHRD superfamily | 1 | |
| DOMON_like superfamily | 1 | |
| VOC superfamily | 1 | |
| LamG superfamily | 3 | |
| Trypsin superfamily | 1 | |
| microbial_RNases superfamily | 1 | |
| Glyco_hydro_114 superfamily | 1 | |
| Abhydrolase superfamily | 3 | |
| MhpC superfamily | 1 | |
| RNase_T2 superfamily | 2 | |
| DUF3455 superfamily | 3 | |
| YdcF-like superfamily | 1 | |
| Fasciclin superfamily | 1 | |
| PLN00052 superfamily | 1 | |
| YoaJ superfamily | 2 | |
| Hydrophobin superfamily | 1 | |
| SodA superfamily | 1 | |
| DPBB_1 superfamily | 1 | |
| M35_like superfamily | 1 | |
| cysteine_hydrolases superfamily | 1 | |
| CM_2 superfamily | 1 | |
| Cupin_5 superfamily | 1 | |
| Lyz_like superfamily | 1 | |
| DUF3237 superfamily | 1 | |
| Fimbrial superfamily | 1 | |
| Domain | CVNH | 2 |
| Hce2 | 2 | |
| LicD | 1 | |
| Pectate_lyase | 4 | |
| NPP1 | 3 | |
| Glyco_hydro_61 | 4 | |
| Cerato-platanin | 3 | |
| Cutinase | 6 | |
| mannanase_GH134 | 1 | |
| PAN_1 | 1 | |
| Glyco_hydro_11 | 4 | |
| Methyltransf_23 | 1 | |
| EthD | 2 | |
| TenA_C_Bt3146-like | 1 | |
| Hydrophobin_2 | 2 | |
| CBM_4_9 | 1 | |
| TNT | 1 | |
| WSC | 1 | |
| Peroxidase_2 | 1 | |
| HsbA | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.; Li, H.; Zhou, Y.; Bao, Y.; Duan, Z.; Wang, C.; Powell, C.A.; Chen, B.; Zhang, M.; Yao, W. Predication of the Effector Proteins Secreted by Fusarium sacchari Using Genomic Analysis and Heterogenous Expression. J. Fungi 2022, 8, 59. https://doi.org/10.3390/jof8010059
Huang Z, Li H, Zhou Y, Bao Y, Duan Z, Wang C, Powell CA, Chen B, Zhang M, Yao W. Predication of the Effector Proteins Secreted by Fusarium sacchari Using Genomic Analysis and Heterogenous Expression. Journal of Fungi. 2022; 8(1):59. https://doi.org/10.3390/jof8010059
Chicago/Turabian StyleHuang, Zhen, Huixue Li, Yuming Zhou, Yixue Bao, Zhenzhen Duan, Caixia Wang, Charles A. Powell, Baoshan Chen, Muqing Zhang, and Wei Yao. 2022. "Predication of the Effector Proteins Secreted by Fusarium sacchari Using Genomic Analysis and Heterogenous Expression" Journal of Fungi 8, no. 1: 59. https://doi.org/10.3390/jof8010059
APA StyleHuang, Z., Li, H., Zhou, Y., Bao, Y., Duan, Z., Wang, C., Powell, C. A., Chen, B., Zhang, M., & Yao, W. (2022). Predication of the Effector Proteins Secreted by Fusarium sacchari Using Genomic Analysis and Heterogenous Expression. Journal of Fungi, 8(1), 59. https://doi.org/10.3390/jof8010059







