Identification of a Bidirectional Promoter from Trichoderma reesei and Its Application in Dual Gene Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Construction of Plasmids and T. reesei Mutant Strains
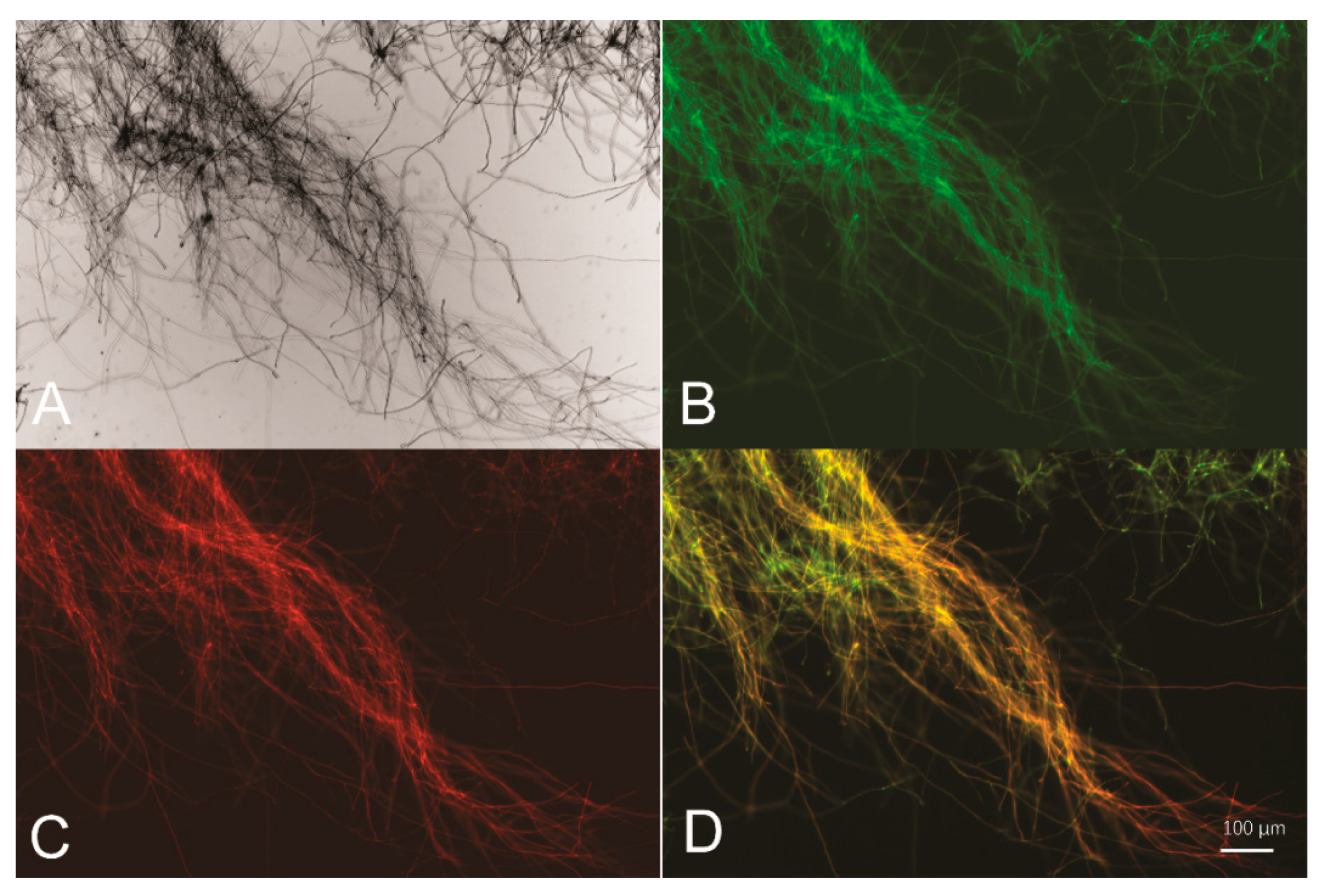
2.3. Fluorescence Microscopic Analysis
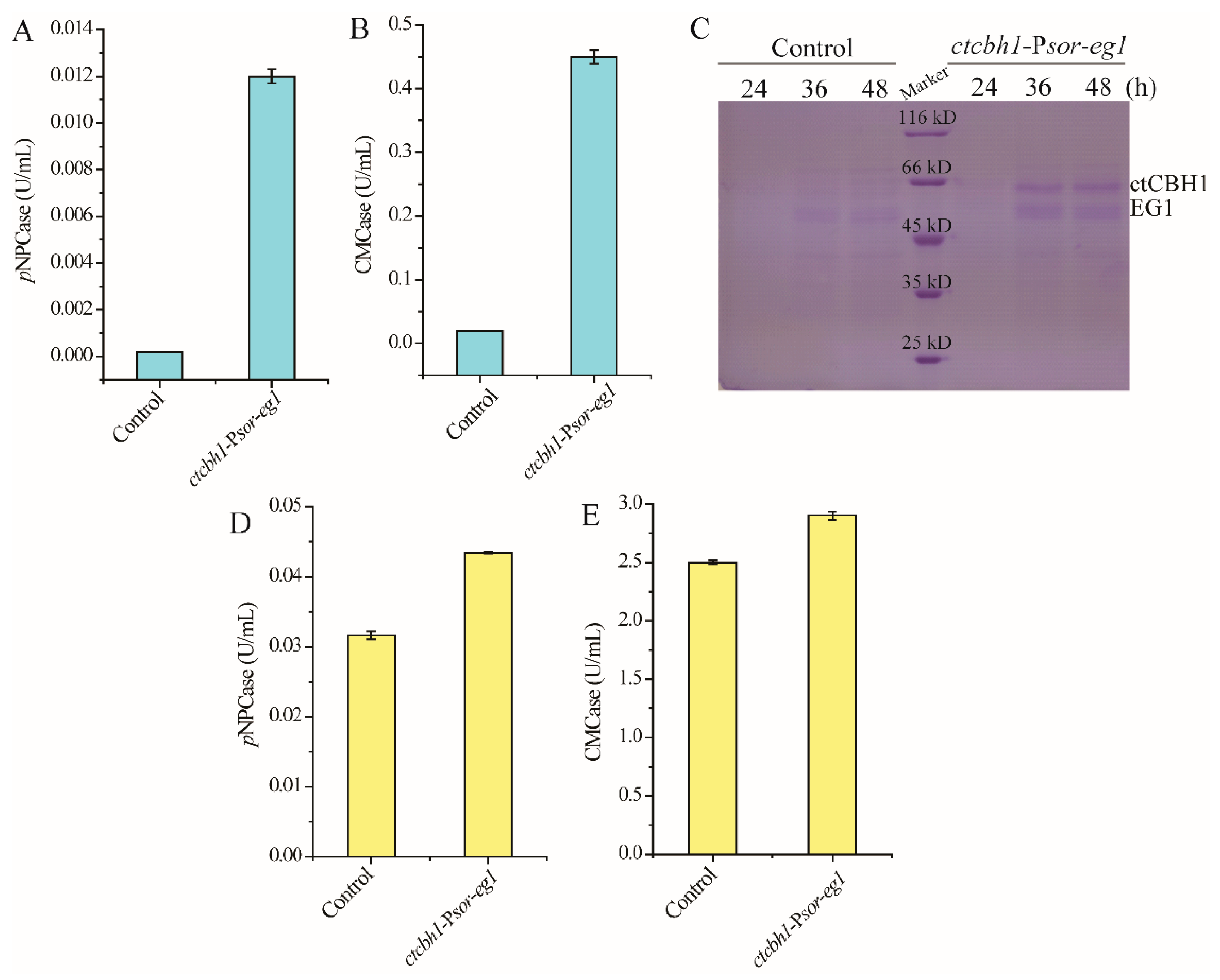
2.4. Enzyme Activity and Protein Analyses
2.5. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
2.6. Sequence Analysis
3. Results
3.1. The Intergenic Region between sor1 and sor2 Functions as a Bidirectional Promoter in T. reesei
3.2. Quantitative Determination of Promoter Strength of Psor
3.3. Co-Expression of Two Cellulase Genes Using Psor
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bischof, R.H.; Ramoni, J.; Seiboth, B. Cellulases and beyond: The first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell Fact. 2016, 15, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Taylor, L.E., II; Vander Wall, T.A.; Linger, J.; Himmel, M.E.; Podkaminer, K.; Adney, W.S.; Decker, S.R. Heterologous protein expression in Hypocrea jecorina: A historical perspective and new developments. Biotechnol. Adv. 2015, 33, 142–154. [Google Scholar] [CrossRef] [Green Version]
- Shenouda, M.L.; Ambilika, M.; Skellam, E.; Cox, R.J. Heterologous Expression of Secondary Metabolite Genes in Trichoderma reesei for Waste Valorization. J. Fungi 2022, 8, 355. [Google Scholar] [CrossRef]
- Fitz, E.; Wanka, F.; Seiboth, B. The Promoter Toolbox for Recombinant Gene Expression in Trichoderma reesei. Front. Bioeng. Biotechnol. 2018, 6, 135. [Google Scholar] [CrossRef]
- Nyyssonen, E.; Keranen, S. Multiple roles of the cellulase CBHI in enhancing production of fusion antibodies by the filamentous fungus Trichoderma reesei. Curr. Genet. 1995, 28, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Zheng, F.; Li, C.; Zhang, W.; Chen, G.; Liu, W. Characterization of a copper responsive promoter and its mediated overexpression of the xylanase regulator 1 results in an induction-independent production of cellulases in Trichoderma reesei. Biotechnol. Biofuels 2015, 8, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, W.; Pelechano, V.; Jarvelin, A.I.; Steinmetz, L.M. Functional consequences of bidirectional promoters. Trends Genet. 2011, 27, 267–276. [Google Scholar] [CrossRef] [Green Version]
- Dash, A.; Gurdaswani, V.; D’Souza, J.S.; Ghag, S.B. Functional characterization of an inducible bidirectional promoter from Fusarium oxysporum f. sp. cubense. Sci. Rep. 2020, 10, 2323. [Google Scholar] [CrossRef] [Green Version]
- Bergh, K.T.; Litzka, O.; Brakhage, A.A. Identification of a major cis-acting DNA element controlling the bidirectionally transcribed penicillin biosynthesis genes acvA (pcbAB) and ipnA (pcbC) of Aspergillus nidulans. J. Bacteriol. 1996, 178, 3908–3916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, J.F. Molecular control of expression of penicillin biosynthesis genes in fungi: Regulatory proteins interact with a bidirectional promoter region. J. Bacteriol. 2000, 182, 2355–2362. [Google Scholar] [CrossRef] [PubMed]
- Muro-Pastor, M.I.; Strauss, J.; Ramon, A.; Scazzocchio, C. A paradoxical mutant GATA factor. Eukaryot. Cell 2004, 3, 393–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiemann, P.; Soukup, A.A.; Folz, J.S.; Wang, P.M.; Noack, A.; Keller, N.P. CoIN: Co-inducible nitrate expression system for secondary metabolites in Aspergillus nidulans. Fungal. Biol. Biotechnol. 2018, 5, 6. [Google Scholar] [CrossRef] [Green Version]
- Rendsvig, J.K.H.; Workman, C.T.; Hoof, J.B. Bidirectional histone-gene promoters in Aspergillus: Characterization and application for multi-gene expression. Fungal. Biol. Biotechnol. 2019, 6, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derntl, C.; Rassinger, A.; Srebotnik, E.; Mach, R.L.; Mach-Aigner, A.R. Identification of the Main Regulator Responsible for Synthesis of the Typical Yellow Pigment Produced by Trichoderma reesei. Appl. Environ. Microbiol. 2016, 82, 6247–6257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; An, N.; Guo, J.; Wang, Z.; Meng, X.; Liu, W. Influences of genetically perturbing synthesis of the typical yellow pigment on conidiation, cell wall integrity, stress tolerance, and cellulase production in Trichoderma reesei. J. Microbiol. 2021, 59, 426–434. [Google Scholar] [CrossRef]
- Wang, L.; Lv, X.; Cao, Y.; Zheng, F.; Meng, X.; Shen, Y.; Chen, G.; Liu, W.; Zhang, W. A novel transcriptional regulator RXE1 modulates the essential transactivator XYR1 and cellulase gene expression in Trichoderma reesei. Appl. Microbiol. Biotechnol. 2019, 103, 4511–4523. [Google Scholar] [CrossRef]
- Mandels, M.; Parrish, F.W.; Reese, E.T. Sophorose as an inducer of cellulase in Trichoderma viride. J. Bacteriol. 1962, 83, 400–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Yang, R.; Cao, Y.; Zheng, F.; Meng, X.; Zhong, Y.; Chen, G.; Zhang, W.; Liu, W. CLP1, a Novel Plant Homeo Domain Protein, Participates in Regulating Cellulase Gene Expression in the Filamentous Fungus Trichoderma reesei. Front. Microbiol. 2019, 10, 1700. [Google Scholar] [CrossRef] [Green Version]
- Heckman, K.L.; Pease, L.R. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2007, 2, 924–932. [Google Scholar] [CrossRef]
- Zhang, S.; Fan, Y.; Xia, Y.X.; Keyhani, N.O. Sulfonylurea resistance as a new selectable marker for the entomopathogenic fungus Beauveria bassiana. Appl. Microbiol. Biotechnol. 2010, 87, 1151–1156. [Google Scholar] [CrossRef]
- Penttila, M.; Nevalainen, H.; Ratto, M.; Salminen, E.; Knowles, J. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 1987, 61, 155–164. [Google Scholar] [CrossRef]
- Bailey, M.J.; Biely, P.; Poutanen, K. Interlaboratory Testing of Methods for Assay of Xylanase Activity. J. Biotechnol. 1992, 23, 257–270. [Google Scholar] [CrossRef]
- Scruggs, B.S.; Gilchrist, D.A.; Nechaev, S.; Muse, G.W.; Burkholder, A.; Fargo, D.C.; Adelman, K. Bidirectional Transcription Arises from Two Distinct Hubs of Transcription Factor Binding and Active Chromatin. Mol. Cell 2015, 58, 1101–1112. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Du, J.; He, R.; Zhang, Z.; Qi, F.; Huang, J.; Qin, L. Improved Production of Majority Cellulases in Trichoderma reesei by Integration of cbh1 Gene from Chaetomium thermophilum. Front. Microbiol. 2020, 11, 1633. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Li, F.; Yang, R.; Meng, X.; Zhang, W.; Liu, W. Identification of a Bidirectional Promoter from Trichoderma reesei and Its Application in Dual Gene Expression. J. Fungi 2022, 8, 1059. https://doi.org/10.3390/jof8101059
Wu X, Li F, Yang R, Meng X, Zhang W, Liu W. Identification of a Bidirectional Promoter from Trichoderma reesei and Its Application in Dual Gene Expression. Journal of Fungi. 2022; 8(10):1059. https://doi.org/10.3390/jof8101059
Chicago/Turabian StyleWu, Xiaoxiao, Fuzhe Li, Renfei Yang, Xiangfeng Meng, Weixin Zhang, and Weifeng Liu. 2022. "Identification of a Bidirectional Promoter from Trichoderma reesei and Its Application in Dual Gene Expression" Journal of Fungi 8, no. 10: 1059. https://doi.org/10.3390/jof8101059
APA StyleWu, X., Li, F., Yang, R., Meng, X., Zhang, W., & Liu, W. (2022). Identification of a Bidirectional Promoter from Trichoderma reesei and Its Application in Dual Gene Expression. Journal of Fungi, 8(10), 1059. https://doi.org/10.3390/jof8101059





