Identification of Key Regulatory Pathways of Basidiocarp Formation in Pleurotus spp. Using Modeling, Simulation and System Biology Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Development of Model
2.2. Kinetic Rate Reaction Calculation and Model Simulation
2.3. Network Analysis
3. Results
3.1. Dynamic Behavior Studies and Model Simulations
3.2. Topological Analysis of Fruit Body Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Royse, D.J.; Baars, J.; Tan, Q. Current overview of mushroom production in the world. In Edible and Medicinal Mushrooms: Technology and Applications; Zied, D.C., Pardo-Giménez, A., Eds.; John Wiley & Sons Ltd.: New York, NY, USA, 2017; pp. 5–13. [Google Scholar]
- Singh, M.; Kamal, S.; Sharma, V.P. Species and Region-wise Mushroom Production in Leading Mushroom Producing Countries—China, Japan, USA, Canada and India. Mushroom Res. 2021, 30, 99–108. [Google Scholar] [CrossRef]
- Reis, F.S.; Barros, L.; Martins, A.; Ferreira, I.C.F.R. Chemical composition and nutritional value of the most widely appreciated cultivated mushrooms: An inter-species comparative study. Food Chem. Toxicol. 2012, 50, 191–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopes, R.D.O.; Pereira, P.M.; Pereira, A.R.B.; Fernandes, K.V.; Carvalho, J.F.; da Silva de França, A.; Valente, R.H.; da Silva, M.; Ferreira-Leitão, V.S. Atrazine, desethylatrazine (DEA) and desisopropylatrazine (DIA) degradation by Pleurotus ostreatus INCQS 40310. Biocatal. Biotransform. 2020, 38, 415–430. [Google Scholar] [CrossRef]
- Pozdnyakova, N.N.; Jarosz-Wilkolazka, A.; Polak, J.; Grąz, M.; Turkovskaya, O.V. Decolourisation of anthraquinone-and anthracene-type dyes by versatile peroxidases from Bjerkandera fumosa and Pleurotus ostreatus D1. Biocatal. Biotransform. 2015, 33, 69–80. [Google Scholar] [CrossRef]
- Wu, Y.; Choi, M.-H.; Li, J.; Yang, H.; Shin, H.-J. Mushroom Cosmetics: The Present and Future. Cosmetics 2016, 3, 22. [Google Scholar] [CrossRef]
- Singh, M.; Kamal, S. Genetic Aspects and Strategies for Obtaining Hybrids. In Edible and Medicinal Mushrooms; John Wiley & Sons, Ltd.: Chichester, UK, 2017; pp. 35–87. ISBN 9781119149446. [Google Scholar]
- Wösten, H.A.B.; Wessels, J.G.H. The Emergence of Fruiting Bodies in Basidiomycetes. In Growth, Differentiation and Sexuality; Springer: Berlin/Heidelberg, Germany, 2006; pp. 393–414. [Google Scholar]
- Stamets, P. Growing Gourmet and Medicinal Mushrooms, 3rd ed.; Ten speed press: Toronto, ON, Canada, 2000. [Google Scholar]
- Zhang, J.; Ren, A.; Chen, H.; Zhao, M.; Shi, L.; Chen, M.; Wang, H.; Feng, Z. Transcriptome Analysis and Its Application in Identifying Genes Associated with Fruiting Body Development in Basidiomycete Hypsizygus marmoreus. PLoS ONE 2015, 10, e0123025. [Google Scholar] [CrossRef] [Green Version]
- Wessels, J.G.H. Tansley Review No. 45 Wall growth, protein excretion and morphogenesis in fungi. New Phytol. 1993, 123, 397–413. [Google Scholar] [CrossRef]
- Barh, A.; Sharma, V.P.; Annepu, S.K.; Kamal, S.; Sharma, S.; Bhatt, P. Genetic improvement in Pleurotus (oyster mushroom): A review. 3 Biotech 2019, 9, 322. [Google Scholar] [CrossRef]
- Han, C.-H.; Zhang, G.-Q.; Wang, H.-X.; Ng, T.B. Schizolysin, a hemolysin from the split gill mushroom Schizophyllum commune. FEMS Microbiol. Lett. 2010, 309, 115–121. [Google Scholar] [CrossRef]
- Kamada, T.; Sano, H.; Nakazawa, T.; Nakahori, K. Regulation of fruiting body photomorphogenesis in Coprinopsis cinerea. Fungal Genet. Biol. 2010, 47, 917–921. [Google Scholar] [CrossRef]
- Herzog, R.; Solovyeva, I.; Bölker, M.; Lugones, L.G.; Hennicke, F. Exploring molecular tools for transformation and gene expression in the cultivated edible mushroom Agrocybe aegerita. Mol. Genet. Genom. 2019, 294, 663–677. [Google Scholar] [CrossRef]
- Krizsán, K.; Almási, É.; Merényi, Z.; Sahu, N.; Virágh, M.; Kószó, T.; Mondo, S.; Kiss, B.; Bálint, B.; Kües, U.; et al. Transcriptomic atlas of mushroom development reveals conserved genes behind complex multicellularity in fungi. Proc. Natl. Acad. Sci. USA 2019, 116, 7409–7418. [Google Scholar] [CrossRef] [Green Version]
- Pelkmans, J.F.; Lugones, L.G.; Wösten, H.A.B. Fruiting Body Formation in Basidiomycetes. In Growth, Differentiation and Sexuality; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 387–405. [Google Scholar]
- Berne, S.; Križaj, I.; Pohleven, F.; Turk, T.; Maček, P.; Sepčić, K. Pleurotus and Agrocybe hemolysins, new proteins hypothetically involved in fungal fruiting. Biochim. Biophys. Acta-Gen. Subj. 2002, 1570, 153–159. [Google Scholar] [CrossRef]
- Peñas, M.M.; Ásgeirsdóttir, S.A.; Lasa, I.; Culiañez-Macià, F.A.; Pisabarro, A.G.; Wessels, J.G.H.; Ramírez, L. Identification, characterization, and in situ detection of a fruit-body-specific hydrophobin of Pleurotus ostreatus. Appl. Environ. Microbiol. 1998, 64, 4028–4034. [Google Scholar] [CrossRef] [Green Version]
- Muraguchi, H.; Kamada, T. A mutation in the eln2 gene encoding a cytochrome P450 of Coprinus cinereus affects mushroom morphogenesis. Fungal Genet. Biol. 2000, 29, 49–59. [Google Scholar] [CrossRef]
- Suzuki, H.; Vuong, T.V.; Gong, Y.; Chan, K.; Ho, C.Y.; Master, E.R.; Kondo, A. Sequence diversity and gene expression analyses of expansin-related proteins in the white-rot basidiomycete, Phanerochaete carnosa. Fungal Genet. Biol. 2014, 72, 115–123. [Google Scholar] [CrossRef]
- Bhatt, P.; Barh, A. Bioinformatic Tools to Study the Soil Microorganisms: An In Silico Approach for Sustainable Agriculture. In In Silico Approach for Sustainable Agriculture; Springer: Singapore, 2018; pp. 169–182. [Google Scholar]
- Funahashi, A.; Morohashi, M.; Matsuoka, Y.; Jouraku, A.; Kitano, H. CellDesigner: A graphical biological network editor and workbench interfacing simulator. In Introduction to Systems Biology; Humana Press: Totowa, NJ, USA, 2007; pp. 422–434. ISBN 9781588297068. [Google Scholar]
- Hucka, M.; Bergmann, F.T.; Dräger, A.; Hoops, S.; Keating, S.M.; Le Novère, N.; Myers, C.J.; Olivier, B.G.; Sahle, S.; Schaff, J.C.; et al. The Systems Biology Markup Language (SBML): Language Specification for Level 3 Version 2 Core. J. Integr. Bioinform. 2018, 15, 20170081. [Google Scholar] [CrossRef]
- Pathak, R.K.; Baunthiyal, M.; Pandey, N.; Pandey, D.; Kumar, A. Modeling of the jasmonate signaling pathway in Arabidopsis thaliana with respect to pathophysiology of Alternaria blight in Brassica. Sci. Rep. 2017, 7, 16790. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, P.; Sethi, K.; Gangola, S.; Bhandari, G.; Verma, A.; Adnan, M.; Singh, Y.; Chaube, S. Modeling and simulation of atrazine biodegradation in bacteria and its effect in other living systems. J. Biomol. Struct. Dyn. 2020, 40, 3285–3295. [Google Scholar] [CrossRef]
- Machné, R.; Finney, A.; Müller, S.; Lu, J.; Widder, S.; Flamm, C. The SBML ODE Solver Library: A native API for symbolic and fast numerical analysis of reaction networks. Bioinformatics 2006, 22, 1406–1407. [Google Scholar] [CrossRef] [Green Version]
- Assenov, Y.; Ramírez, F.; Schelhorn, S.-E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinform. Appl. 2008, 24, 282–284. [Google Scholar] [CrossRef] [Green Version]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological Network Exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Zinovyev, A.; Viara, E.; Calzone, L.; Barillot, E. BiNoM: A Cytoscape plugin for manipulating and analyzing biological networks. Bioinformatics 2008, 24, 876–877. [Google Scholar] [CrossRef] [Green Version]
- Suderman, M.; Hallett, M. Tools for visually exploring biological networks. Bioinformatics 2007, 23, 2651–2659. [Google Scholar] [CrossRef]
- Bhatt, P.; Pal, K.; Bhandari, G.; Barh, A. Modelling of the methyl halide biodegradation in bacteria and its effect on environmental systems. Pestic. Biochem. Physiol. 2019, 158, 88–100. [Google Scholar] [CrossRef]
- Max Planck Institute for Informatics NetworkAnalyzer. Available online: https://med.bioinf.mpi-inf.mpg.de/netanalyzer/help/2.5/#refBarabasi04 (accessed on 18 January 2021).
- Maslov, S.; Sneppen, K. Specificity and stability in topology of protein networks. Science 2002, 296, 910–913. [Google Scholar] [CrossRef] [Green Version]
- Kohl, M.; Wiese, S.; Warscheid, B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol. Biol. 2011, 696, 291–303. [Google Scholar] [CrossRef]
- Barabási, A.L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef]
- Chun, S.C.; Muthu, M.; Hasan, N.; Tasneem, S.; Gopal, J. Mycoremediation of PCBs by Pleurotus ostreatus: Possibilities and Prospects. Appl. Sci. 2019, 9, 4185. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Kim, B.G.; Kim, K.J.; Lee, J.S.; Yun, D.W.; Hahn, J.H.; Kim, G.H.; Lee, K.H.; Suh, D.S.; Kwon, S.T.; et al. Comparative analysis of sequences expressed during the liquid-cultured mycelia and fruit body stages of Pleurotus ostreatus. Fungal Genet. Biol. 2002, 35, 115–134. [Google Scholar] [CrossRef]
- Berne, S.; Pohleven, J.; Vidic, I.; Rebolj, K.; Pohleven, F.; Turk, T.; Ek, P.M.; Sonnenberg, A.; Sepčić, K.; Eastwood, D.C. Ostreolysin enhances fruiting initiation in the oyster mushroom (Pleurotus ostreatus). Mycol. Res. 2007, 111, 1431–1436. [Google Scholar] [CrossRef] [PubMed]
- Wösten, H.A.B.; Wessels, J.G.H. Hydrophobins, from molecular structure to multiple functions in fungal development. Mycoscience 1997, 38, 363–374. [Google Scholar] [CrossRef]
- Lugones, L.; Wosten, H.B.; Wessels, J.H. A hydrophobin (ABH3) specifically secreted by vegetatively growing hyphae of Agaricus bisporus (common white button mushroom). Microbiology 1998, 144, 2345–2353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peñas, M.M.; Aranguren, J.; Ramírez, L.; Pisabarro, A.G. Structure of gene coding for the fruit body-specific hydrophobia Fbh1 of the edible basidiomycete Pleurotus ostreatus. Mycologia 2004, 96, 75–82. [Google Scholar] [CrossRef]
- Peñas, M.M.; Rust, B.; Larraya, L.M.; Ramírez, L.; Pisabarro, A.G. Differentially regulated, vegetative-mycelium-specific hydrophobins of the edible basidiomycete Pleurotus ostreatus. Appl. Environ. Microbiol. 2002, 68, 3891–3898. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.; Chen, H.; Zhang, M.; Wen, Q.; Qiu, L.; Shen, J. Identification and expression analysis of Pofst3 suggests a role during Pleurotus ostreatus primordia formation. Fungal Biol. 2019, 123, 200–208. [Google Scholar] [CrossRef]
- Wang, H.; Tong, X.; Tian, F.; Jia, C.; Li, C.; Li, Y. Transcriptomic profiling sheds light on the blue-light and red-light response of oyster mushroom (Pleurotus ostreatus). AMB Express 2020, 10, 10. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Gong, W.; Zhu, Z.; Yan, L.; Hu, Z.; Peng, Y. Comparative transcriptomics of Pleurotus eryngii reveals blue-light regulation of carbohydrate-active enzymes (CAZymes) expression at primordium differentiated into fruiting body stage. Genomics 2018, 110, 201–209. [Google Scholar] [CrossRef]
- Lettera, V.; Piscitelli, A.; Leo, G.; Birolo, L.; Pezzella, C.; Sannia, G. Identification of a new member of Pleurotus ostreatus laccase family from mature fruiting body. Fungal Biol. 2010, 114, 724–730. [Google Scholar] [CrossRef]
- Liu, X.B.; Xia, E.H.; Li, M.; Cui, Y.Y.; Wang, P.M.; Zhang, J.X.; Xie, B.G.; Xu, J.P.; Yan, J.J.; Li, J.; et al. Transcriptome data reveal conserved patterns of fruiting body development and response to heat stress in the mushroomforming fungus Flammulina filiformis. PLoS ONE 2020, 15, e0239890. [Google Scholar] [CrossRef]
- Du, F.; Zou, Y.; Hu, Q.; Zhang, H.; Ye, D. Comparative transcriptomic analysis reveals molecular processes involved in pileus morphogenesis in Pleurotus eryngii under different light conditions. Genomics 2020, 112, 1707–1715. [Google Scholar] [CrossRef]
- Ohm, R.A.; De Jong, J.F.; Lugones, L.G.; Aerts, A.; Kothe, E.; Stajich, J.E.; De Vries, R.P.; Record, E.; Levasseur, A.; Baker, S.E.; et al. Genome sequence of the model mushroom Schizophyllum commune. Nat. Biotechnol. 2010, 28, 957–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muraguchi, H.; Umezawa, K.; Niikura, M.; Yoshida, M.; Kozaki, T.; Ishii, K.; Sakai, K.; Shimizu, M.; Nakahori, K.; Sakamoto, Y.; et al. Strand-Specific RNA-Seq Analyses of Fruiting Body Development in Coprinopsis cinerea. PLoS ONE 2015, 10, e0141586. [Google Scholar] [CrossRef] [Green Version]
- Varga, T.; Krizsán, K.; Földi, C.; Dima, B.; Sánchez-García, M.; Sánchez-Ramírez, S.; Szöllősi, G.J.; Szarkándi, J.G.; Papp, V.; Albert, L.; et al. Megaphylogeny resolves global patterns of mushroom evolution. Nat. Ecol. Evol. 2019, 3, 668–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ota, K.; Butala, M.; Viero, G.; Serra, M.D.; Sepčić, K.; Maček, P. Fungal MACPF-like proteins and aegerolysins: Bi-component pore-forming proteins? In MACPF/CDC Proteins—Agents of Defence, Attack and Invasion; Anderluh, G., Gilbert, R., Eds.; Springer Dordrecht: Berlin/Heidelberg, Germany, 2014; Volume 80, pp. 271–291. [Google Scholar]
- Tomita, T.; Noguchi, K.; Mimuro, H.; Ukaji, F.; Ito, K.; Sugawara-Tomita, N.; Hashimoto, Y. Pleurotolysin, a novel sphingomyelin-specific two-component cytolysin from the edible mushroom Pleurotus ostreatus, assembles into a transmembrane pore complex. J. Biol. Chem. 2004, 279, 26975–26982. [Google Scholar] [CrossRef] [Green Version]
- Sepcic, K.; Berne, S.; Potrich, C.; Turk, T.; Macek, P.; Menestrina, G. Interaction of ostreolysin, a cytolytic protein from the edible mushroom Pleurotus ostreatus, with lipid membranes and modulation by lysophospholipids. Eur. J. Biochem. 2003, 270, 1199–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurahashi, A.; Sato, M.; Kobayashi, T.; Nishibori, K.; Fujimori, F. Homologous genes, Pe. pleurotolysin A and Pe. ostreolysin, are both specifically and highly expressed in primordia and young fruiting bodies of Pleurotus eryngii. Mycoscience 2014, 55, 113–117. [Google Scholar] [CrossRef]
- Kulkarni, S.S.; Nene, S.N.; Joshi, K.S. A comparative study of production of hydrophobin like proteins (HYD-LPs) in submerged liquid and solid state fermentation from white rot fungus Pleurotus ostreatus. Biocatal. Agric. Biotechnol. 2020, 23, 101440. [Google Scholar] [CrossRef]
- Schuren, F.H.J.; Wessels, J.G.H. Two genes specifically expressed in fruiting dikaryons of Schizophyllum commune: Homologies with a gene not regulated by mating-type genes. Gene 1990, 90, 199–205. [Google Scholar] [CrossRef]
- Kershaw, M.J.; Talbot, N.J. Hydrophobins and Repellents: Proteins with Fundamental Roles in Fungal Morphogenesis. Fungal Genet. Biol. 1998, 23, 18–33. [Google Scholar] [CrossRef]
- Ásgeirsdóttir, S.A.; De Vries, O.M.H.; Wessels, J.G.H. Identification of three differentially expressed hydrophobins in Pleurotus ostreatus (oyster mushroom). Microbiology 1998, 144, 2961–2969. [Google Scholar] [CrossRef] [Green Version]
- Van Wetter, M.A.; Wösten, H.A.B.; Wessels, J.G.H. SC3 and SC4 hydrophobins have distinct roles in formation of aerial structures in dikaryons of Schizophyllum commune. Mol. Microbiol. 2000, 36, 201–210. [Google Scholar] [CrossRef]
- De Groot, P.W.J.; Schaap, P.J.; Sonnenberg, A.S.M.; Visser, J.; Van Griensven, L.J.L.D. The Agaricus bisporus hypA gene encodes a hydrophobin and specifically accumulates in peel tissue of mushroom caps during fruit body development. J. Mol. Biol. 1996, 257, 1008–1018. [Google Scholar] [CrossRef]
- Chakraborty, T.K.; Das, N.; Mukherjee, M. Evidences of high carbon catabolic enzyme activities during sporulation of Pleurotus ostreatus (Florida). J. Basic Microbiol. 2003, 43, 462–467. [Google Scholar] [CrossRef]
- Schwalb, M.N.; Shanler, A. Phototropic and geotropic responses during the development of normal and mutant fruit bodies of the basidiomycete Schizophyllum commune. J. Gen. Microbiol. 1974, 82, 209–212. [Google Scholar] [CrossRef] [Green Version]
- Kojima, M.; Kimura, N.; Miura, R. Regulation of primary metabolic pathways in oyster mushroom mycelia induced by blue light stimulation: Accumulation of shikimic acid. Sci. Rep. 2015, 5, 8630. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; Ma, F.; Zhang, X.; Zhang, J. Carbohydrate changes during growth and fruiting in Pleurotus ostreatus. Fungal Biol. 2016, 120, 852–861. [Google Scholar] [CrossRef]
- Lee, S.; Tak, E.; Lee, J.; Rashid, M.; Murphy, M.P.; Ha, J.; Kim, S.S. Mitochondrial H2O2 generated from electron transport chain complex i stimulates muscle differentiation. Cell Res. 2011, 21, 817–834. [Google Scholar] [CrossRef] [Green Version]
- Jiao, X.; Li, G.; Wang, Y.; Nie, F.; Cheng, X.; Abdullah, M.; Lin, Y.; Cai, Y. Systematic analysis of the Pleurotus ostreatus laccase gene (PoLac) Family and functional characterization of PoLac2 involved in the degradation of cotton-straw lignin. Molecules 2018, 23, 880. [Google Scholar] [CrossRef] [Green Version]
- Barh, A.; Kumari, B.; Sharma, S.; Annepu, S.K.; Kumar, A.; Kamal, S.; Sharma, V.P. Mushroom mycoremediation: Kinetics and mechanism. In Smart Bioremediation Technologies: Microbial Enzymes; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–22. ISBN 9780128183076. [Google Scholar]
- Park, M.; Kim, M.; Kim, S.; Ha, B.; Ro, H.-S. Differential Expression of Laccase Genes in Pleurotus ostreatus and Biochemical Characterization of Laccase Isozymes Produced in Pichia pastoris. Mycobiology 2015, 43, 280–287. [Google Scholar] [CrossRef]
- Pezzella, C.; Autore, F.; Giardina, P.; Piscitelli, A.; Sannia, G.; Faraco, V. The Pleurotus ostreatus laccase multi-gene family: Isolation and heterologous expression of new family members. Curr. Genet. 2009, 55, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Zhang, M.; Qu, J.; Zhang, J. iTRAQ-Based Quantitative Proteomic Analysis Reveals Proteomic Changes in Mycelium of Pleurotus ostreatus in Response to Heat Stress and Subsequent Recovery. Front. Microbiol. 2018, 9, 2368. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Wang, L.; Wu, X.; Gao, W.; Zhang, J.; Huang, C. Expression patterns of two pal genes of Pleurotus ostreatus across developmental stages and under heat stress. BMC Microbiol. 2019, 19, 231. [Google Scholar] [CrossRef] [PubMed]


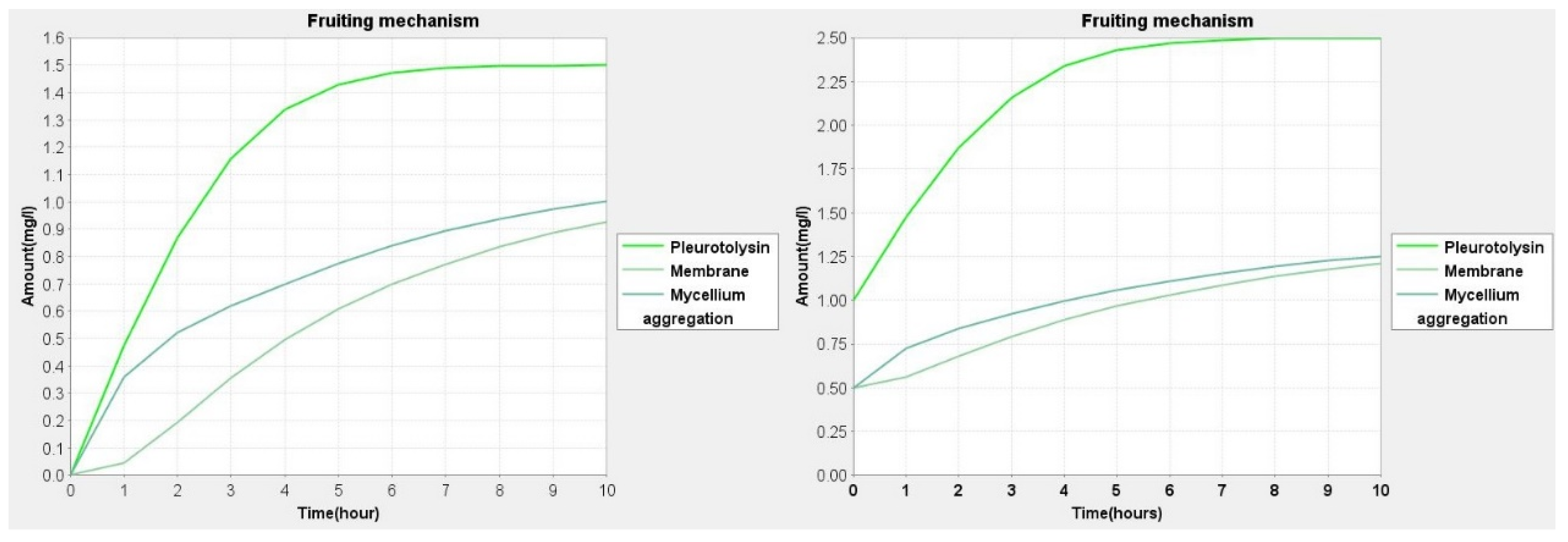

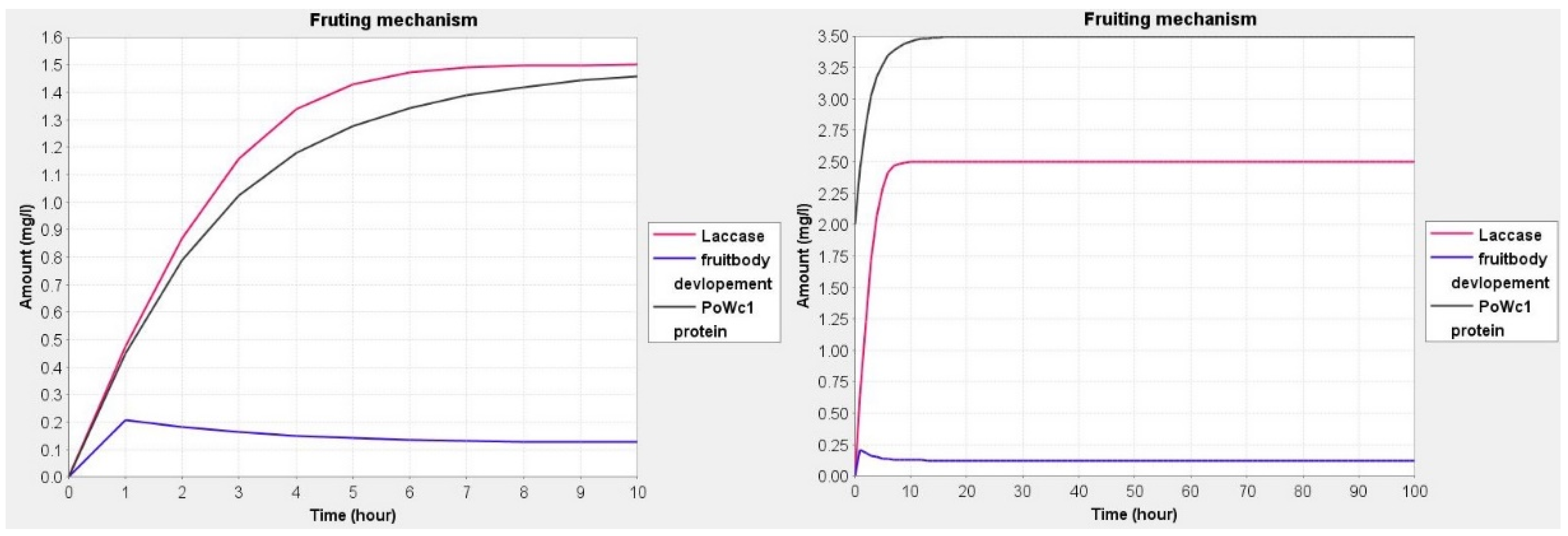
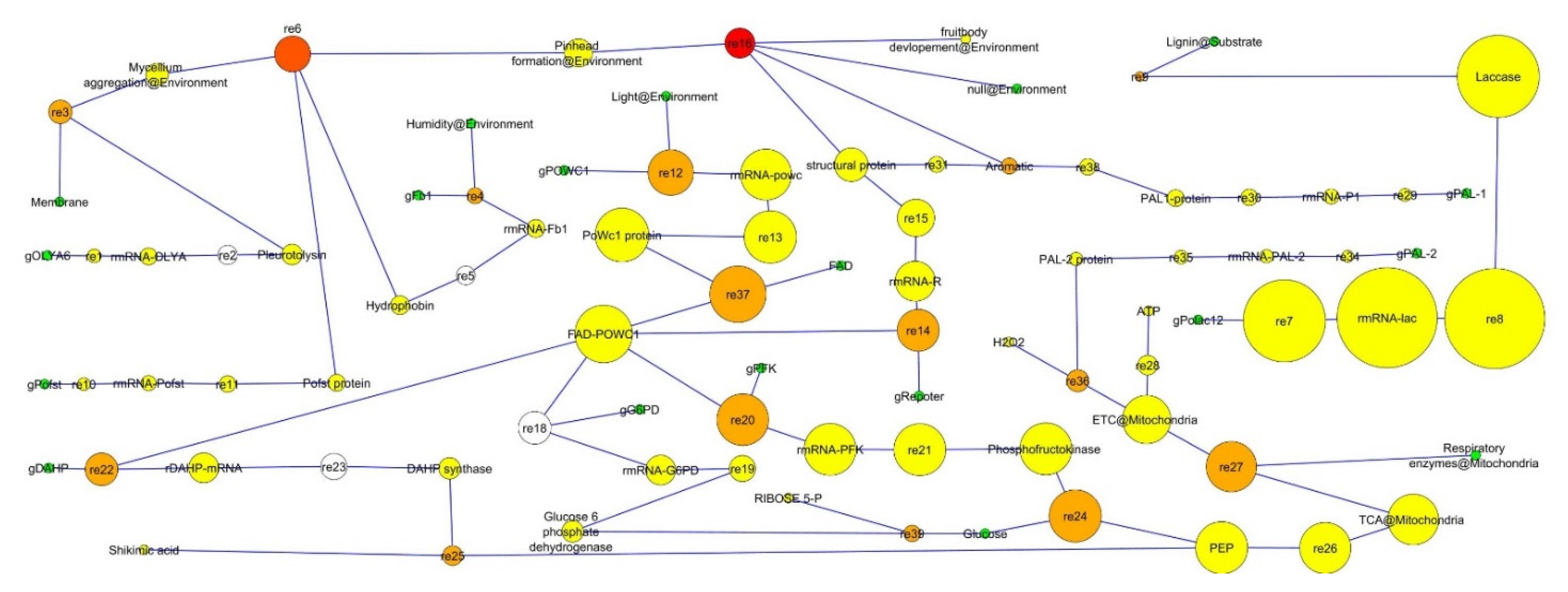

| Sl. No. | Simple Topology Parameters | Values |
|---|---|---|
| 1. | Clustering coefficient | 0.0 |
| 2. | Connected components | 2.0 |
| 3. | Network diameter (largest distance between two nodes) | 18.0 |
| 4. | Network radius | 1 |
| 5. | Shortest paths | 747(%) |
| 6. | Characteristic path length/average shortest path length | 5.63 |
| 7. | Average number of neighbors/average connectivity | 2.02 |
| 8. | Network density | 0.0 |
| Sl. No. | Protein | Role | Reference |
|---|---|---|---|
| 1 | Pleurotolysin | Pore-forming protein having role in fruiting | [39] |
| 2 | Hydrophobins | Initiate pinhead growth makes aerial structure-aerial hyphae and fruit body | [40,41,42,43] |
| 3 | Pofst protein | Regulation of fruiting by inhibiting the excessive cluster formation | [44] |
| 4 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | Role in glycolysis/gluconeogenesis and pentose phosphate pathway to promote fruitbody growth | [45] |
| 5 | 6-phosphogluconate dehydrogenase (6PGD) | Role in glycolysis/gluconeogenesis and pentose phosphate pathway to promote fruitbody growth | [45] |
| 6 | Phosphoenol pyruvate carboxykinase (PEPCK) | Role in glycolysis/gluconeogenesis and pentose phosphate pathway to promote fruitbody growth | [45] |
| 7 | Pleurotus White Collar protein | It induces transcription and expression of respiratory genes | [46] |
| 8 | Carbohydrate-active enzymes | Primordial differentiation to fruiting body with help of blue light | [46] |
| 9 | Laccases | Role in fruit body formation | [47] |
| 10 | PAL genes | Expression upregulated during primodia fruiting body and spore development. These genes along with tyrosinases involved in pileus pigment formation | [48,49] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barh, A.; Sharma, K.; Bhatt, P.; Annepu, S.K.; Nath, M.; Shirur, M.; Kumari, B.; Kaundal, K.; Kamal, S.; Sharma, V.P.; et al. Identification of Key Regulatory Pathways of Basidiocarp Formation in Pleurotus spp. Using Modeling, Simulation and System Biology Studies. J. Fungi 2022, 8, 1073. https://doi.org/10.3390/jof8101073
Barh A, Sharma K, Bhatt P, Annepu SK, Nath M, Shirur M, Kumari B, Kaundal K, Kamal S, Sharma VP, et al. Identification of Key Regulatory Pathways of Basidiocarp Formation in Pleurotus spp. Using Modeling, Simulation and System Biology Studies. Journal of Fungi. 2022; 8(10):1073. https://doi.org/10.3390/jof8101073
Chicago/Turabian StyleBarh, Anupam, Kanika Sharma, Pankaj Bhatt, Sudheer Kumar Annepu, Manoj Nath, Mahantesh Shirur, Babita Kumari, Kirti Kaundal, Shwet Kamal, Ved Parkash Sharma, and et al. 2022. "Identification of Key Regulatory Pathways of Basidiocarp Formation in Pleurotus spp. Using Modeling, Simulation and System Biology Studies" Journal of Fungi 8, no. 10: 1073. https://doi.org/10.3390/jof8101073
APA StyleBarh, A., Sharma, K., Bhatt, P., Annepu, S. K., Nath, M., Shirur, M., Kumari, B., Kaundal, K., Kamal, S., Sharma, V. P., Gupta, S., Sharma, A., Gupta, M., & Dutta, U. (2022). Identification of Key Regulatory Pathways of Basidiocarp Formation in Pleurotus spp. Using Modeling, Simulation and System Biology Studies. Journal of Fungi, 8(10), 1073. https://doi.org/10.3390/jof8101073






