Sgh1, an SR-like Protein, Is Involved in Fungal Development, Plant Infection, and Pre-mRNA Processing in Fusarium graminearum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analyses
2.2. Fungal Strains and Culture Conditions
2.3. Generation of Δsgh1 Mutants
2.4. Generation of the SGH1-, SGH1ΔRS-, SGH1ΔRRM1-, SGH1ΔRRM2-, and SGH1ΔRRM3-GFP
2.5. Plant Infection and DON Production Assays
2.6. Yeast Two-Hybrid and Bimolecular Fluorescence Complementation (BiFC) Assays
2.7. RNA-Seq Analysis
2.8. CFW and DAPI Staining
2.9. Quantification of Nuclear/Cytoplasmic Intensity Ratio of Fluorescence
3. Results
3.1. FGRAMPH1_01T26155 Encodes a Conserved SR-like Protein in Filamentous Ascomycetes
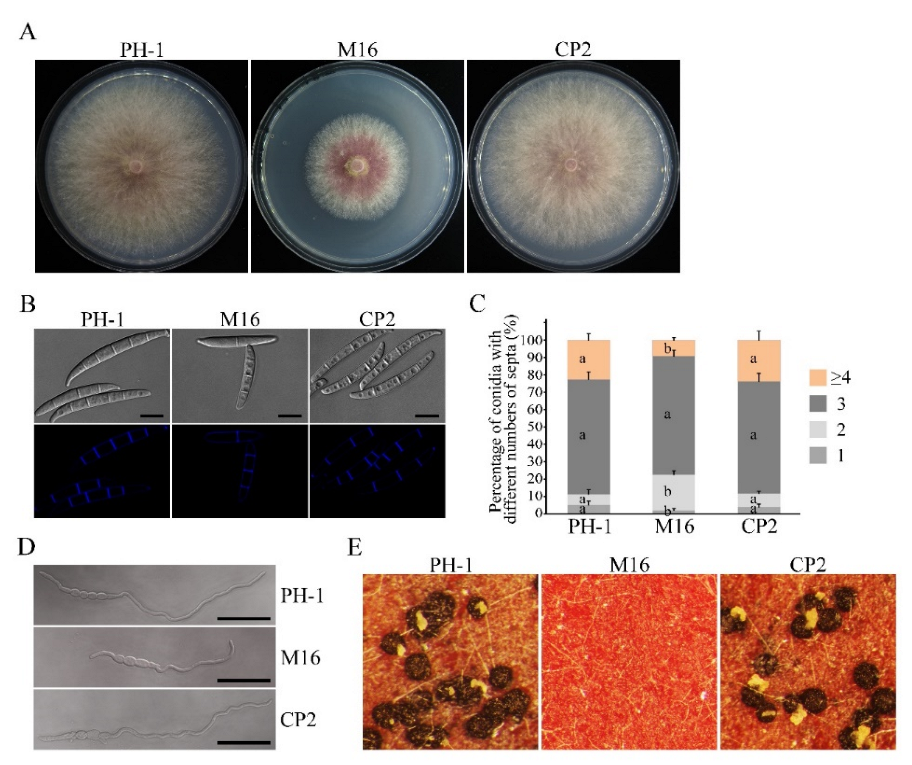
3.2. The Δsgh1 Mutant Is Defective in Vegetative Growth, Conidiogenesis, and Sexual Reproduction
| Strain | Brief Description | Reference |
|---|---|---|
| PH-1 | Wild-type strain | [43] |
| M6, M14, M16 | SGH1 deletion mutant of PH-1 | This study |
| CP2 | Transformant of M16 expressing SGH1-GFP construct | This study |
| DRS2 | Transformant of M16 expressing SGH1ΔRS-GFP construct | This study |
| DRRM1-1 | Transformant of M16 expressing SGH1ΔRRM1-GFP construct | This study |
| DRRM2-2 | Transformant of M16 expressing SGH1ΔRRM2-GFP construct | This study |
| RRRM3-2 | Transformant of M16 expressing SGH1ΔRRM3-GFP construct | This study |
| BFSS-5 | Transformant of PH-1 expressing SRK1ΔS-YFPN and SGH1-YFPC constructs | This study |
| DSSG-3 | Transformant of Δsrk1 mutant expressing SGH1-GFP construct | This study |
| Strain | Growth Rate | Conidiation | Perithecia Formation | DON (μg/g) d |
|---|---|---|---|---|
| (mm/d) a | (105 Spores/mL) b | (Perithecia/cm2) c | ||
| PH-1 | 7.5 ± 0.1 a | 24.41 ± 2.79 a | 665.7 ± 88.8 a | 1436.36 ± 19.53 a |
| M16 | 4.5 ± 0.1 b | 6.81 ± 1.53 b | 0 b | 585.91 ± 55.50 b |
| CP2 | 7.5 ± 0.3 a | 25.87 ± 2.6 a | 676.0 ± 83.7 a | 1497.24 ± 23.76 a |
| DRS2 | 7.4 ± 0.1 a | 21.86 ± 1.29 a | 708.8 ± 123.4 a | 1381.97 ± 284.16 ac |
| DRRM1-1 | 7.3 ± 0.2 a | 22.41 ± 0.65 a | 713.4 ± 93.1 a | 1191.93 ± 209.51 c |
| DRRM2-2 | 7.4 ± 0.2 a | 20.93 ± 0.22 a | 670.3 ± 80.8 a | 911.96 ± 251.08 c |
| DRRM3-2 | 5.6 ± 0.1 c | 11.16 ± 0.61 c | 340.2 ± 85.9 c | 637.99 ± 38.06 b |
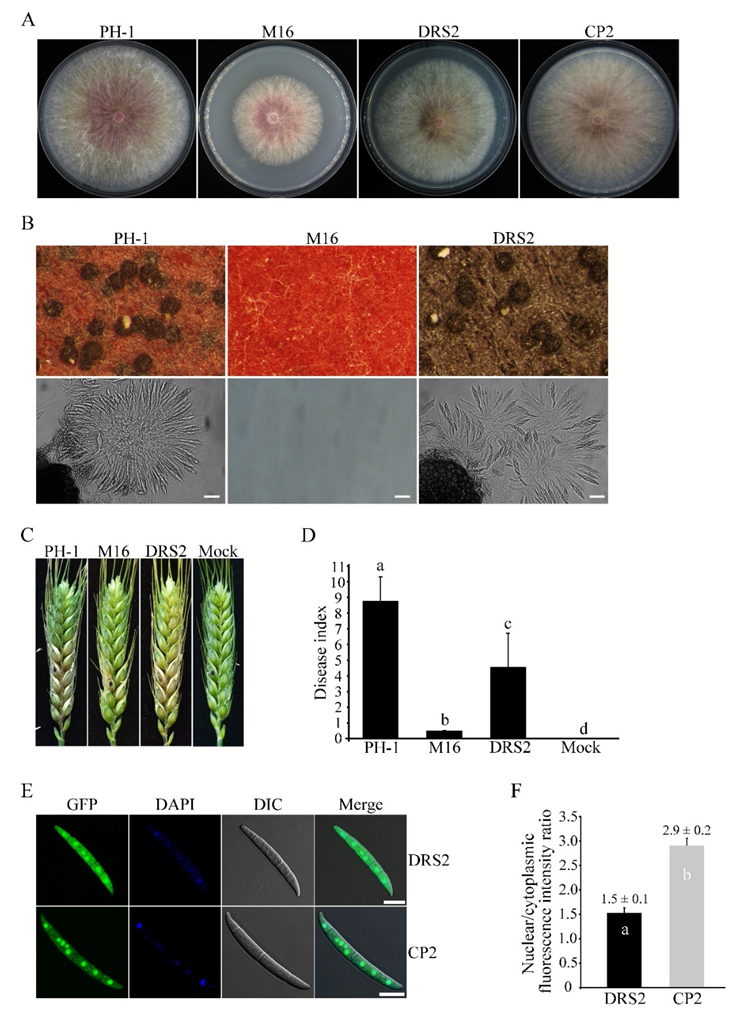
3.3. SGH1 Plays a Critical Role in Plant Infection
3.4. The Δsgh1 Deletion Mutant Showed Increased Sensitivity to Osmotic and Cell Wall Stresses
3.5. Subcellular Localization of Sgh1-GFP Fusion Protein
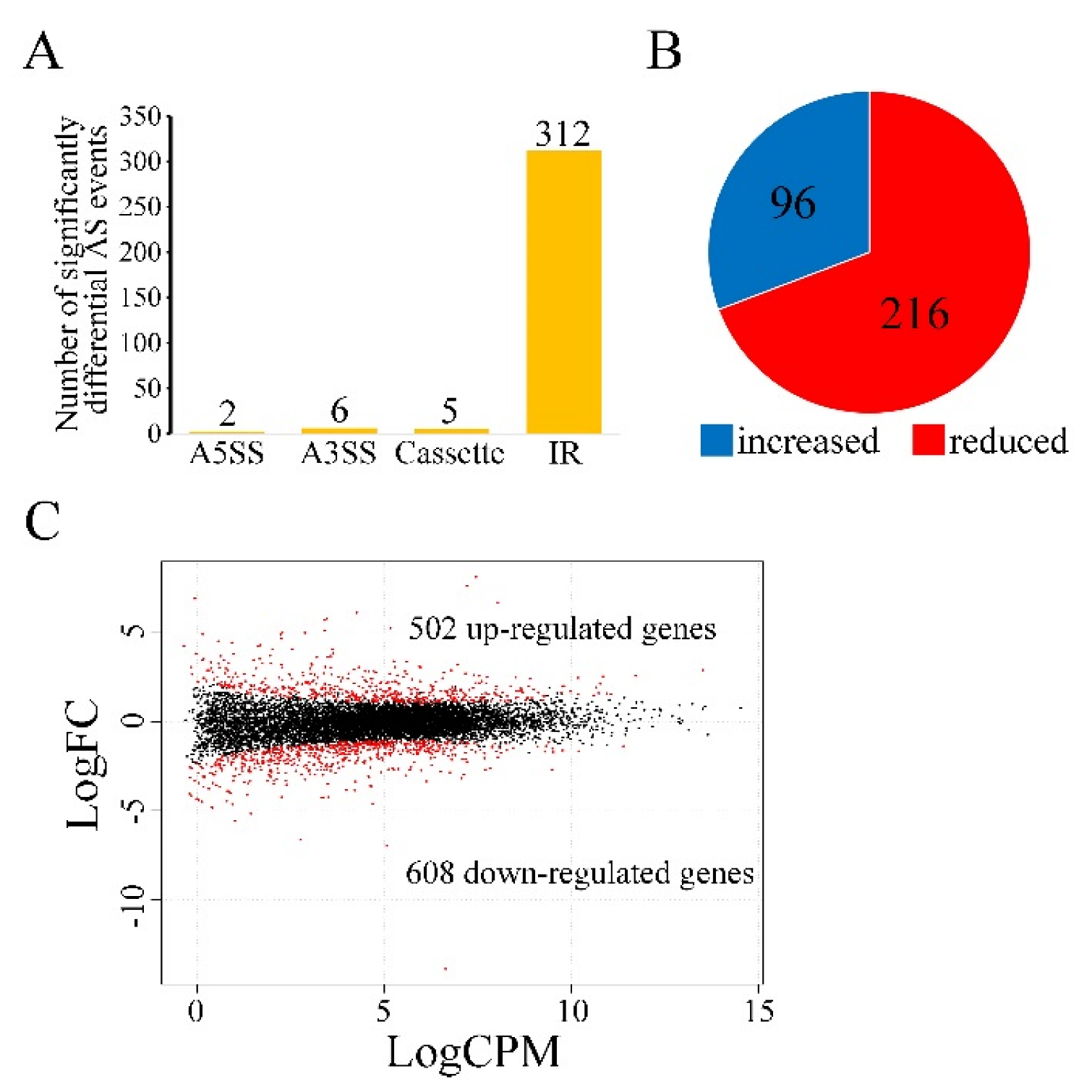
3.6. Sgh1 Regulates RNA Splicing and Gene Transcription
3.7. The RS Domain Is Important for Both Functions and Nuclear Localization of Sgh1
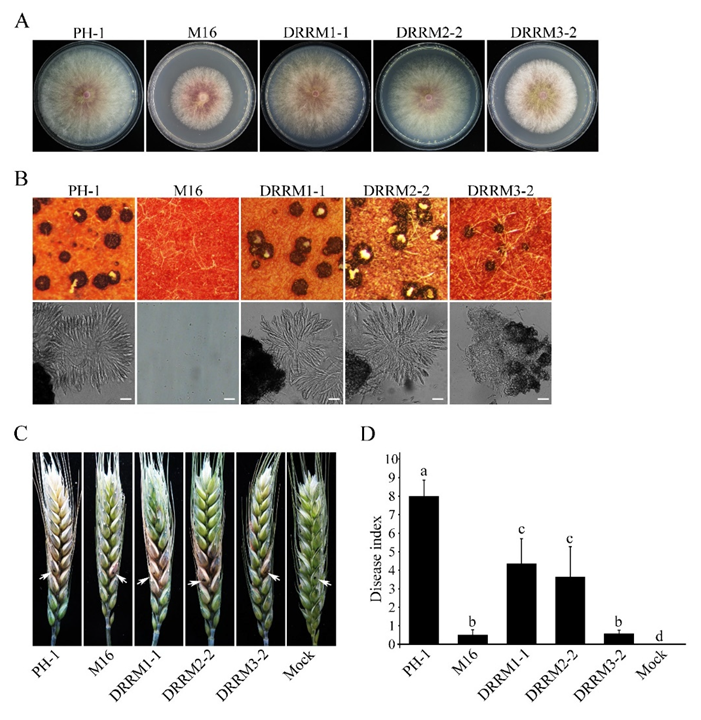
3.8. Functional Characterization of the Three RRM Domains of Sgh1
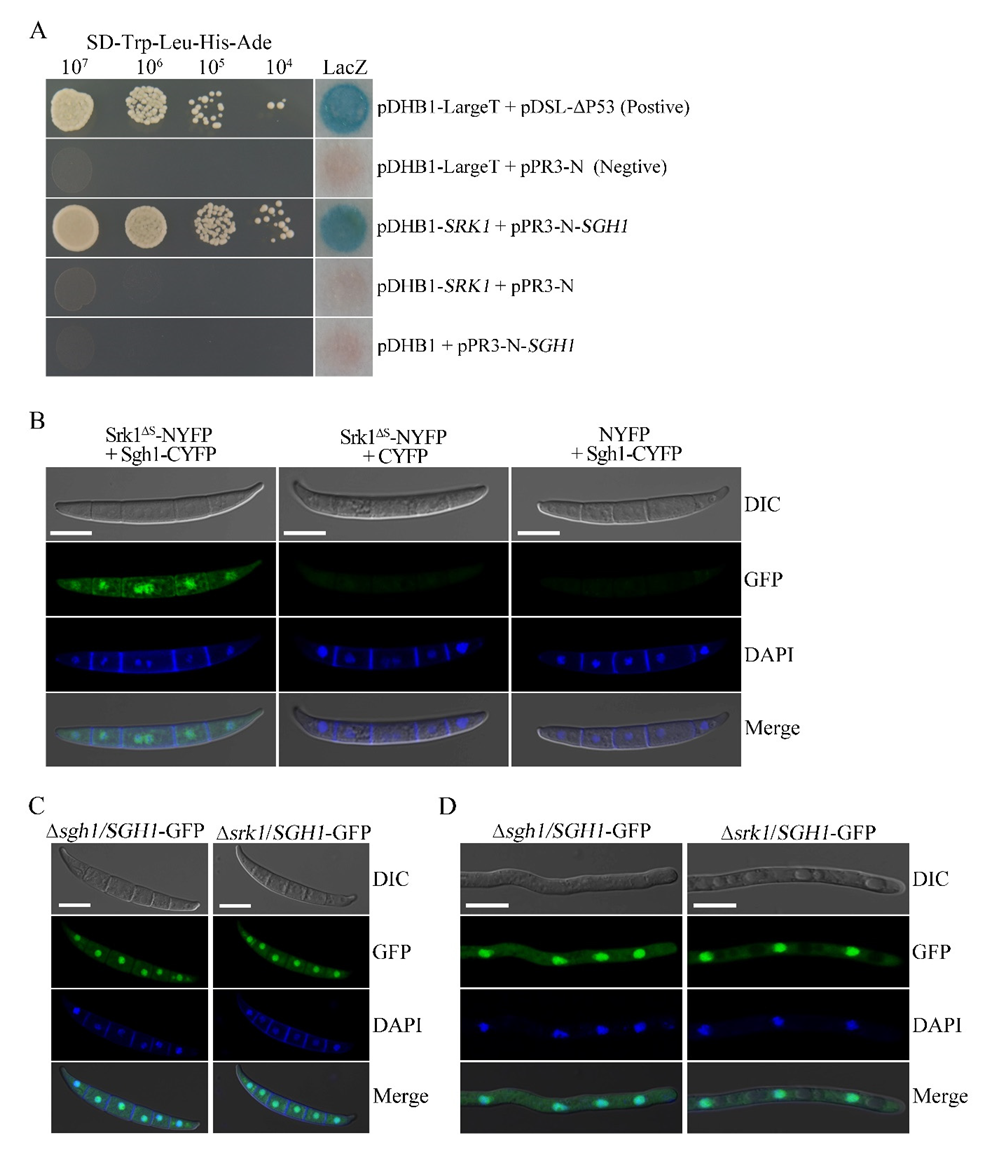
3.9. Sgh1 Physically Interacts with the SR Protein-Specific Kinase Srk1
3.10. Deletion of SRK1 Kinase Does Not Affect the Subcellular Localization of Sgh1-GFP
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goswami, R.S.; Kistler, H.C. Heading for Disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004, 5, 515–525. [Google Scholar] [CrossRef]
- Zhang, Y.; He, J.; Jia, L.-J.; Yuan, T.-L.; Zhang, D.; Guo, Y.; Wang, Y.; Tang, W.-H. Cellular tracking and gene profiling of Fusarium graminearum during maize stalk rot disease development elucidates its strategies in confronting phosphorus limitation in the host apoplast. PLoS Pathog. 2016, 12, e1005485. [Google Scholar] [CrossRef] [Green Version]
- Brown, D.W.; Dyer, R.B.; McCormick, S.P.; Kendra, D.F.; Plattner, R.D. Functional demarcation of the Fusarium core trichothecene gene cluster. Fungal Genet. Biol. 2004, 41, 454–462. [Google Scholar] [CrossRef] [PubMed]
- McMullen, M.; Jones, R.; Gallenberg, D. Scab of wheat and barley: A re-emerging disease of devastating Impact. Plant Dis. 1997, 81, 1340–1348. [Google Scholar] [CrossRef] [Green Version]
- Proctor, R.H.; Hohn, T.M.; McCormick, S.P. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant Microbe Interact. 1995, 8, 593–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maldonado-Ramirez, S.L.; Schmale, D.G., III; Shields, E.J.; Bergstrom, G.C. The relative abundance of viable spores of Gibberella zeae in the planetary boundary layer suggests the role of long-distance transport in regional epidemics of Fusarium head blight. Agric. For. Meteorol. 2005, 132, 20–27. [Google Scholar] [CrossRef]
- Trail, F.; Gaffoor, I.; Vogel, S. Ejection mechanics and trajectory of the ascospores of Gibberella zeae (anamorph Fusarium graminearum). Fungal Genet. Biol. 2005, 42, 528–533. [Google Scholar] [CrossRef]
- Boenisch, M.J.; Schäfer, W. Fusarium graminearum forms mycotoxin producing infection structures on wheat. BMC Plant Biol. 2011, 11, 110. [Google Scholar] [CrossRef] [Green Version]
- Brown, N.A.; Urban, M.; van de Meene, A.M.; Hammond-Kosack, K.E. The infection biology of Fusarium graminearum: Defining the pathways of spikelet to spikelet colonisation in wheat ears. Fungal Biol. 2010, 114, 555–571. [Google Scholar] [CrossRef]
- Will, C.L.; Lührmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3, a003707. [Google Scholar] [CrossRef] [PubMed]
- Wan, R.; Bai, R.; Zhan, X.; Shi, Y. How is precursor messenger RNA spliced by the spliceosome? Annu. Rev. Biochem. 2019, 89, 333–358. [Google Scholar] [CrossRef]
- Busch, A.; Hertel, K.J. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip. Rev. RNA 2012, 3, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David, C.J.; Manley, J.L. Alternative pre-mRNA splicing regulation in cancer: Pathways and programs unhinged. Genes Dev. 2010, 24, 2343–2364. [Google Scholar] [CrossRef] [Green Version]
- Cartegni, L.; Chew, S.L.; Krainer, A. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 2002, 3, 285–298. [Google Scholar] [CrossRef]
- Long, J.C.; Caceres, J.F. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2009, 417, 15–27. [Google Scholar] [CrossRef] [Green Version]
- Jeong, S. SR Proteins: Binders, regulators, and connectors of RNA. Mol. Cells 2017, 40, 613. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Fu, X.-D. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 2013, 122, 191–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Steitz, J.A. SRprises along a messenger’s journey. Mol. Cell 2005, 17, 613–615. [Google Scholar] [PubMed]
- Shepard, P.J.; Hertel, K.J. The SR protein family. Genome Biol. 2009, 10, 242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannakouros, T.; Nikolakaki, E.; Mylonis, I.; Georgatsou, E. Serine-arginine protein kinases: A small protein kinase family with a large cellular presence. FEBS J. 2011, 278, 570–586. [Google Scholar] [CrossRef]
- Plass, M.; Agirre, E.; Reyes, D.; Camara, F.; Eyras, E. Co-evolution of the branch site and SR proteins in eukaryotes. Trends Genet. 2008, 24, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Tsurumi, A.; Alaei, S.; Wilson, C.; Chiu, C.; Oya, J.; Ngo, B. Dsk1p kinase phosphorylates SR proteins and regulates their cellular localization in fission yeast. Biochem. J. 2007, 405, 21–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Gao, X.; Sun, M.; Liu, H.; Xu, J.R. The FgSRP1 SR-protein gene is important for plant infection and pre-mRNA processing in Fusarium graminearum. Environ. Microbiol. 2017, 19, 4065–4079. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dai, Y.; Huang, Y.; Wang, K.; Lu, P.; Xu, H.; Xu, J.R.; Liu, H. The SR-protein FgSrp2 regulates vegetative growth, sexual reproduction and pre-mRNA processing by interacting with FgSrp1 in Fusarium graminearum. Curr. Genet. 2020, 66, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Xue, C.; Peng, Y.; Katan, T.; Kistler, H.C.; Xu, J.R. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant Microbe Interact. 2002, 15, 1119–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Zhang, S.; Hou, R.; Zhao, Z.; Zheng, Q.; Xu, Q.; Zheng, D.; Wang, G.; Liu, H.; Gao, X.; et al. Functional Analysis of the Kinome of the Wheat Scab Fungus Fusarium graminearum. PLoS Pathog. 2011, 7, e1002460. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Sun, P.; Gong, Z.; Gu, L.; Lou, Y.; Fang, W.; Zhang, L.; Su, L.; Yang, T.; Wang, B.; et al. Srk1 kinase, a SR protein-specific kinase, is important for sexual reproduction, plant infection and pre-mRNA processing in Fusarium graminearum. Environ. Microbiol. 2018, 20, 3261–3277. [Google Scholar] [CrossRef]
- Ren, J.; Li, C.; Gao, C.; Xu, J.-R.; Jiang, C.; Wang, G. Deletion of FgHOG1 is suppressive to the mgv1 mutant by stimulating Gpmk1 activation and avoiding intracellular turgor elevation in Fusarium graminearum. Front. Microbiol. 2019, 10, 1073. [Google Scholar] [CrossRef] [Green Version]
- Catlett, N.L.; Lee, B.-N.; Yoder, O.C.; Turgeon, B. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. Rep. 2003, 50, 9–11. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Wang, C.; Hou, R.; Zhou, X.; Li, G.; Zhang, S.; Xu, J.-R. The AMT1 Arginine methyltransferase gene is important for plant infection and normal hyphal growth in Fusarium graminearum. PLoS ONE 2012, 7, e38324. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Heyer, C.; Choi, Y.-E.; Mehrabi, R.; Xu, J.-R. The CID1 cyclin C-like gene is important for plant infection in Fusarium graminearum. Fungal Genet. Biol. 2010, 47, 143–151. [Google Scholar] [CrossRef]
- Bluhm, B.H.; Zhao, X.; Flaherty, J.E.; Xu, J.R.; Dunkle, L.D. RAS2 regulates growth and pathogenesis in Fusarium graminearum. Mol. Plant Microbe Interact. 2007, 20, 627–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, C.; Cao, S.; Wang, Z.; Xu, H.; Liang, J.; Liu, H.; Wang, G.; Ding, M.; Wang, Q.; Gong, C.; et al. An expanded subfamily of G-protein-coupled receptor genes in Fusarium graminearum required for wheat infection. Nat. Microbiol. 2019, 4, 1582–1591. [Google Scholar] [CrossRef]
- Jiang, C.; Hei, R.; Yang, Y.; Zhang, S.; Wang, Q.; Wang, W.; Zhang, Q.; Yan, M.; Zhu, G.; Huang, P.; et al. An orphan protein of Fusarium graminearum modulates host immunity by mediating proteasomal degradation of TaSnRK1α. Nat. Commun. 2020, 11, 4382. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Lin, Y.; Fang, W.; Zhao, X.; Lou, Y.; Wang, G.; Zheng, H.; Liang, Q.; Abubakar, Y.S.; Olsson, S.; et al. The endosomal recycling of FgSnc1 by FgSnx41-FgSnx4 heterodimer is essential for polarized growth and pathogenicity in Fusarium graminearum. New Phytol. 2018, 219, 654–671. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Dimont, E.; Shi, J.; Kirchner, R.; Hide, W. edgeRun: An R package for sensitive, functionally relevant differential expression discovery using an unconditional exact test. Bioinformatics 2015, 31, 2589–2590. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Wang, Q.; He, Y.; Chen, L.; Hao, C.; Jiang, C.; Li, Y.; Dai, Y.; Kang, Z.; Xu, J.-R. Genome-wide A-to-I RNA editing in fungi independent of ADAR enzymes. Genome Res. 2016, 26, 499–509. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Melesse, M.; Zhang, S.; Hao, C.; Wang, C.; Zhang, H.; Hall, M.C.; Xu, J.R. FgCDC14 regulates cytokinesis, morphogenesis, and pathogenesis in Fusarium graminearum. Mol. Microbiol. 2015, 98, 770–786. [Google Scholar] [CrossRef]
- Liang, J.; Fu, X.; Hao, C.; Bian, Z.; Liu, H.; Xu, J.; Wang, G. FgBUD14 is important for ascosporogenesis and involves both stage-specific alternative splicing and RNA editing during sexual reproduction. Environ. Microbiol. 2021, 23, 5052–5068. [Google Scholar] [CrossRef] [PubMed]
- Ba, A.N.N.; Pogoutse, A.; Provart, N.; Moses, A.M. NLStradamus: A simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinform. 2009, 10, 202. [Google Scholar] [CrossRef] [Green Version]
- Cuomo, C.A.; Güldener, U.; Xu, J.-R.; Trail, F.; Turgeon, B.G.; Di Pietro, A.; Walton, J.D.; Ma, L.-J.; Baker, S.E.; Rep, M.; et al. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 2007, 317, 1400–1402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Liu, H.; Li, G.; Liu, M.; Yun, Y.; Wang, C.; Ma, Z.; Xu, J.-R. The MADS-box transcription factor FgMcm1 regulates cell identity and fungal development in Fusarium graminearum. Environ. Microbiol. 2015, 17, 2762–2776. [Google Scholar] [CrossRef] [PubMed]
- Son, H.; Seo, Y.-S.; Min, K.; Park, A.R.; Lee, J.; Jin, J.-M.; Lin, Y.; Cao, P.; Hong, S.-Y.; Kim, E.-K.; et al. A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum. PLoS Pathog. 2011, 7, e1002310. [Google Scholar] [CrossRef] [PubMed]
- Son, H.; Lee, J.; Park, A.R.; Lee, Y.-W. ATP citrate lyase is required for normal sexual and asexual development in Gibberella zeae. Fungal Genet. Biol. 2011, 48, 408–417. [Google Scholar] [CrossRef]
- Twyffels, L.; Gueydan, C.; Kruys, V. Shuttling SR proteins: More than splicing factors. FEBS J. 2011, 278, 3246–3255. [Google Scholar] [CrossRef]
- Gilbert, W.; Siebel, C.W.; Guthrie, C. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA 2001, 7, 302–313. [Google Scholar] [CrossRef] [Green Version]
- James, S.W.; Banta, T.; Barra, J.; Ciraku, L.; Coile, C.; Cuda, Z.; Day, R.; Dixit, C.; Eastlack, S.; Giang, A.; et al. Restraint of the G2/M transition by the SR/RRM family mRNA shuttling binding protein SNXAHRB1 in Aspergillus nidulans. Genetics 2014, 198, 617–633. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Jin, Q.; Jiang, C.; Li, Y.; Li, C.; Liu, H.; Kang, Z.; Xu, J.-R. FgPrp4 Kinase Is Important for Spliceosome B-Complex Activation and Splicing Efficiency in Fusarium graminearum. PLoS Genet. 2016, 12, e1005973. [Google Scholar] [CrossRef]
- Hackmann, A.; Wu, H.; Schneider, U.-M.; Meyer, K.; Jung, K.; Krebber, H. Quality control of spliced mRNAs requires the shuttling SR proteins Gbp2 and Hrb1. Nat. Commun. 2014, 5, 3123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Middleton, R.; Gao, D.; Thomas, A.; Singh, B.; Au, A.; Wong, J.J.-L.; Bomane, A.; Cosson, B.; Eyras, E.; Rasko, J.E.J.; et al. IRFinder: Assessing the impact of intron retention on mammalian gene expression. Genome Biol. 2017, 18, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Windgassen, M.; Sturm, D.; Cajigas, I.J.; González, C.I.; Seedorf, M.; Bastians, H.; Krebber, H. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol. Cell Biol. 2004, 24, 10479–10491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedley, M.L.; Amrein, H.; Maniatis, T. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc. Natl. Acad. Sci. USA 1995, 92, 11524–11528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aubol, B.E.; Plocinik, R.M.; Hagopian, J.C.; Ma, C.-T.; McGlone, M.L.; Bandyopadhyay, R.; Fu, X.-D.; Adams, J.A. Partitioning RS Domain Phosphorylation in an SR Protein through the CLK and SRPK Protein Kinases. J. Mol. Biol. 2013, 425, 2894–2909. [Google Scholar] [CrossRef] [Green Version]
- Maris, C.; Dominguez, C.; Allain, F.H.-T. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005, 272, 2118–2131. [Google Scholar] [CrossRef]
- Cléry, A.; Blatter, M.; Allain, F.H.-T. RNA recognition motifs: Boring? Not quite. Curr. Opin. Struct. Biol. 2008, 18, 290–298. [Google Scholar] [CrossRef]
- Martínez-Lumbreras, S.; Taverniti, V.; Zorrilla, S.; Séraphin, B.; Pérez-Cañadillas, J.M. Gbp2 interacts with THO/TREX through a novel type of RRM domain. Nucleic Acids Res. 2015, 44, 437–448. [Google Scholar] [CrossRef] [Green Version]
- Hurt, E.; Luo, M.-J.; Röther, S.; Reed, R.; Sträßer, K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc. Natl. Acad. Sci. USA 2004, 101, 1858–1862. [Google Scholar] [CrossRef] [Green Version]
- Porat, Z.; Erez, O.; Kahana, C. Cellular localization and phosphorylation of Hrb1p is independent of Sky1p. Biochim. Biophys. Acta 2006, 1763, 207–213. [Google Scholar] [CrossRef]
- Yun, C.Y.; Fu, X.-D. Conserved Sr Protein Kinase Functions in Nuclear Import and Its Action Is Counteracted by Arginine Methylation in Saccharomyces cerevisiae. J. Cell Biol. 2000, 150, 707–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariyachet, C.; Beißel, C.; Li, X.; Lorrey, S.; Mackenzie, O.; Martin, P.M.; O’Brien, K.; Pholcharee, T.; Sim, S.; Krebber, H.; et al. Post-translational modification directs nuclear and hyphal tip localization of Candida albicans mRNA-binding protein Slr1. Mol. Microbiol. 2017, 104, 499–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Côté, J.; Boisvert, F.-M.; Boulanger, M.-C.; Bedford, M.T.; Richard, S. Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1. Mol. Biol. Cell 2003, 14, 274–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, W.A.; Schurter, B.T.; Wong-Staal, F.; David, M. Arginine methylation of RNA helicase a determines its subcellular localization. J. Biol. Chem. 2004, 279, 22795–22798. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Sun, P.; Sun, Z.; Zhu, J.; Yu, D.; Tang, Z.; Wang, Z.; Wang, C.; Zheng, H. Sgh1, an SR-like Protein, Is Involved in Fungal Development, Plant Infection, and Pre-mRNA Processing in Fusarium graminearum. J. Fungi 2022, 8, 1056. https://doi.org/10.3390/jof8101056
Wang G, Sun P, Sun Z, Zhu J, Yu D, Tang Z, Wang Z, Wang C, Zheng H. Sgh1, an SR-like Protein, Is Involved in Fungal Development, Plant Infection, and Pre-mRNA Processing in Fusarium graminearum. Journal of Fungi. 2022; 8(10):1056. https://doi.org/10.3390/jof8101056
Chicago/Turabian StyleWang, Guanghui, Peng Sun, Zhongjuan Sun, Jindong Zhu, Dan Yu, Zhe Tang, Zonghua Wang, Chenfang Wang, and Huawei Zheng. 2022. "Sgh1, an SR-like Protein, Is Involved in Fungal Development, Plant Infection, and Pre-mRNA Processing in Fusarium graminearum" Journal of Fungi 8, no. 10: 1056. https://doi.org/10.3390/jof8101056







