Suppression of Chitin-Triggered Immunity by a New Fungal Chitin-Binding Effector Resulting from Alternative Splicing of a Chitin Deacetylase Gene
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plants, Fungi, Bacteria, and Culture Conditions
2.2. Sequence Analysis and Protein Modelling
2.3. RNA Extraction and cDNA Synthesis
2.4. Plasmid Construction
2.5. Protein Expression and Purification
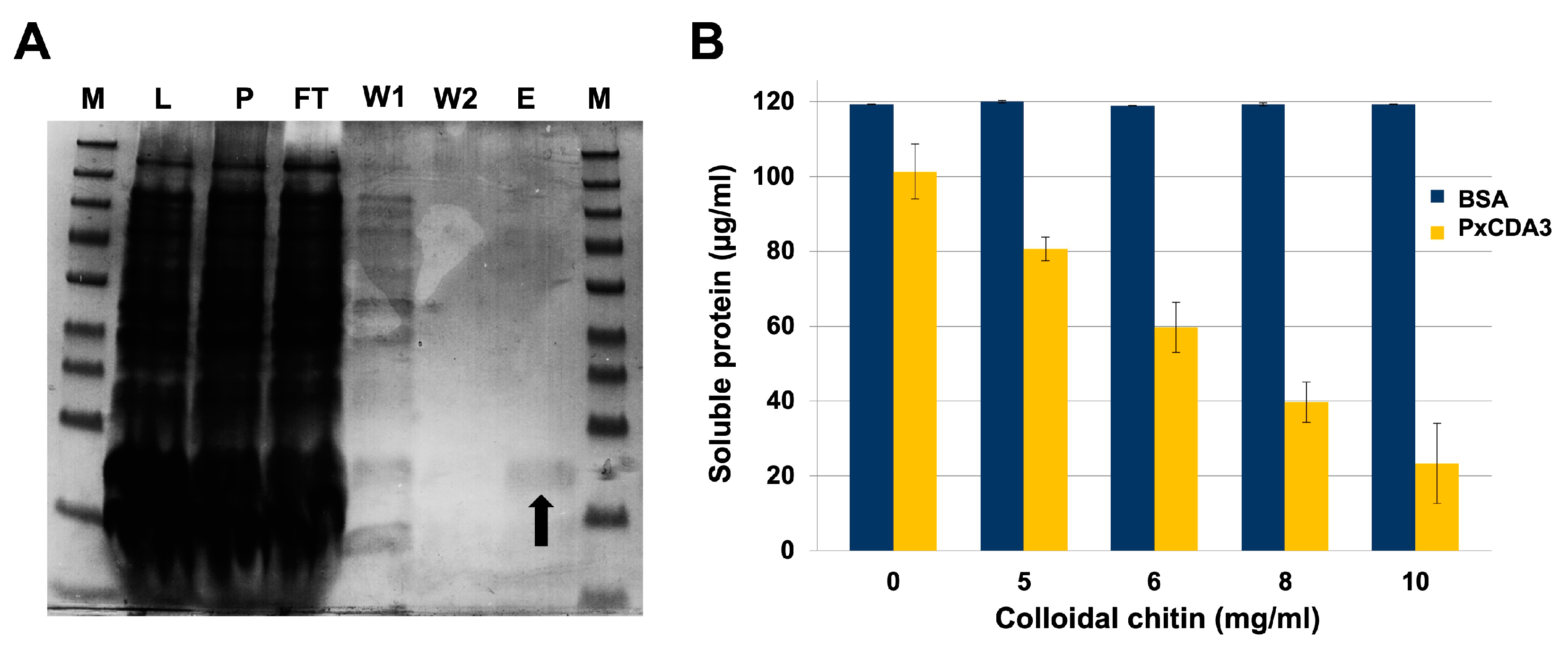
2.6. Chitin-Binding Activity Assays
2.7. Chitinase Activity and Chitin-Triggered Oxidative Burst Assay
2.8. Transformation by Growth onto Agroinfiltrated Tissues (TGAT) and Confocal Laser Scanning Microscopy (CLSM)
2.9. Real Time-Quantitative Polymerase Chain Reaction (RT-qPCR)
3. Results
3.1. Alternative Splicing of the PxCDA Gene Results in a Chitin-Binding Protein
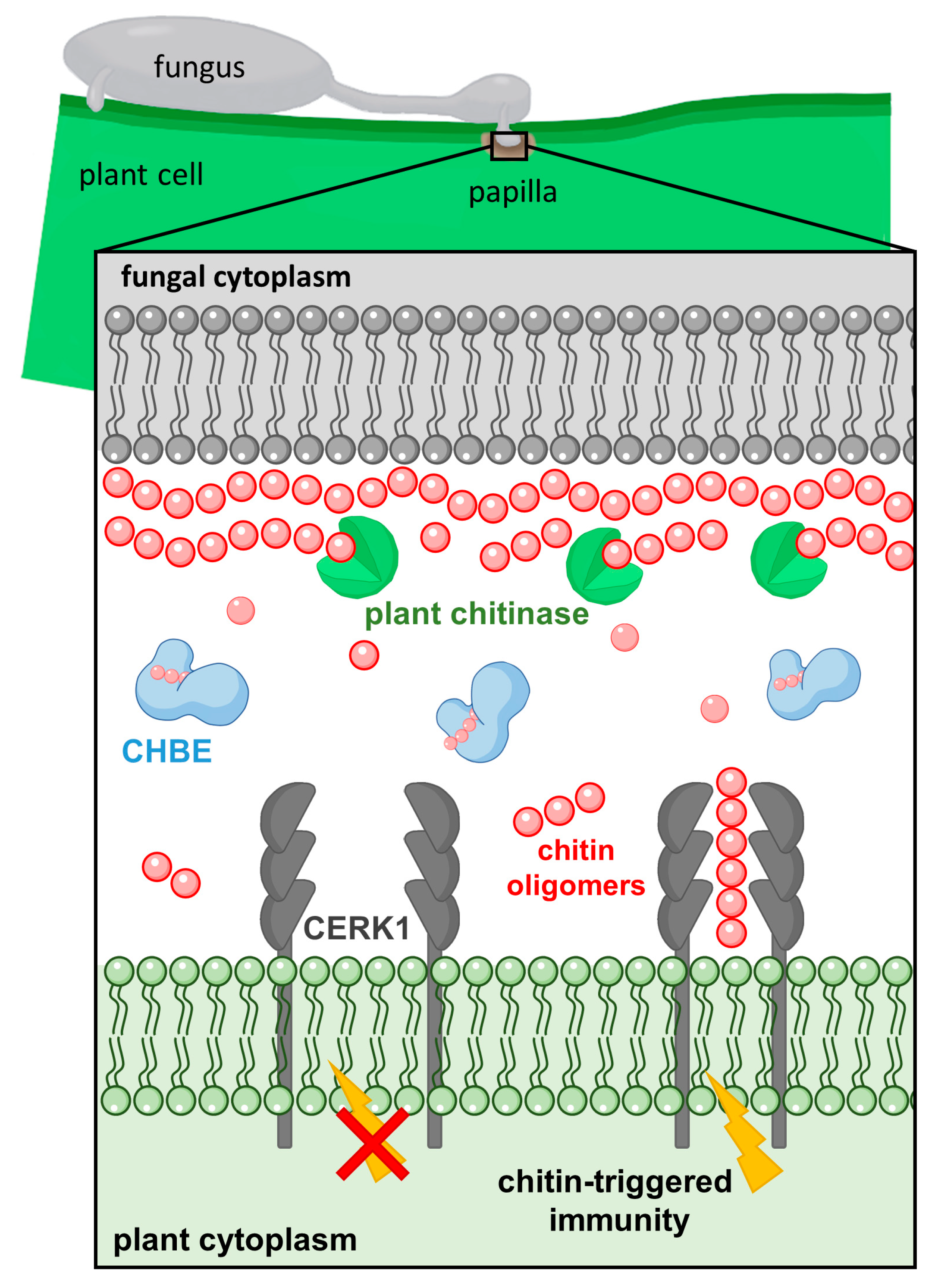
3.2. PxCDA3 Protein Prevents Chitin Recognition by Sequestering Immunogenic Oligomers
3.3. PxCHBE Protein Is Deployed at the Plant Papilla Where Chitin Is Densely Accumulated
3.4. CHBE Is a Novel Fungal Effector That May Be Present in Other Powdery Mildew Fungi
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Glawe, D.A. The powdery mildews: A review of the world’s most familiar (yet poorly known) plant pathogens. Annu. Rev. Phythopathol. 2008, 46, 27–51. [Google Scholar] [CrossRef] [PubMed]
- Ridout, C.J. Profiles in pathogenesis and mutualism: Powdery mildews. In The Mycota, Plant Relationships, 2nd ed.; Deising, H.B., Ed.; Springer: Berlin/Heidelberg, Germany, 2009; Volume 5, pp. 50–67. [Google Scholar]
- Micali, C.O.; Neumann, U.; Grunewald, D.; Panstruga, R.; O’Connell, R. Biogenesis of a specialized plant–fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell. Microbiol. 2011, 13, 210–226. [Google Scholar] [CrossRef] [PubMed]
- Weßling, R.; Schmidt, S.M.; Micali, C.O.; Knaust, F.; Reinhardt, R.; Naumann, U.; Ver Loren van Themaat, E.; Panstruga, R. Transcriptome analysis of enriched Golovinomyces orontii haustoria by deep 454 pyrosequencing. Fungal Genet. Biol. 2012, 49, 470–482. [Google Scholar] [CrossRef] [PubMed]
- Pérez-García, A.; Romero, D.; Fernández-Ortuño, D.; López-Ruiz, F.; de Vicente, A.; Torés, J.A. The powdery mildew fungus Podosphaera fusca (synonym Podosphaera xanthii), a constant threat to cucurbits. Mol. Plant Pathol. 2009, 10, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Vela-Corcía, D.; Bautista, R.; de Vicente, A.; Spanu, P.D.; Pérez-García, A. De novo analysis of the epiphytic transcriptome of the cucurbit powdery mildew fungus Podosphaera xanthii and identification of candidate secreted effector proteins. PLoS ONE 2016, 11, e0163379. [Google Scholar] [CrossRef] [PubMed]
- Polonio, A.; Seoane, P.; Claros, M.G.; Pérez-García, A. The haustorial transcriptome of the cucurbit pathogen Podosphaera xanthii reveals new insights into the biotrophy and pathogenesis of powdery mildew fungi. BMC Genom. 2019, 20, 543. [Google Scholar] [CrossRef]
- Polonio, A.; Pineda, M.; Bautista, R.; Martínez-Cruz, J.; Pérez-Bueno, M.L.; Barón, M.; Pérez-García, A. RNA-seq analysis and fluorescence imaging of melon powdery mildew disease reveal an orchestrated reprogramming of host physiology. Sci. Rep. 2019, 9, 7978. [Google Scholar] [CrossRef]
- Polonio, A.; Díaz-Martínez, L.; Fernández-Ortuño, D.; de Vicente, A.; Romero, D.; López-Ruiz, F.J.; Pérez-García, A. A hybrid genome assembly resource for Podosphaera xanthii, the main causal agent of powdery mildew disease in cucurbits. Mol. Plant-Microbe Interact. 2021, 34, 319–324. [Google Scholar] [CrossRef]
- Martínez-Cruz, J.; Romero, D.; de Vicente, A.; Pérez-García, A. Transformation of the cucurbit powdery mildew pathogen Podosphaera xanthii by Agrobacterium tumefaciens. New Phytol. 2017, 213, 1961–1973. [Google Scholar] [CrossRef]
- Martínez-Cruz, J.; Romero, D.; de Vicente, A.; Pérez-García, A. Transformation by growth onto agro-infiltrated tissues (TGAT), a simple and efficient alternative for transient transformation of the cucurbit powdery mildew pathogen Podosphaera xanthii. Mol. Plant Pathol. 2018, 19, 2502–2515. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Cruz, J.; Romero, D.; de la Torre, F.N.; Fernández-Ortuño, D.; Torés, J.A.; de Vicente, A.; Pérez-García, A. The functional characterization of Podosphaera xanthii candidate effector genes reveals novel target functions for fungal pathogenicity. Mol. Plant-Microbe Interact. 2018, 31, 914–931. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Jiménez, L.; Polonio, A.; Vielba-Fernández, A.; Pérez-García, A.; Fernández-Ortuño, D. Gene mining for conserved, non-annotated proteins of Podosdphaera xanthii identifies novel target candidates for controlling powdery mildews by spray-induced gene silencing. J. Fungi 2021, 7, 735. [Google Scholar] [CrossRef] [PubMed]
- Young, V.L.; Simpson, R.M.; Ward, V.K. Characterization of anexochitinase from Epiphyas postvittana nucleopolyhedrovirus (family Baculoviridae). J. Gen. Virol. 2005, 86, 3253–3261. [Google Scholar] [CrossRef]
- Wan, J.; Zhang, X.; Stacey, G. Chitin signaling and plant disease resistance. Plant Signal. Behav. 2008, 3, 831–833. [Google Scholar] [CrossRef] [PubMed]
- de Jonge, R.; van Esse, H.P.; Kombrink, A.; Shinya, T.; Desaki, Y.; Bours, R.; van der Krol, S.; Shibuya, N.; Joosten, M.H.A.J.; Thomma, B.P.H.J. Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 2010, 329, 953–955. [Google Scholar] [CrossRef]
- Liu, B.; Li, J.-F.; Ao, Y.; Qu, J.; Li, Z.; Su, J.; Zhang, Y.; Liu, J.; Feng, D.; Qi, K.; et al. Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 2012, 24, 3406–3419. [Google Scholar] [CrossRef]
- Kombrink, A.; Thomma, B.P.H.J. LysM effectors secreted proteins supporting fungal life. PLoS Pathog. 2013, 9, e1003769. [Google Scholar] [CrossRef]
- Tanaka, K.; Nguyen, C.T.; Liang, Y.; Cao, Y.; Stacey, G. Role of LysM receptors in chitin-triggered plant innate immunity. Plant Signal. Behav. 2013, 8, e22598. [Google Scholar] [CrossRef]
- Mentlak, T.A.; Kombrink, A.; Shinya, T.; Ryder, L.S.; Otomo, I.; Saitoh, H.; Terauchi, R.; Nishizawa, Y.; Shibuya, N.; Thomma, B.P.H.J.; et al. Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 2012, 24, 322–335. [Google Scholar] [CrossRef]
- van den Burg, H.A.; Harrison, S.J.; Joosten, M.H.A.J.; Vervoort, J.; de Wit, P.J.G.M. Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant Microbe Interact. 2006, 19, 1420–1430. [Google Scholar] [CrossRef] [Green Version]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Minami, E.; Shibuya, N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef] [PubMed]
- Doehlemann, G.; Hemetsberger, C. Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol. 2013, 198, 1001–1016. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Vallet, A.; Mesters, J.R.; Thomma, B.P. The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol. Rev. 2015, 39, 171–183. [Google Scholar] [CrossRef]
- Fiorin, G.L.; Sanchéz-Vallet, A.; Thomazella, D.P.T.; do Prado, P.F.V.; do Nascimento, L.C.; Figueira, A.V.O.; Thomma, B.P.H.J.; Pereira, G.A.G.; Teixeira, P.J.P.L. Suppression of plant immunity by fungal chitinase-like effectors. Curr. Biol. 2018, 28, 3023–3030. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.; Kombrink, A.; Motteram, J.; Loza-Reyes, E.; Lucas, J.; Hammond-Kosack, K.E.; Thomma, B.P.; Rudd, J.J. Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol. 2011, 156, 756–769. [Google Scholar] [CrossRef]
- Martínez-Cruz, J.; Romero, D.; Hierrezuelo, J.; Thon, M.; de Vicente, A.; Pérez-García, A. Effectors with chitinase activity (EWCAs), a family of conserved, secreted fungal chitinases that suppress chitin-triggered immunity. Plant Cell 2021, 33, 1319–1340. [Google Scholar] [CrossRef]
- Polonio, A.; Fernández-Ortuño, D.; de Vicente, A.; Pérez-García, A. A haustorial-expressed lytic polysaccharide monooxygenase from the cucurbit powdery mildew pathogen Podosphaera xanthii contributes to the suppression of chitin-triggered immunity. Mol. Plant Pathol. 2021, 22, 580–601. [Google Scholar] [CrossRef]
- Blair, D.E.; Hekmat, O.; Schüttelkopf, A.W.; Shrestha, B.; Tokuyasu, K.; Withers, S.G.; van Aalten, D.M.F. Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum. Biochemistry 2006, 45, 9416–9426. [Google Scholar] [CrossRef]
- Gao, F.; Zhang, B.-S.; Zhao, J.-H.; Huang, J.-F.; Jia, P.-S.; Wang, S.; Zhang, J.; Zhou, J.-M.; Guo, H.-S. Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat. Plants 2019, 5, 1167–1176. [Google Scholar] [CrossRef]
- Rizzi, Y.S.; Happel, P.; Lenz, S.; Mounashree, J.U.; Bonin, M.; Cord-Landwehr, S.; Singh, R.; Moerschbacher, B.M.; Kahmann, R. Chitosan and chitin deacetylase activity are necessary for development and virulence of Ustilago maydis. mBio 2021, 12, e03419-20. [Google Scholar] [CrossRef]
- Martínez-Cruz, J.M.; Polonio, Á.; Zanni, R.; Romero, D.; Gálvez, J.; Fernández-Ortuño, D.; Pérez-García, A. Chitin deacetylase, a novel target for the design of agricultural fungicides. J. Fungi 2021, 7, 1009. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, B.; Torés, J.A. Cultivo in vitro de Sphaerotheca fuliginea (Schlecht. ex Fr.), efecto de diferentes fuentes de carbono sobre su desarrollo. Bol. San. Veg. Plagas 1997, 23, 283–288. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef]
- Nakagawa, T.; Kurose, T.; Hino, T.; Tanaka, K.; Kawamukai, M.; Niwa, Y.; Toyooka, K.; Jinbo, T.; Timura, T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007, 104, 34–41. [Google Scholar] [CrossRef]
- Khan, M.A.; Hamid, R.; Ahmad, M.; Abdin, M.Z.; Javed, S. Optimization of culture media for enhanced chitinase production from a novel strain of Stenotrophomonas maltophilia using response surface methodology. J. Microbiol. Biotechnol. 2010, 20, 1597–1602. [Google Scholar] [CrossRef]
- Martínez-Cruz, J.; Romero, D.; Dávila, J.C.; Pérez-García, A. The Podosphaera xanthii haustorium, the fungal Trojan horse of cucurbit-powdery mildew interactions. Fungal Genet. Biol. 2014, 71, 21–31. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef]
- Vela-Corcía, D.; Bellón-Gómez, D.; López-Ruiz, F.; Torés, J.A.; Perez-García, A. The Podosphaera fusca TUB2 gene, a molecular “Swiss Army knife” with multiple applications in powdery mildew research. Fungal Biol. 2014, 118, 228–241. [Google Scholar] [CrossRef]
- Gong, B.-Q.; Wang, F.-Z.; Li, J.-F. Hide-and-seek: Chitin-triggered plant immunity and fungal counterstrategies. Trends Plant Sci. 2020, 8, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Dixit, R.; Arakane, Y.; Specht, C.A.; Richard, C.; Kramer, K.J.; Beeman, R.W.; Muthukrishnan, S. Domain organization and phylogenetic analysis of proteins from the chitin deacetylase gene family of Tribolium castaneum and three other species of insects. Insect Biochem. Mol. Biol. 2008, 38, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Yen, L.L. Investigations on the lgx-1 Gene in Caenorhabditis elegans. Ph.D. Thesis, Tufts University, Medford, MA, USA, 2010. [Google Scholar]
- de Jonge, R.; Thomma, B.P.H.J. Fungal LysM effectors: Extinguishers of host immunity? Trends Microbiol. 2009, 17, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Bork, P.; Sander, C.; Valencia, A. Convergent evolution of similar enzymatic function on different protein folds: The hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 1993, 2, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Stebbins, C.E.; Galán, J.E. Structural mimicry in bacterial virulence. Nature 2001, 412, 701–705. [Google Scholar] [CrossRef]
- Xia, Y. Proteases in pathogenesis and plant defence. Cell. Microbiol. 2004, 6, 905–913. [Google Scholar] [CrossRef]
- Shabab, M.; Shindo, T.; Gu, C.; Kaschani, F.; Pansuriya, T.; Chintha, R.; Harzen, A.; Colby, T.; Kamoun, S.; van der Hoorn, R.A.L. Fungal effector protein AVR2 targets diversifying defense-related Cys proteases of tomato. Plant Cell 2008, 20, 1169–1183. [Google Scholar] [CrossRef]
- Hacquard, S.; Joly, D.L.; Lin, Y.C.; Tisserant, E.; Feau, N.; Delaruelle, C.; Legué, V.; Kohler, A.; Tanguay, P.; Petre, B.; et al. A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici-populina (poplar leaf rust). Mol. Plant Microbe Interact. 2012, 25, 279–293. [Google Scholar] [CrossRef]
- Kruger, W.M.; Szabo, L.J.; Zeyen, R.J. Transcription of the defense response genes chitinase IIb, PAL and peroxidase is induced by the barley powdery mildew fungus and is only indirectly modulated by R genes. Physiol. Mol. Plant Pathol. 2003, 63, 167–178. [Google Scholar] [CrossRef]
- Fung, R.W.M.; Gonzalo, M.; Fekete, C.; Kovacs, L.G.; He, Y.; Marsh, E.; McIntyre, L.M.; Schachtman, D.P.; Qiu, W. Powdery mildew induces defense-oriented reprograming of the transcriptome in a susceptible but not in a resistant grapevine. Plant Physiol. 2008, 146, 236–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-García, A.; Olalla, L.; Rivera, E.; del Pino, D.; Cánovas, I.; de Vicente, A.; Torés, J.A. Development of Sphaerotheca fusca on susceptible, resistant, and temperature-sensitive resistant melon cultivars. Mycol. Res. 2001, 105, 1216–1222. [Google Scholar] [CrossRef]
- van Esse, H.P.; Bolton, M.D.; Stergiopoulos, I.; de Wit, P.J.G.M.; Thomma, B.P.H.J. The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant Microbe Interact. 2007, 20, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Nish, S.; Medzhitov, R. Host defense pathways: Role of redundancy and compensation in infectious disease phenotypes. Immunity 2011, 34, 629–636. [Google Scholar] [CrossRef]
- Frénal, K.; Soldati-Favre, D. Plasticity and redundancy in proteins important for Toxoplasma invasion. PLoS Pathog. 2015, 11, e1005069. [Google Scholar] [CrossRef] [Green Version]






| Assay | Substrate a | Products (µg mL−1) b | ||||||
|---|---|---|---|---|---|---|---|---|
| Mono | Di | Tri | Tetra | Penta | Hexa | Hepta | ||
| Experiment 1 | (GlcNAc)1–7 | - c | - | - | - | 57 | 235 | Nd d |
| Experiment 2 | (GlcNAc)1–7 | - | - | - | - | 50 | 186 | nd |
| Experiment 3 | (GlcNAc)1–7 | - | - | - | - | 49 | 193 | nd |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Cruz, J.M.; Polonio, Á.; Ruiz-Jiménez, L.; Vielba-Fernández, A.; Hierrezuelo, J.; Romero, D.; de Vicente, A.; Fernández-Ortuño, D.; Pérez-García, A. Suppression of Chitin-Triggered Immunity by a New Fungal Chitin-Binding Effector Resulting from Alternative Splicing of a Chitin Deacetylase Gene. J. Fungi 2022, 8, 1022. https://doi.org/10.3390/jof8101022
Martínez-Cruz JM, Polonio Á, Ruiz-Jiménez L, Vielba-Fernández A, Hierrezuelo J, Romero D, de Vicente A, Fernández-Ortuño D, Pérez-García A. Suppression of Chitin-Triggered Immunity by a New Fungal Chitin-Binding Effector Resulting from Alternative Splicing of a Chitin Deacetylase Gene. Journal of Fungi. 2022; 8(10):1022. https://doi.org/10.3390/jof8101022
Chicago/Turabian StyleMartínez-Cruz, Jesús M., Álvaro Polonio, Laura Ruiz-Jiménez, Alejandra Vielba-Fernández, Jesús Hierrezuelo, Diego Romero, Antonio de Vicente, Dolores Fernández-Ortuño, and Alejandro Pérez-García. 2022. "Suppression of Chitin-Triggered Immunity by a New Fungal Chitin-Binding Effector Resulting from Alternative Splicing of a Chitin Deacetylase Gene" Journal of Fungi 8, no. 10: 1022. https://doi.org/10.3390/jof8101022
APA StyleMartínez-Cruz, J. M., Polonio, Á., Ruiz-Jiménez, L., Vielba-Fernández, A., Hierrezuelo, J., Romero, D., de Vicente, A., Fernández-Ortuño, D., & Pérez-García, A. (2022). Suppression of Chitin-Triggered Immunity by a New Fungal Chitin-Binding Effector Resulting from Alternative Splicing of a Chitin Deacetylase Gene. Journal of Fungi, 8(10), 1022. https://doi.org/10.3390/jof8101022







