Identifying the Main Drivers in Microbial Diversity for Cabernet Sauvignon Cultivars from Europe to South Africa: Evidence for a Cultivar-Specific Microbial Fingerprint
Abstract
:1. Introduction
2. Materials and Methods
2.1. Grape Sampling and DNA Extraction
2.2. Sequencing Library Construction
2.3. Data Analysis
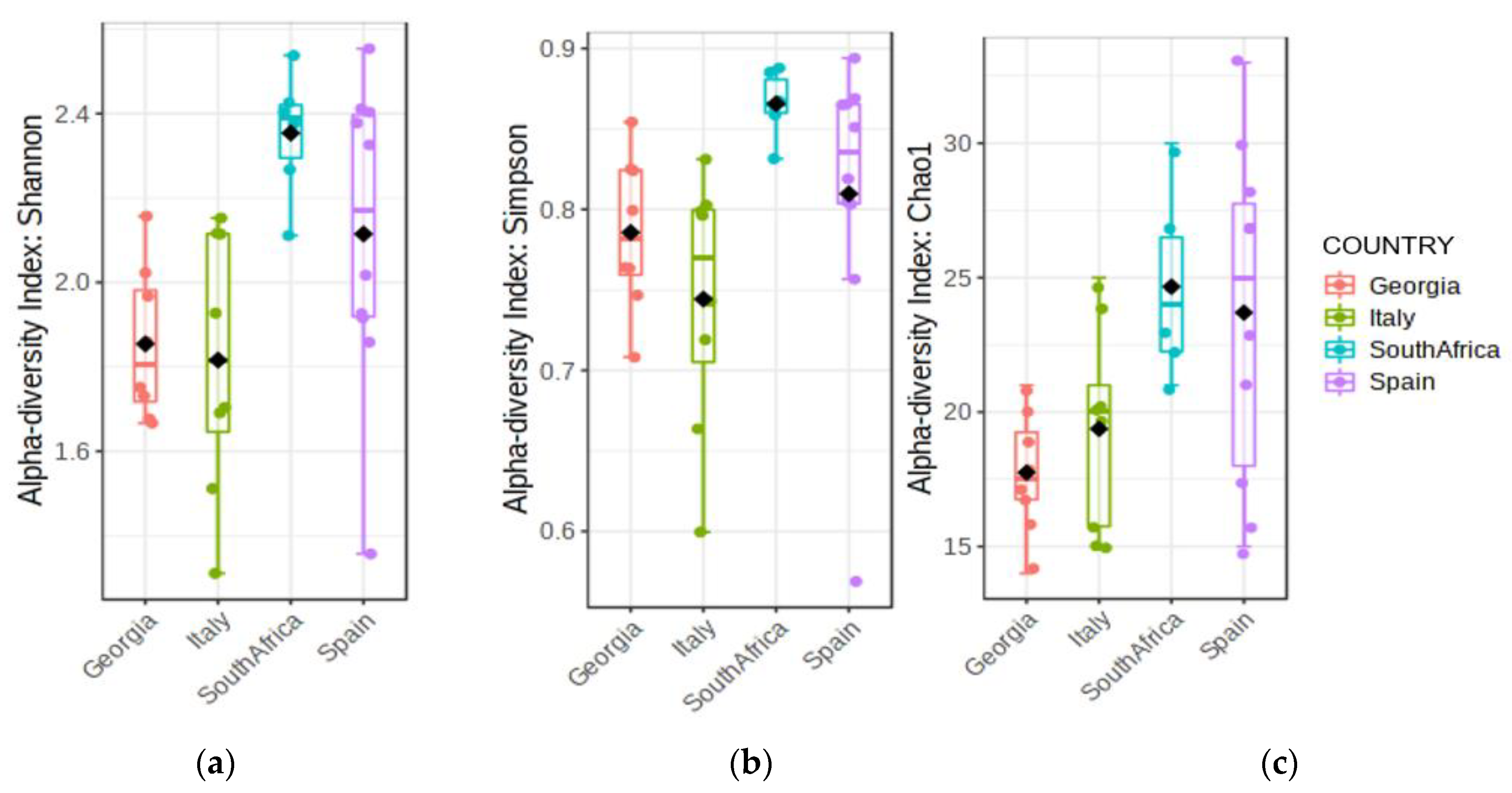
3. Results
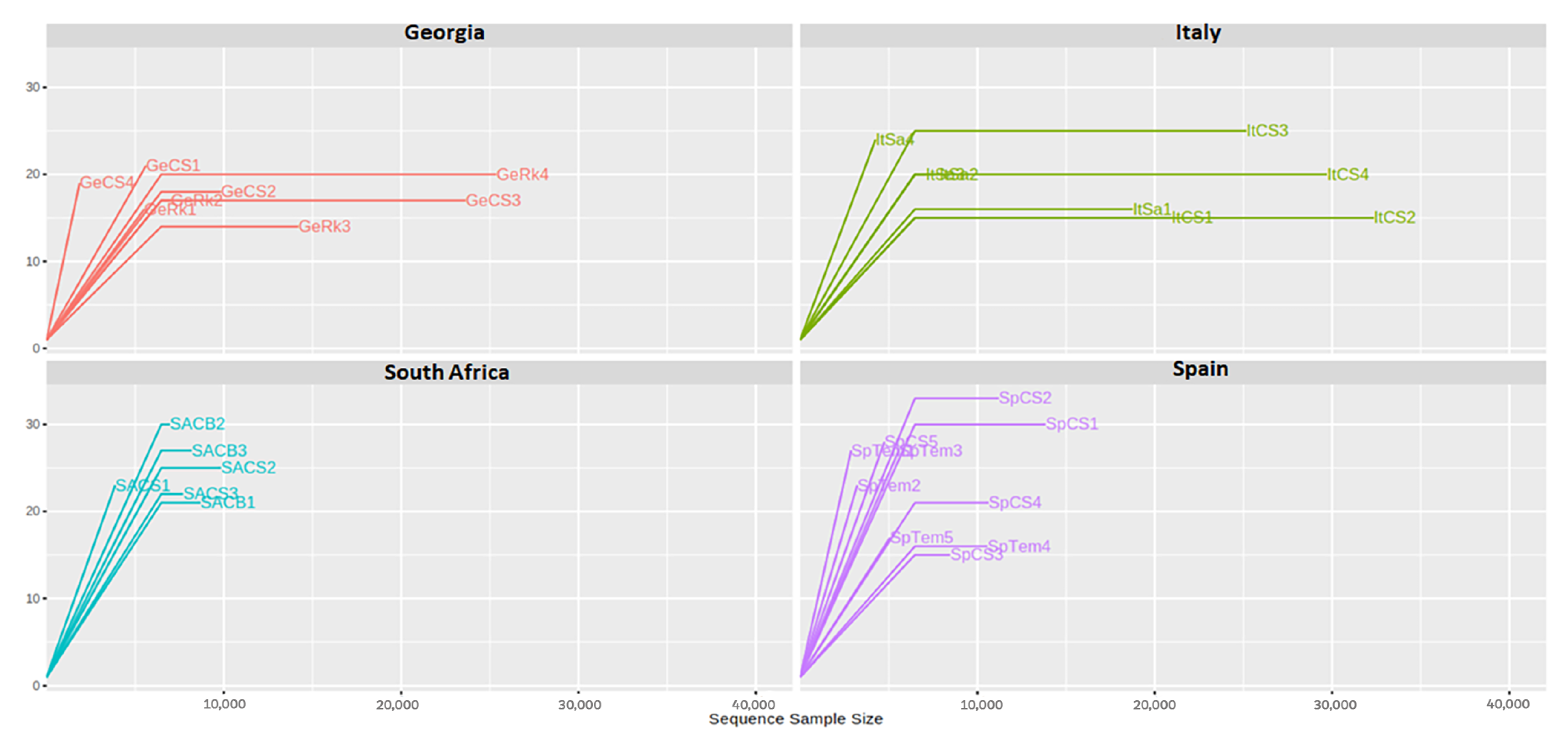
3.1. Design of Experiments and Sequence Analysis
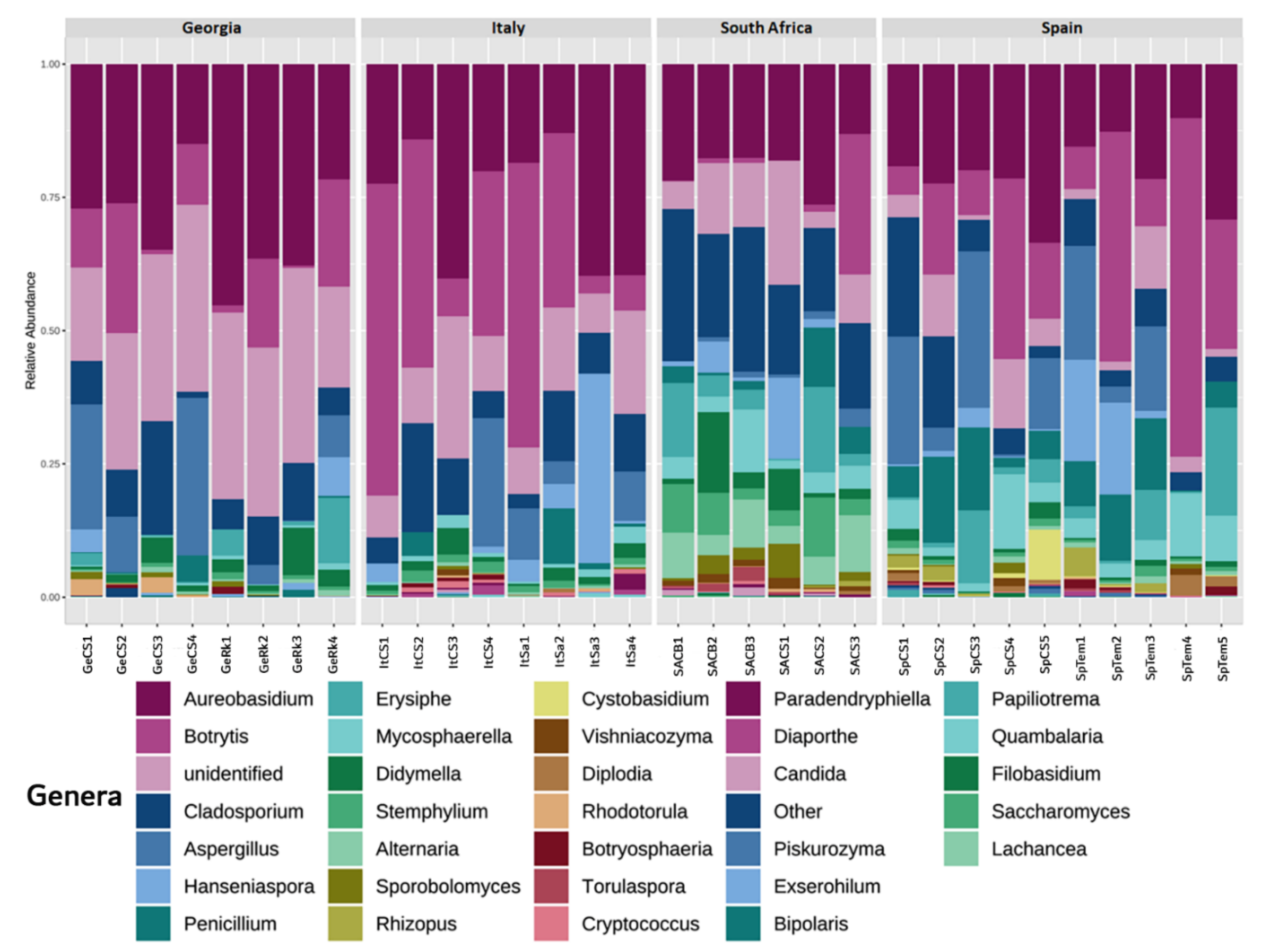
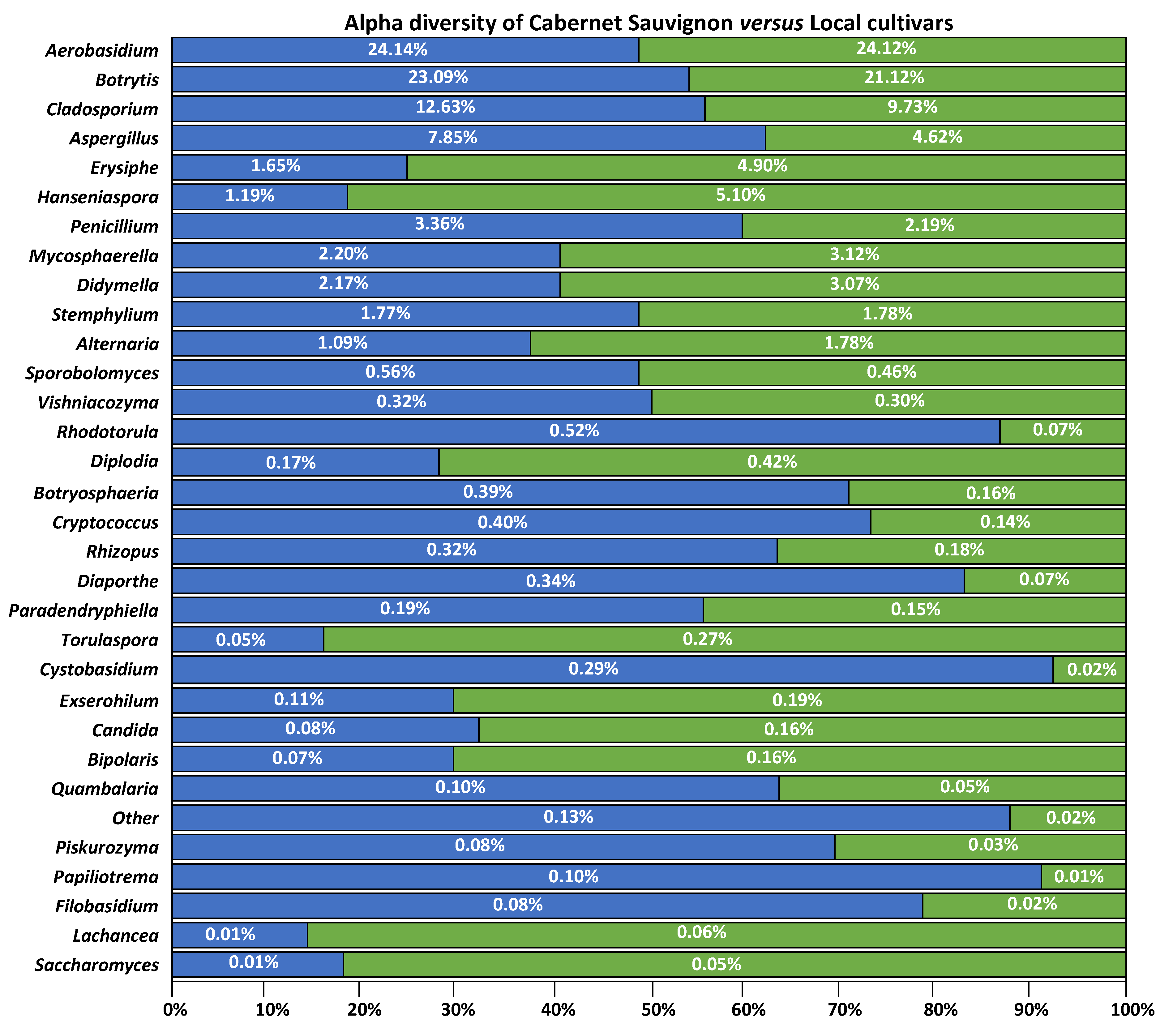
3.2. Fungal Distribution
3.3. Deconstructing the Drivers Defining Microbial Population
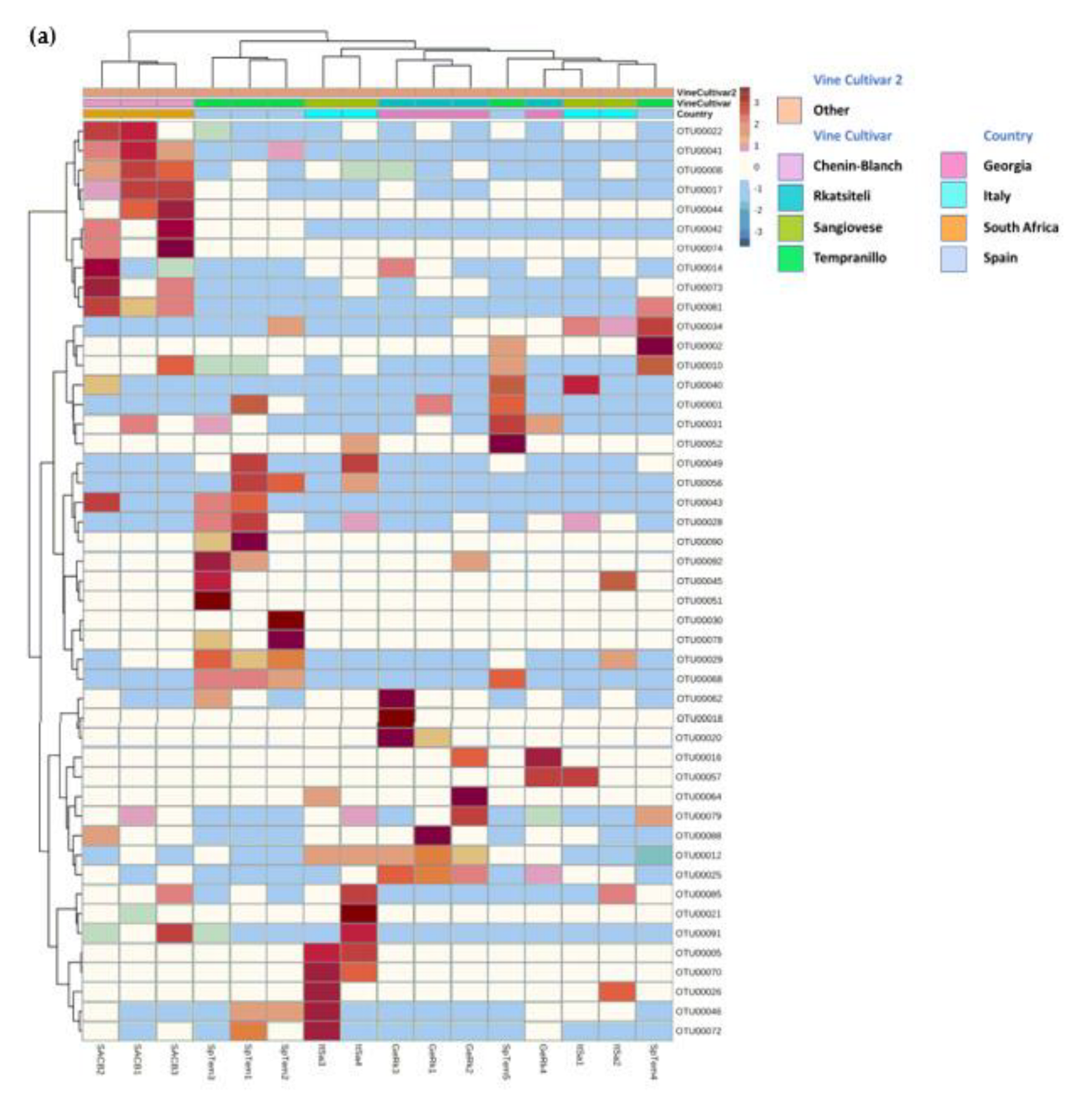
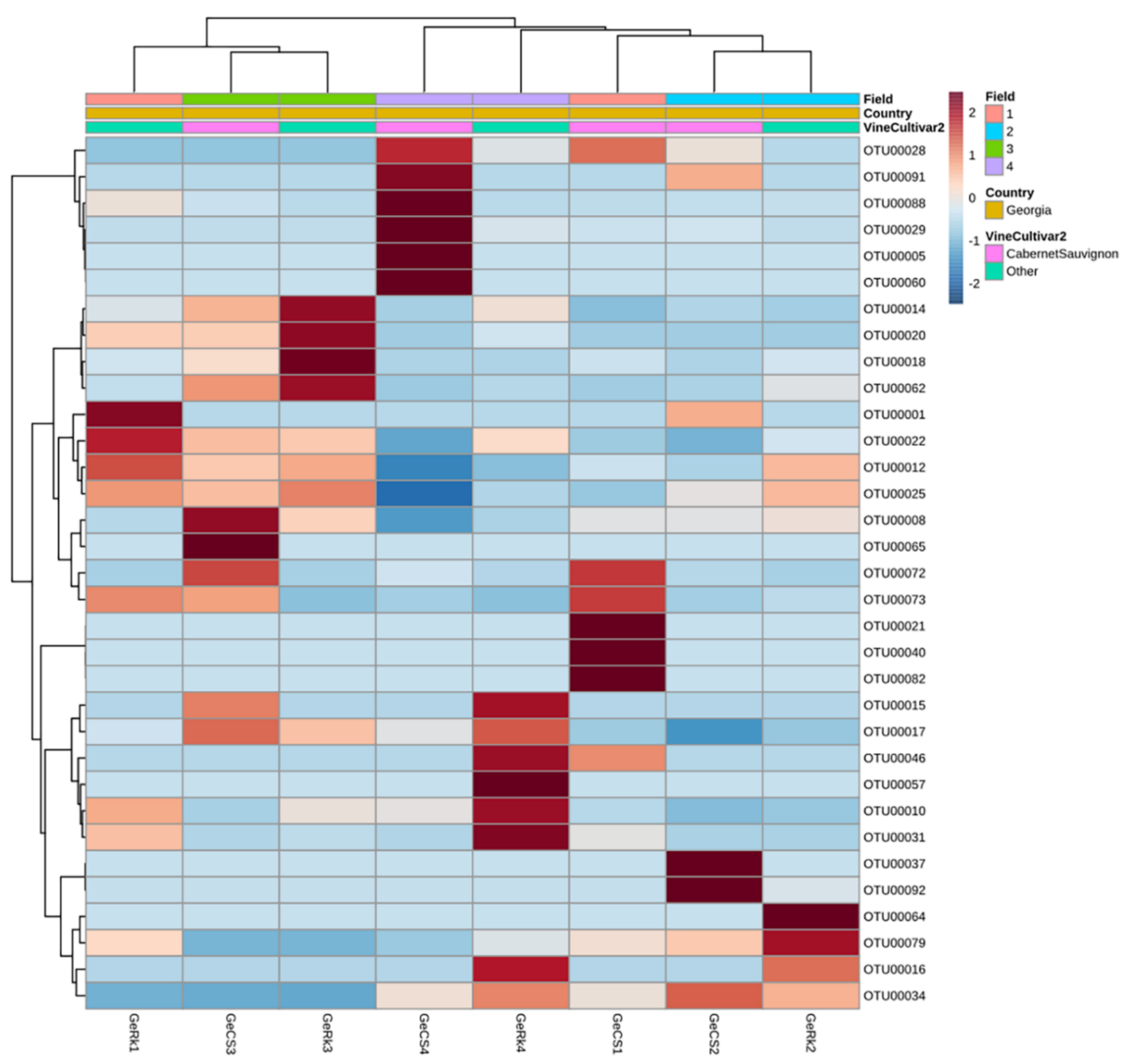
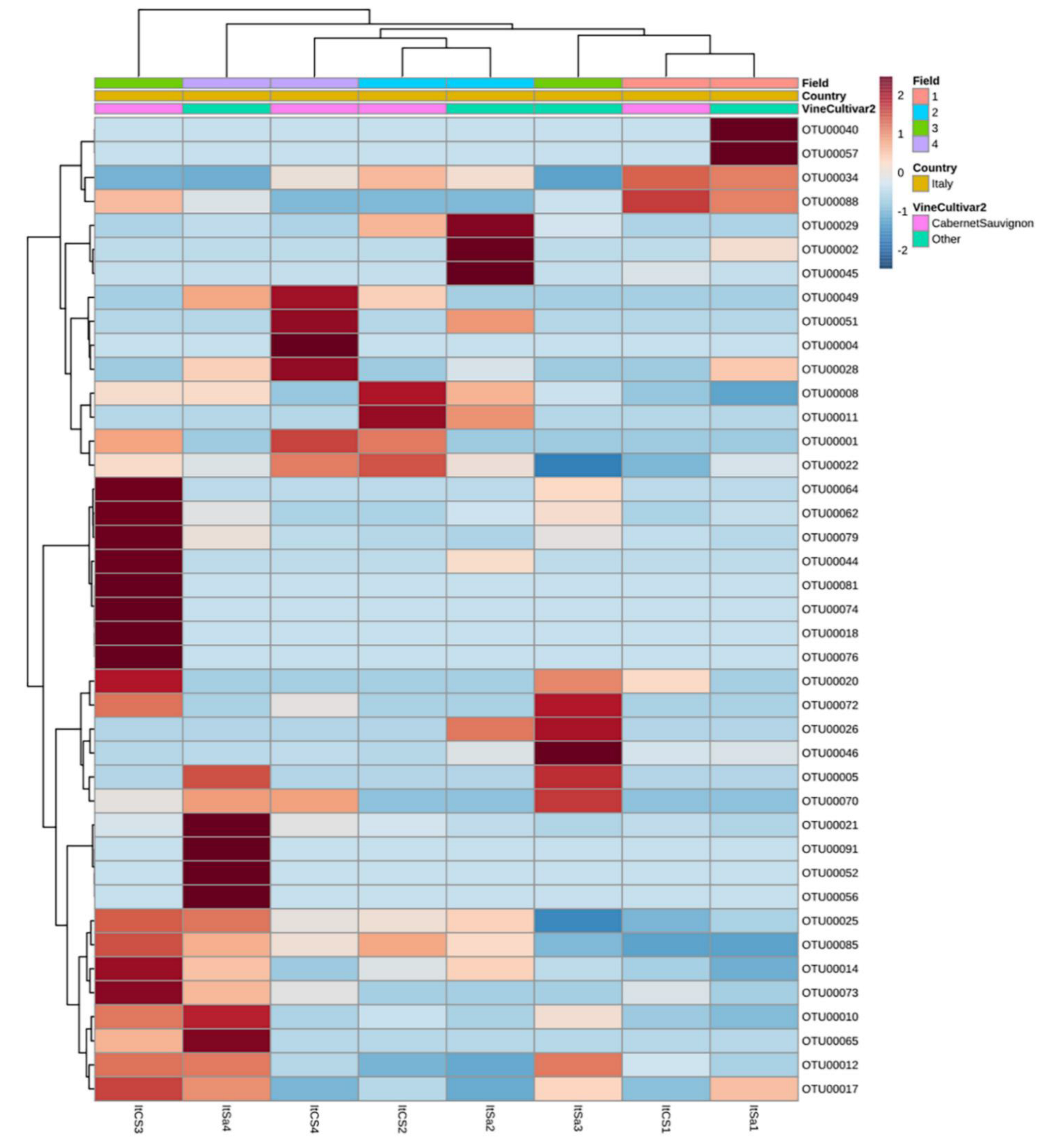
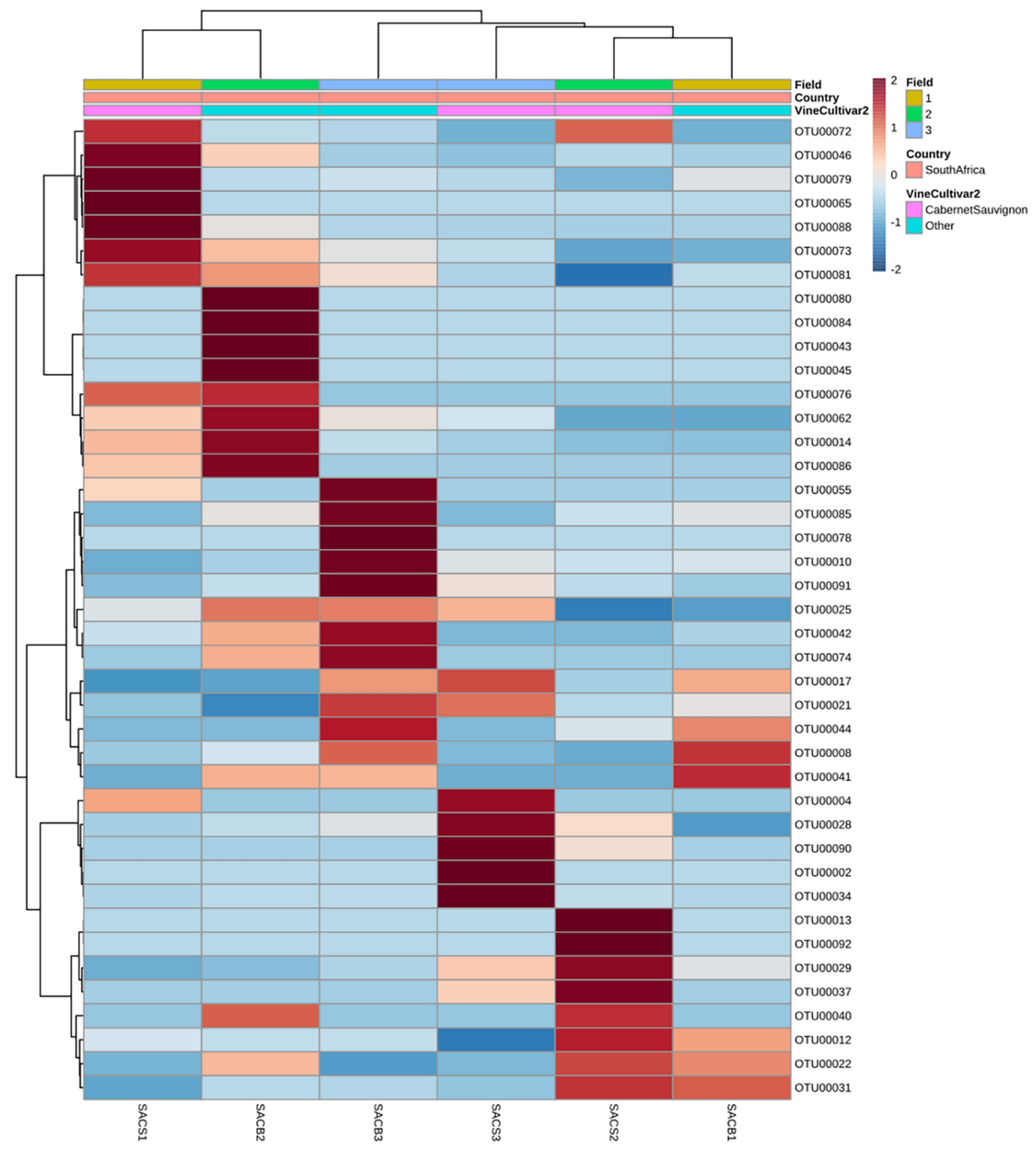
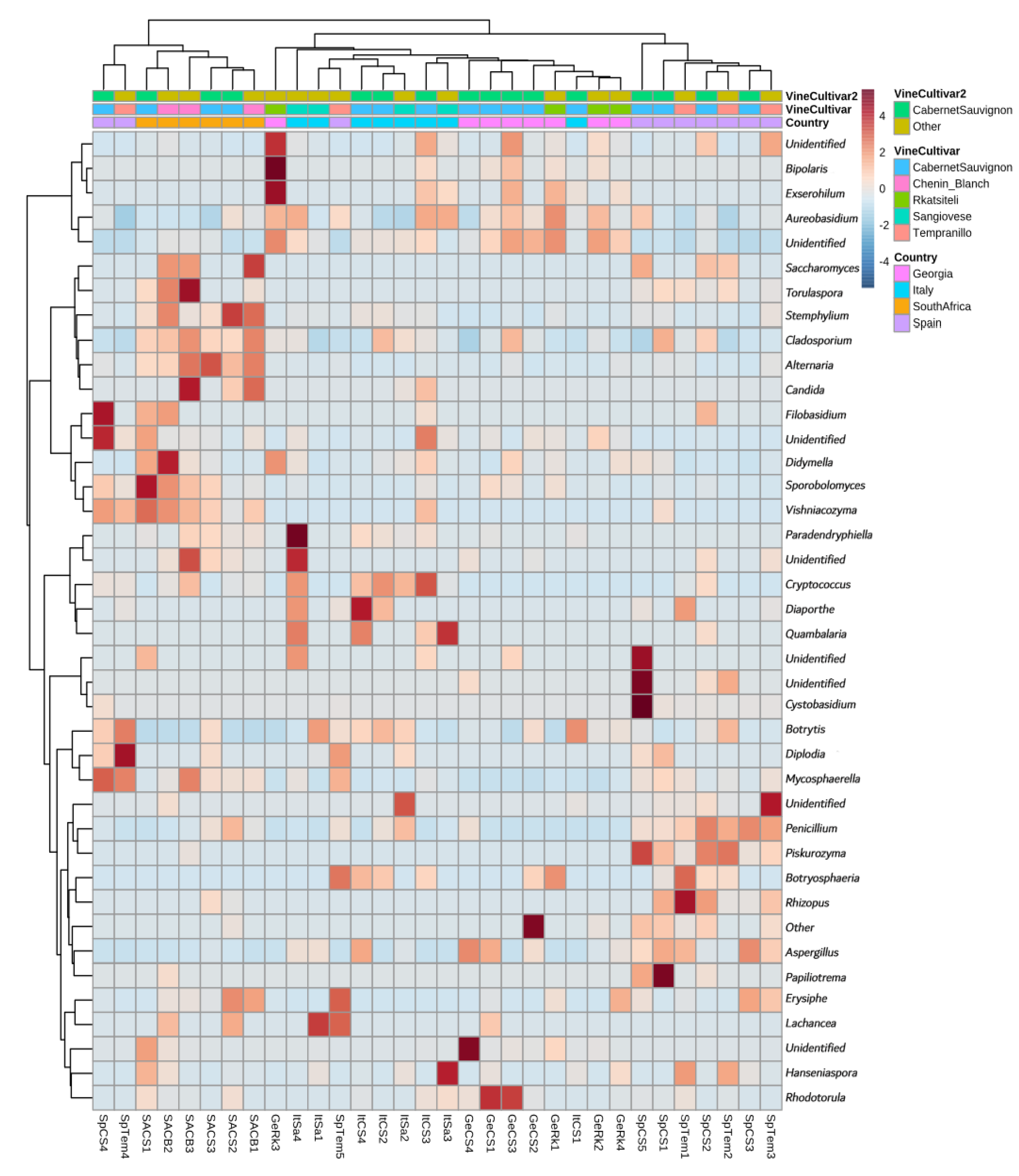
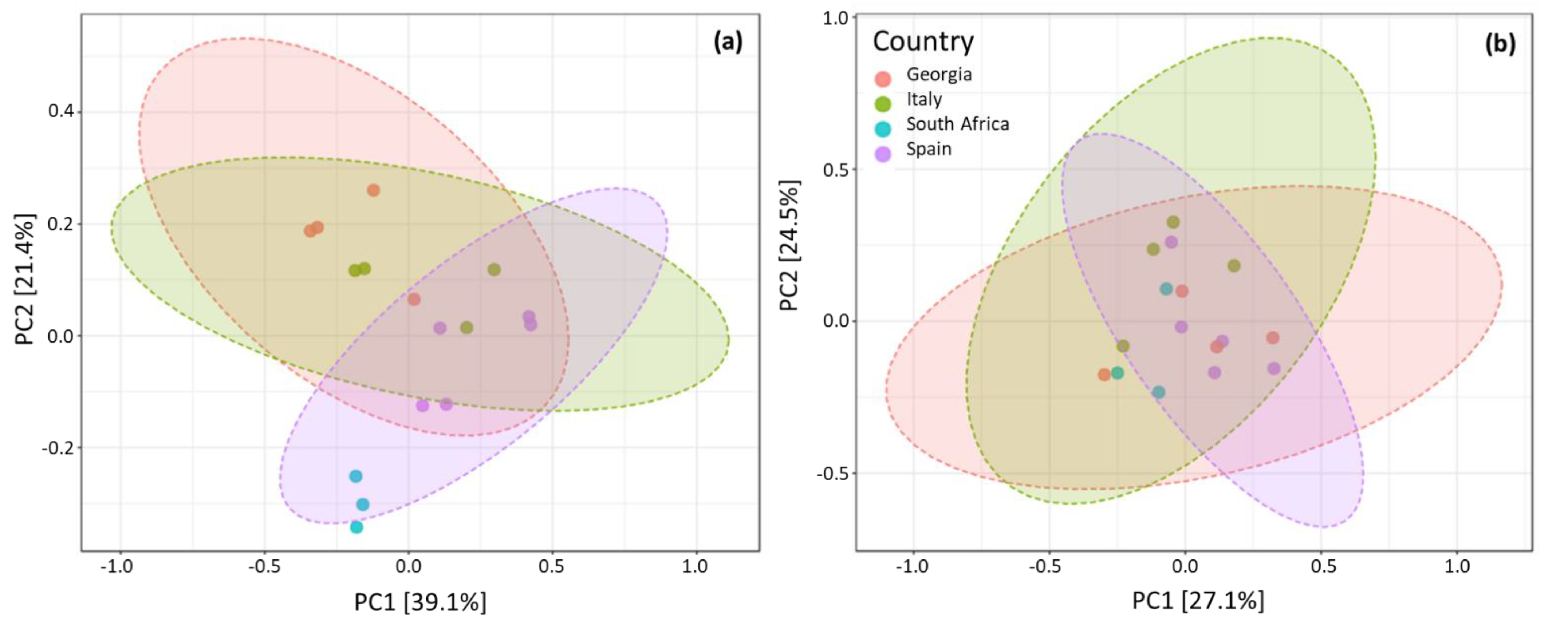
3.3.1. Country of Isolation Is the Primary Driver
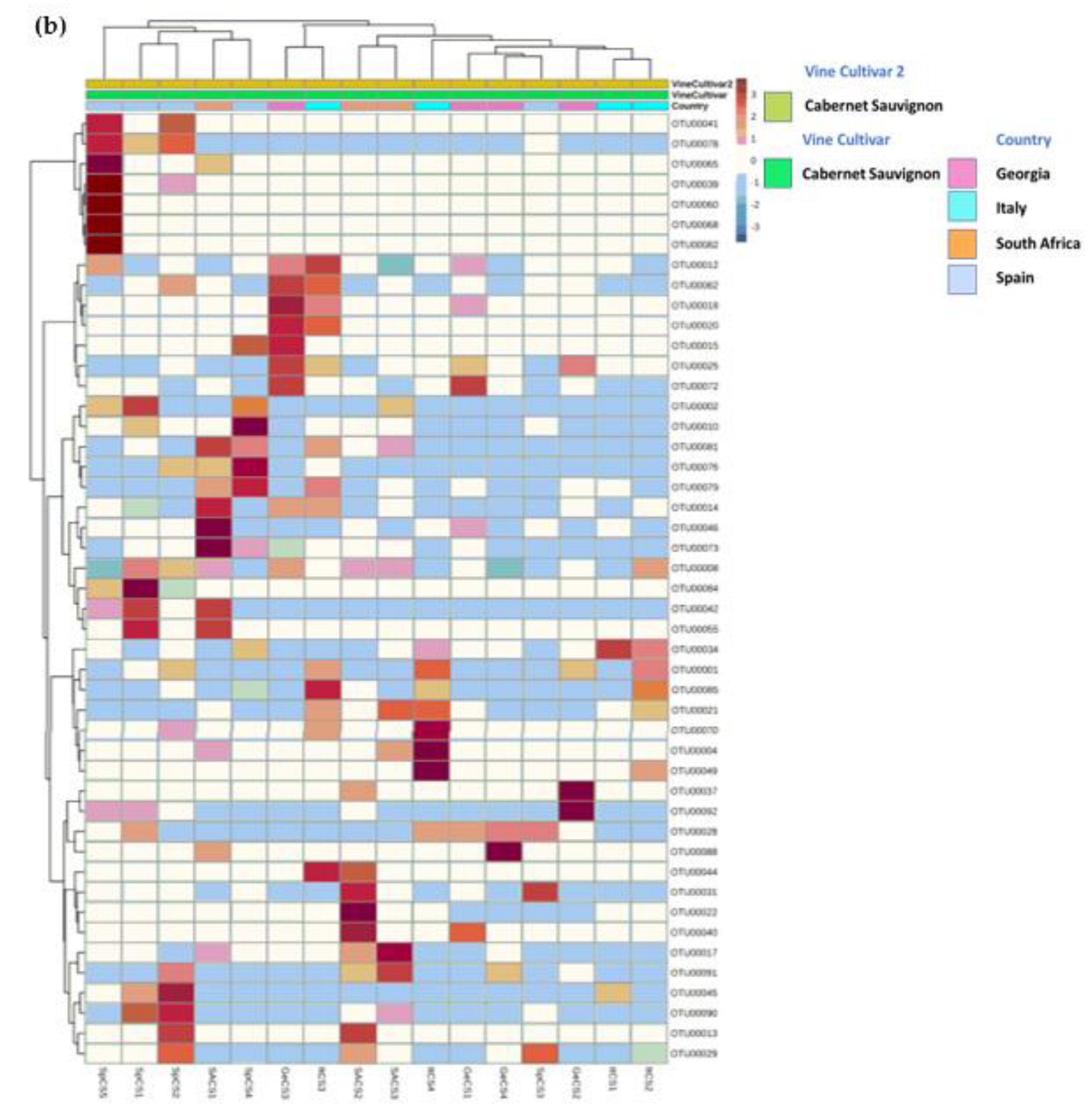
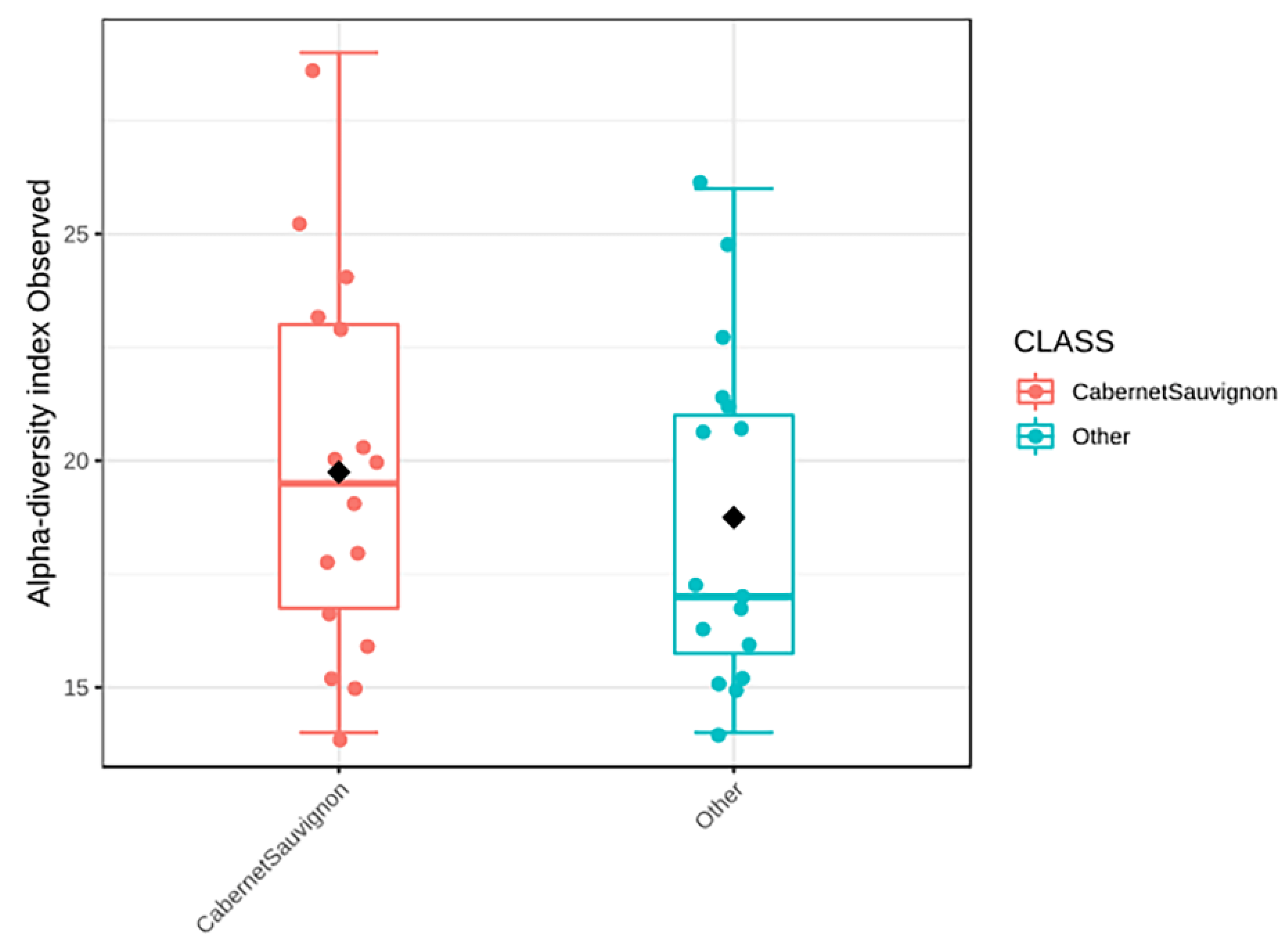
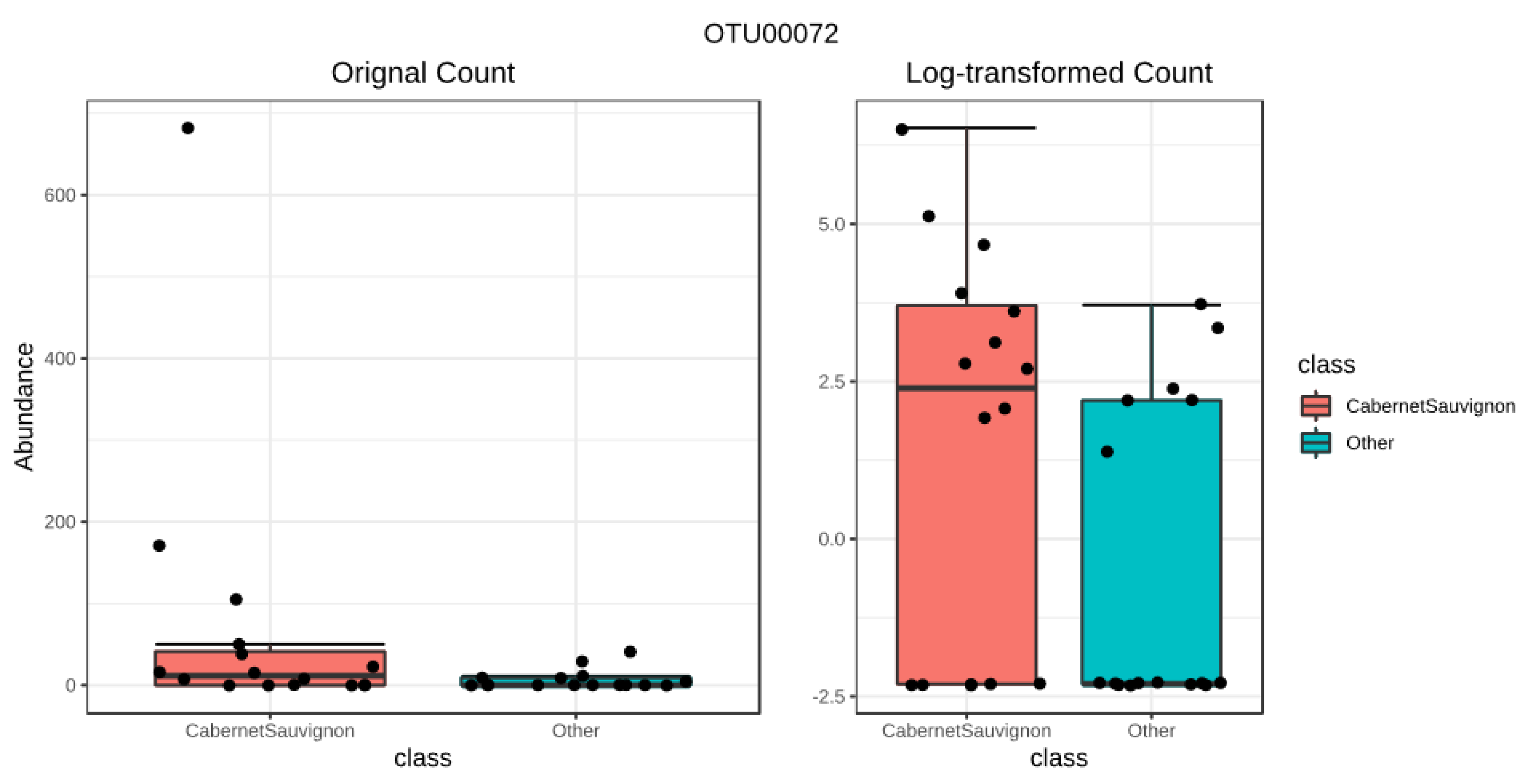
3.3.2. Cabernet Sauvignon Vine Cultivar Shows a Specific Microbial Fingerprint
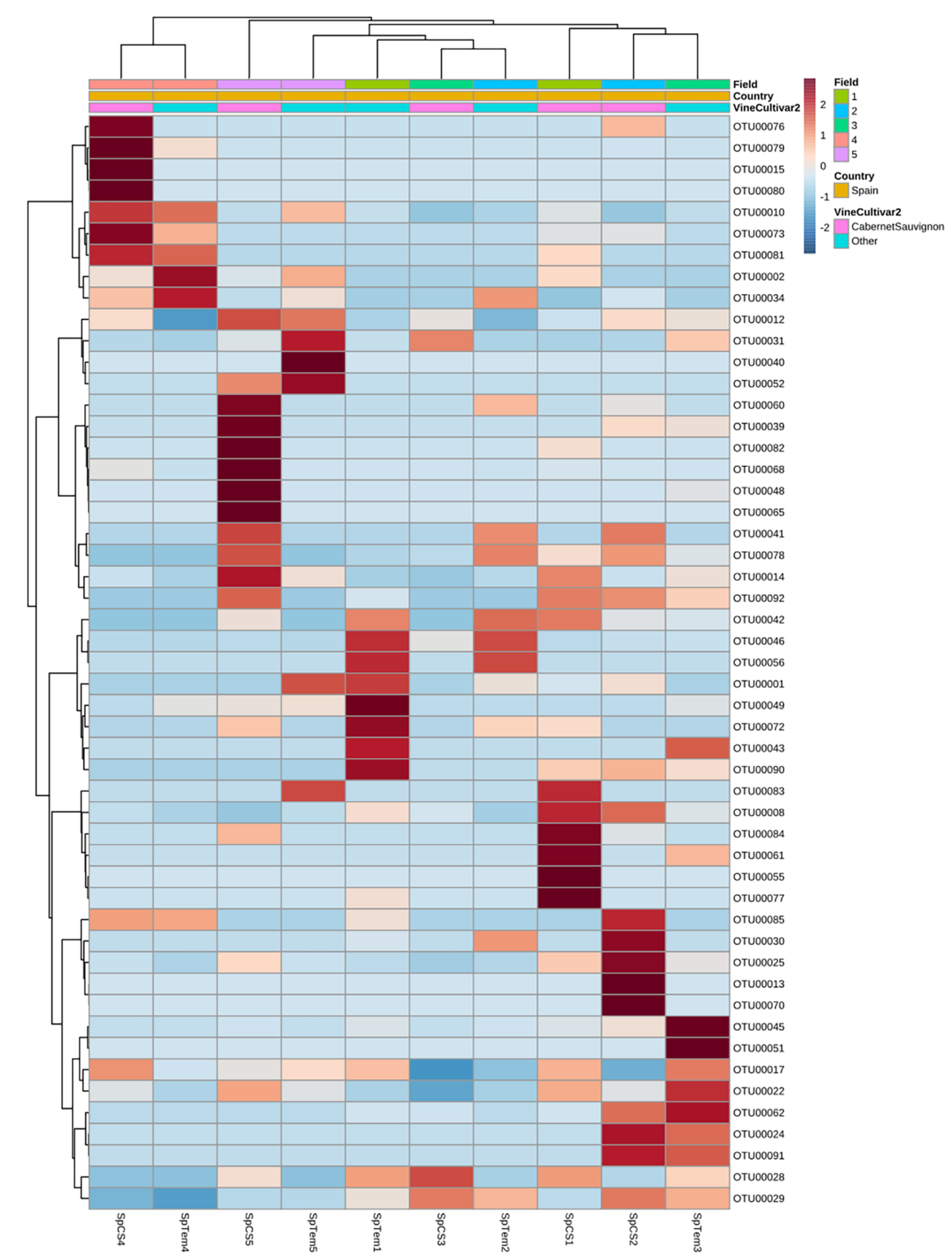
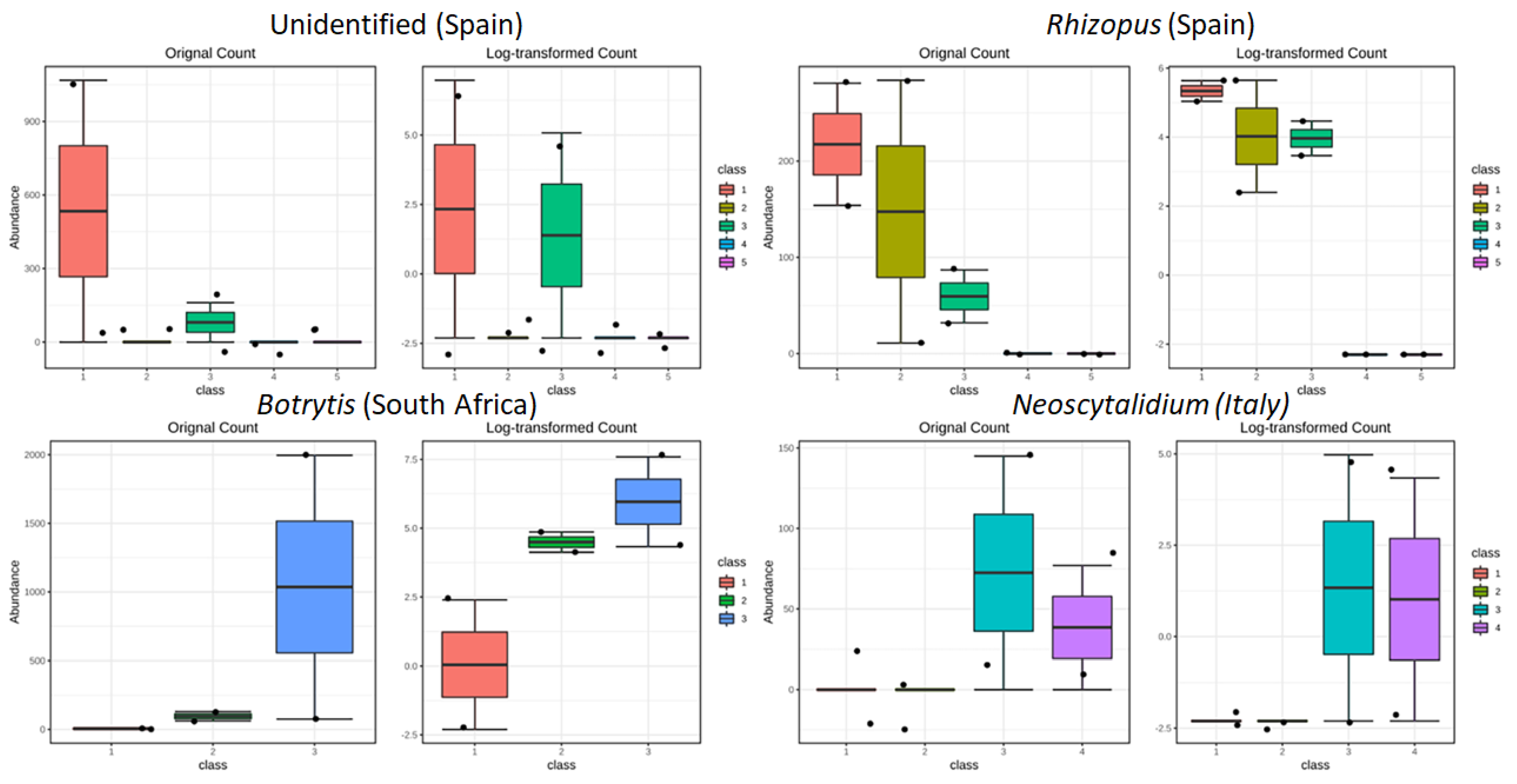
3.3.3. Drivers of Differentiation at a Local Level
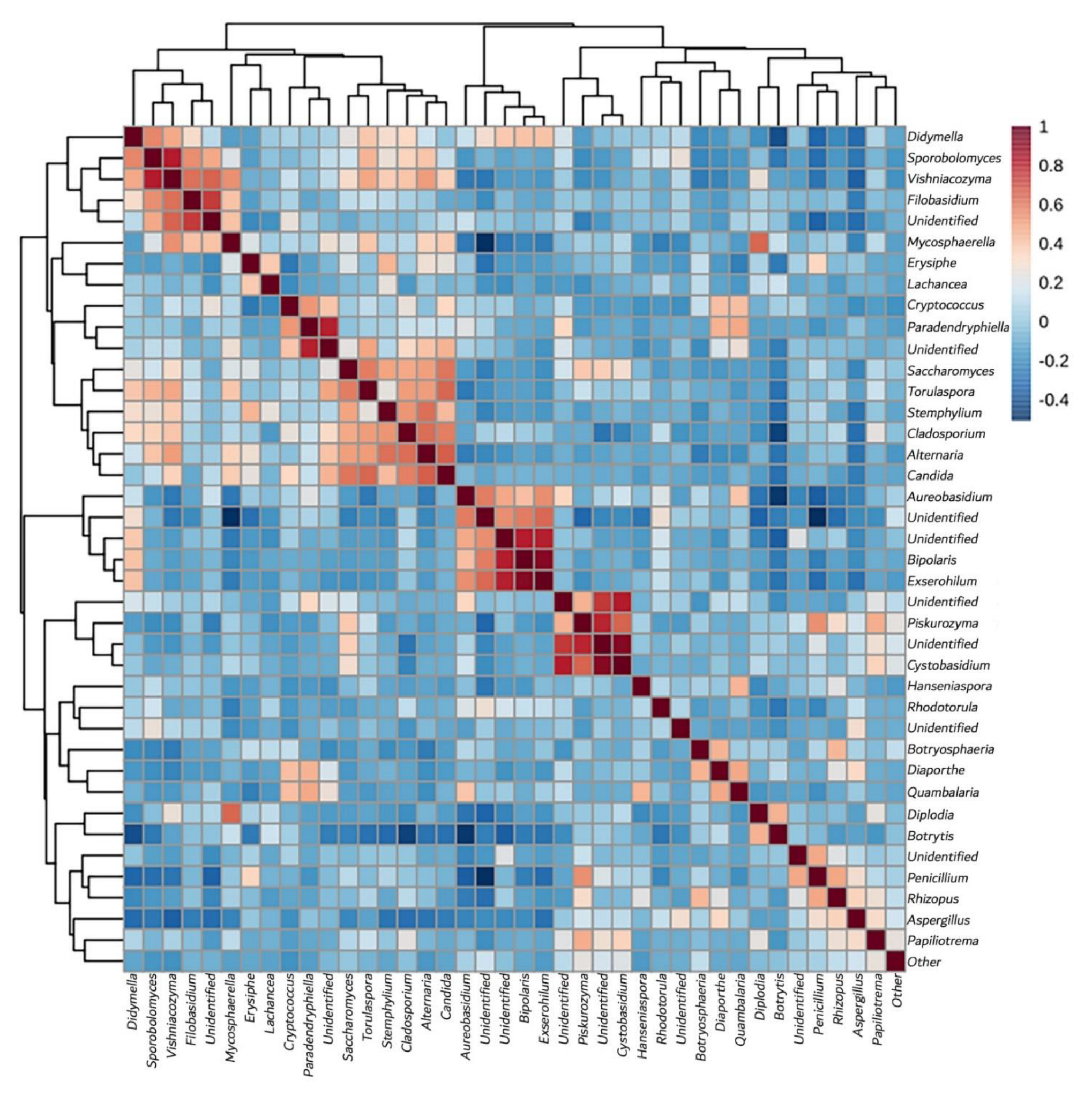
3.3.4. Yeast Genera May Shape Population Diversity
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Country | Cultivar | Field Number | Sample Name | Winery Name | Wine District | GPS Coordinates |
|---|---|---|---|---|---|---|
| Georgia (Ge) | Cabernet Sauvignon (CS) | 1 | GeCS1 | Khareba | Gurjaani, Kakheti | 41°53′34.5″ N; 45°42′33.7″ E |
| 2 | GeCS2 | Khareba | Kvareli, Kakheti | 41°57′39.9″ N; 45°39′45.9″ E | ||
| 3 | GeCS3 | Telavi Wine Cellar | Telavi, Kakheti | 41°57′09.0″ N; 45°36′01.6″ E | ||
| 4 | GeCS4 | Besini Company | Telavi, Kakheti | 41°54′51.4″ N; 45°35′12.5″ E | ||
| Rkatsiteli (Rk) | 1 | Ge Rk1 | Khareba | Gurjaani, Kakheti | 41°52′41.7″ N; 45°41′49.3″ E | |
| 2 | Ge Rk2 | Khareba | Kvareli, Kakheti | 41°57′15.2″ N; 45°39′26.4″ E | ||
| 3 | Ge Rk3 | Telavi Wine Cellar | Telavi, Kakheti | 41°57′27.8″ N; 45°36′10.6″ E | ||
| 4 | Ge Rk4 | Besini Company | Telavi, Kakheti | 41°54′54.4″ N; 45°35′36.4″ E | ||
| Italy (It) | Cabernet Sauvignon (CS) | 1 | ItCS1 | Banfi | Montalcino, Tuscany | 42°57′47.4″ N; 11°25′08.5″ E |
| 2 | ItCS2 | Guado al Melo | Castagneto Carducci, Tuscany | 43°11′37.9″ N; 10°37′34.4″ E | ||
| 3 | ItCS3 | Ruffino | Chianti, Tuscany | 43°40′53.7″ N; 11°21′06.1″ E | ||
| 4 | ItCS4 | Ruffino | Chianti, Tuscany | 43°41′31.6″ N; 11°19′13.9″ E | ||
| Sangiovese (Sa) | 1 | ItSa1 | Banfi | Montalcino, Tuscany | 42°57′48.7″ N; 11°25′16.0″ E | |
| 2 | ItSa2 | Guado al Melo | Castagneto Carducci, Tuscany | 43°11′35.2″ N; 10°37′35.3″ E | ||
| 3 | ItSa3 | Ruffino | Chianti, Tuscany | 43°40′54.1″ N; 11°21′07.1″ E | ||
| 4 | ItSa4 | Ruffino | Chianti, Tuscany | 43°41′31.2″ N; 11°19′12.9″ E | ||
| South Africa (SA) | Cabernet Sauvignon (CS) | 1 | SACS1 | Stark-Condé | Stellenbosch | 33.953790° S; 18.911330° E |
| 2 | SACS2 | Reyneke | Stellenbosch | 33.960188° S; 18.754975° E | ||
| 3 | SACS3 | Villiera | Stellenbosch | 33.835582° S; 18.787778° E | ||
| Chenin Blanc (SB) | 1 | SACB1 | Stark-Condé | Stellenbosch | 33.954752° S; 18.911675° E | |
| 2 | SACB2 | Reyneke | Stellenbosch | 33.959321° S; 18.753102° E | ||
| 3 | SACB3 | Villiera | Stellenbosch | 33.843688° S; 18.785048° E | ||
| Spain (Sp) | Cabernet Sauvignon (CS) | 1 | SpCS1 | Valdegón | Logroño, La Rioja | 42°27′57.5″ N; 2°17′39.1″ W |
| 2 | SpCS2 | Bodegas Olarra | Logroño, La Rioja | 42°28′18.4″ N; 2°24′31.1″ W | ||
| 3 | SpCS3 | Barón de Ley | Mendavia, Navarra | 42°25′36.2″ N; 2°10′00.1″ W | ||
| 4 | SpCS4 | Vivanco | Briones, La Rioja | 42°32′16.3″ N; 2°46′24.2″ W | ||
| 5 | SpCS5 | Don Jacobo-Altos del Corral | Navarretes, La Rioja | 42°24′56.2″ N; 2°33′58.7″ W | ||
| Tempranillo (Tem) | 1 | SpTem1 | Valdegón | Logroño, La Rioja | 42°27′57.5″ N; 2°17′39.1″ W | |
| 2 | SpTem2 | Bodegas Olarra | Logroño, La Rioja | 42°28′18.4″ N; 2°24′31.1″ W | ||
| 3 | SpTem3 | Barón de Ley | Mendavia, Navarra | 42°25′36.2″ N; 2°10′00.1″ W | ||
| 4 | SpTem4 | Vivanco | Briones, La Rioja | 42°32′16.3″ N; 2°46′24.2″ W | ||
| 5 | SpTem5 | Don Jacobo-Altos del Corral | Navarretes, La Rioja | 42°24′56.2″ N; 2°33′58.7″ W |
| OTU Code | Genus Identification | OTU Code | Genus Identification | OTU Code | Genus Identification |
|---|---|---|---|---|---|
| 00001 | Botryosphaeria | 00032 | Articulospora | 00063 | Suillus |
| 00002 | Diplodia | 00033 | Lambertella | 00064 | Vuilleminia |
| 00003 | Dothiorella | 00034 | Botrytis | 00065 | unidentified |
| 00004 | Neofusicoccum | 00035 | Phacidium | 00066 | Stereum |
| 00005 | Neoscytalidium | 00036 | Hyphopichia | 00067 | unidentified |
| 00006 | unidentified | 00037 | Metschnikowia | 00068 | Cystobasidium |
| 00007 | unidentified | 00038 | Kregervanrija | 00069 | Buckleyzyma |
| 00008 | Cladosporium | 00039 | Pichia | 00070 | Quambalaria |
| 00009 | Uwebraunia | 00040 | Lachancea | 00071 | Rhodosporidiobolus |
| 00010 | Mycosphaerella | 00041 | Saccharomyces | 00072 | Rhodotorula |
| 00011 | unidentified | 00042 | Torulaspora | 00073 | Sporobolomyces |
| 00012 | Aureobasidium | 00043 | Zygosaccharomyces | 00074 | Cystofilobasidium |
| 00013 | Hormonema | 00044 | Candida | 00075 | Udeniomyces |
| 00014 | Didymella | 00045 | unidentified | 00076 | Filobasidium |
| 00015 | Neoascochyta | 00046 | Hanseniaspora | 00077 | Naganishia |
| 00016 | Sclerostagonospora | 00047 | Zygoascus | 00078 | Piskurozyma |
| 00017 | Alternaria | 00048 | unidentified | 00079 | unidentified |
| 00018 | Bipolaris | 00049 | Diaporthe | 00080 | Holtermanniella |
| 00019 | Comoclathris | 00050 | unidentified | 00081 | Vishniacozyma |
| 00020 | Exserohilum | 00051 | Phomopsis | 00082 | Kwoniella |
| 00021 | Paradendryphiella | 00052 | unidentified | 00083 | Tsuchiyaea |
| 00022 | Stemphylium | 00053 | Colletotrichum | 00084 | Papiliotrema |
| 00023 | unidentified | 00054 | Verticillium | 00085 | Cryptococcus |
| 00024 | Preussia | 00055 | Cordyceps | 00086 | unidentified |
| 00025 | unidentified | 00056 | unidentified | 00087 | unidentified |
| 00026 | unidentified | 00057 | Hirsutella | 00088 | unidentified |
| 00027 | Valsaria | 00058 | Campylospora | 00089 | Mucor |
| 00028 | Aspergillus | 00059 | Nigrospora | 00090 | Rhizopus |
| 00029 | Penicillium | 00060 | unidentified | 00091 | unidentified |
| 00030 | Punctelia | 00061 | unidentified | 00092 | Other |
| 00031 | Erysiphe | 00062 | unidentified |

References
- Darriaut, R.; Lailheugue, V.; Masneuf-Pomarède, I.; Marguerit, E.; Martins, G.; Company, S.; Ballestra, P.; Upton, S.; Ollat, N.; Lauvergeat, V. Grapevine rootstock and soil microbiome interactions: Keys for a resilient viticulture. Hortic. Res. 2022, 19, uhac019. [Google Scholar] [CrossRef]
- Ranade, Y.; Sawant, I.; Saha, S.; Chandrashekar, M.; Pathak, P. Epiphytic Microbial Diversity of Vitis vinifera Fructosphere: Present Status and Potential Applications. Curr. Microbiol. 2021, 78, 1086–1098. [Google Scholar] [CrossRef] [PubMed]
- Lazcano, C.; Decock, C.; Wilson, S.G. Defining and managing for healthy vineyard soils, intersections with the concept of terroir. Front. Environ. Sci. 2020, 8, 68. [Google Scholar] [CrossRef]
- Mezzasalma, V.; Sandionigi, A.; Bruni, I.; Bruno, A.; Lovicu, G.; Casiraghi, M.; Labra, M. Grape microbiome as a reliable and persistent signature of field origin and environmental conditions in Cannonau wine production. PLoS ONE 2017, 12, e0184615. [Google Scholar] [CrossRef]
- Miura, T.; Sánchez, R.; Castañeda, L.E.; Godoy, K.; Barbosa, O. Is microbial terroir related to geographic distance between vineyards? Environ. Microbiol. 2017, 9, 742–749. [Google Scholar] [CrossRef]
- Setati, M.E.; Jacobson, D.; Bauer, F.F. Sequence-based analysis of the Vitis vinifera L. cv Cabernet Sauvignon grape must mycobiome in three South African vineyards employing distinct agronomic systems. Front. Microbiol. 2015, 6, 1358. [Google Scholar] [CrossRef]
- Cordero-Bueso, G.; Vigentini, I.; Foschino, R.; Maghradze, D.; Ruiz-Muñoz, M.; Benitez-Trujillo, F.; Cantoral, J.M. Culturable Yeast Diversity of Grape Berries from Vitis vinifera ssp. sylvestris (Gmelin) Hegi. J. Fungi. 2022, 8, 410. [Google Scholar] [CrossRef]
- Leveau, J.H.J.; Tech, J.J. Grapevine microbiomics: Bacterial diversity on grape leaves and berries revealed by high-throughput sequence analysis of 16S rRNA amplicons. Acta Hortic. 2011, 905, 31–42. [Google Scholar] [CrossRef]
- Morrison-Whittle, P.; Goddard, M.R. Quantifying the relative roles of selective and neutral processes in defining eukaryotic microbial communities. ISME J. 2015, 9, 2003–2011. [Google Scholar] [CrossRef]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The soil microbiome influences grapevine-associated microbiota. mBio 2015, 6, e02527-14. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139–E148. [Google Scholar] [CrossRef]
- Marzano, M.; Fosso, B.; Manzari, C.; Grieco, F.; Intranuovo, M.; Cozzi, G.; Mulè, G.; Scioscia, G.; Valiente, G.; Tullo, A.; et al. Complexity and dynamics of the winemaking bacterial communities in berries, musts, and wines from Apulian grape cultivars through time and space. PLoS ONE 2016, 11, e0157383. [Google Scholar] [CrossRef] [PubMed]
- Mezzasalma, V.; Sandionigi, A.; Guzzetti, L.; Galimberti, A.; Grando, M.S.; Tardaguila, J.; Labra, M. Geographical and cultivar features differentiate grape microbiota in Northern Italy and Spain vineyards. Front. Microbiol. 2018, 9, 946. [Google Scholar] [CrossRef] [PubMed]
- del Portillo, M.C.; Franquès, J.; Araque, I.; Reguant, C.; Bordons, A. Bacterial diversity of Grenache and Carignan grape surface from different vineyards at Priorat wine region (Catalonia, Spain). Int. J. Food Microbiol. 2016, 219, 56–63. [Google Scholar] [CrossRef]
- Taylor, M.W.; Tsai, P.; Anfang, N.; Ross, H.A.; Goddard, M.R. Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ. Microbiol. 2014, 16, 2848–2858. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Divol, B.; Setati, M.E. A Shotgun Metagenomic Sequencing Exploration of Cabernet Sauvignon Grape Must Reveals Yeast Hydrolytic Enzymes. S. Afr. J. Enol. Vitic. 2021, 42, 213–223. [Google Scholar] [CrossRef]
- Liu, D.; Chen, Q.; Zhang, P.; Chen, D.; Howell, K.S. The fungal microbiome is an important component of vineyard ecosystems and correlates with regional distinctiveness of wine. mSphere 2020, 5, e00534-20. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yang, S.; Lin, M.; Guo, S.; Han, X.; Ren, M.; Du, L.; Song, Y.; You, Y.; Zhan, J.; et al. The Biogeography of Fungal Communities Across Different Chinese Wine-Producing Regions Associated with Environmental Factors and Spontaneous Fermentation Performance. Front. Microbiol. 2022, 12, 636639. [Google Scholar] [CrossRef]
- Setati, M.E.; Jacobson, D.; Andong, U.C.; Bauer, F. The vineyard yeast microbiome, a mixed model microbial map. PLoS ONE 2012, 7, e52609. [Google Scholar] [CrossRef]
- Liu, D.; Legras, J.L.; Zhang, P.; Chen, D.; Howell, K. Diversity and dynamics of fungi during spontaneous fermentations and association with unique aroma profiles in wine. Int. J. Food Microbiol. 2021, 338, 108983. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Mills, D.A. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 2013, 79, 2519–2526. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 3, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J.; Suggests, M.A.S.S. The vegan package. Community Ecol. Packag. 2007, 10, 631–637. [Google Scholar]
- Liu, D.; Howell, K. Community succession of the grapevine fungal microbiome in the annual growth cycle. Environ. Microbiol. 2021, 23, 1842–1857. [Google Scholar] [CrossRef]
- Chen, A.; Mao, X.; Sun, Q.; Wei, Z.; Li, J.; You, Y.; Zhao, J.; Jiang, G.; Wu, Y.; Wang, L.; et al. Alternaria Mycotoxins: An Overview of Toxicity, Metabolism, and Analysis in Food. J. Agric. Food Chem. 2021, 69, 7817–7830. [Google Scholar] [CrossRef]
- Paterson, R.R.M.; Venâncio, A.; Lima, N.; Guilloux-Bénatier, M.; Rousseaux, S. Predominant mycotoxins, mycotoxigenic fungi and climate change related to wine. Food Res. Int. 2018, 103, 478–491. [Google Scholar] [CrossRef]
- Di Canito, A.; Mateo-Vargas, M.A.; Mazzieri, M.; Cantoral, J.; Foschino, R.; Cordero-Bueso, G.; Vigentini, I. The Role of Yeasts as Biocontrol Agents for Pathogenic Fungi on Postharvest Grapes: A Review. Foods 2021, 10, 1650. [Google Scholar] [CrossRef]
- Ribéreau-Gayon, J.; Glories, Y.; Maujean, A.; Dubourdieu, D. Handbook of Enology. In The Microbiology of Wine and Vinifications, 3rd ed.; Wiley: New York, NY, USA, 2006; Volume 2. [Google Scholar]
- Otto, M.; Geml, J.; Hegyi, Á.I.; Hegyi-Kaló, J.; Pierneef, R.; Pogány, M.; Kun, J.; Gyenesei, A.; Váczy, K.Z. Botrytis cinerea expression profile and metabolism differs between noble and grey rot of grapes. Food Microbiol. 2022, 106, 104037. [Google Scholar] [CrossRef]
- Tančinová, D.; Mašková, Z.; Rybárik, L.; Michalová, V. Species of genera Botrytis, Fusarium and Rhizopus on grapes of the Slovak origin. Potravinarstvo 2017, 11, 403–409. [Google Scholar] [CrossRef]
- Kačániová, M.; Eftimová, Z.M.; Brindza, J.; Felšöciová, S.; Ivanišová, E.; Žiarovská, J.; Kluz, M.; Terentjeva, M. Microbiota of Tokaj Grape Berries of Slovak Regions. Erwerbs-Obstbau 2020, 62, 25–33. [Google Scholar] [CrossRef]
- Felšöciová, S.; Miroslava, K.; František, V. Survey of mycobiota on Slovakian wine grapes from Small Carpathians wine region. Potravinarstvo 2020, 14, 721–728. [Google Scholar] [CrossRef]
- Arkam, M.; Alves, A.; Lopes, A.; Čechová, J.; Pokluda, R.; Eichmeier, A.; Zitouni, A.; Mahamedi, A.E.; Berraf-Tebbal, A. Diversity of Botryosphaeriaceae causing grapevine trunk diseases and their spatial distribution under different climatic conditions in Algeria. Eur. J. Plant Pathol. 2021, 161, 933–952. [Google Scholar] [CrossRef]
- Urbez-Torres, J.R. The status of Botryosphaeriaceae species infecting grapevines. Phytopathol. Mediterr. 2011, 50, S5–S45. [Google Scholar] [CrossRef]
- Droby, S.; Lichter, A. Post-harvest Botrytis infection: Etiology, development and management. In Botrytis: Biology, Pathology and Control; Springer: Berlin/Heidelberg, Germany, 2007; pp. 349–367. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tronchoni, J.; Setati, M.E.; Fracassetti, D.; Valdetara, F.; Maghradze, D.; Foschino, R.; Curiel, J.A.; Morales, P.; Gonzalez, R.; Vigentini, I.; et al. Identifying the Main Drivers in Microbial Diversity for Cabernet Sauvignon Cultivars from Europe to South Africa: Evidence for a Cultivar-Specific Microbial Fingerprint. J. Fungi 2022, 8, 1034. https://doi.org/10.3390/jof8101034
Tronchoni J, Setati ME, Fracassetti D, Valdetara F, Maghradze D, Foschino R, Curiel JA, Morales P, Gonzalez R, Vigentini I, et al. Identifying the Main Drivers in Microbial Diversity for Cabernet Sauvignon Cultivars from Europe to South Africa: Evidence for a Cultivar-Specific Microbial Fingerprint. Journal of Fungi. 2022; 8(10):1034. https://doi.org/10.3390/jof8101034
Chicago/Turabian StyleTronchoni, Jordi, Mathabatha Evodia Setati, Daniela Fracassetti, Federica Valdetara, David Maghradze, Roberto Foschino, Jose Antonio Curiel, Pilar Morales, Ramon Gonzalez, Ileana Vigentini, and et al. 2022. "Identifying the Main Drivers in Microbial Diversity for Cabernet Sauvignon Cultivars from Europe to South Africa: Evidence for a Cultivar-Specific Microbial Fingerprint" Journal of Fungi 8, no. 10: 1034. https://doi.org/10.3390/jof8101034







