The Epichloë festucae Antifungal Protein Efe-AfpA Protects Creeping Bentgrass (Agrostis stolonifera) from the Plant Pathogen Clarireedia jacksonii, the Causal Agent of Dollar Spot Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Primer Sequences
2.2. Cloning of Modified E. festucae Antifungal Protein Coding Sequences in Escherichia coli
2.3. Recombinant N-Terminal Modified Efe-AfpA Protein Purification
2.4. Purification of Efe-AfpA from Culture Filtrates of Pichia pastoris
2.5. Cloning and Transformation of Efe-afpA into Penicillium chrysogenum
2.6. Purification of Efe-AfpA from Culture Filtrates of Penicillium chrysogenum and of PAF from an Overexpression Strain of Pe. chrysogenum
2.7. Neurospora crassa Conidial Growth Assays
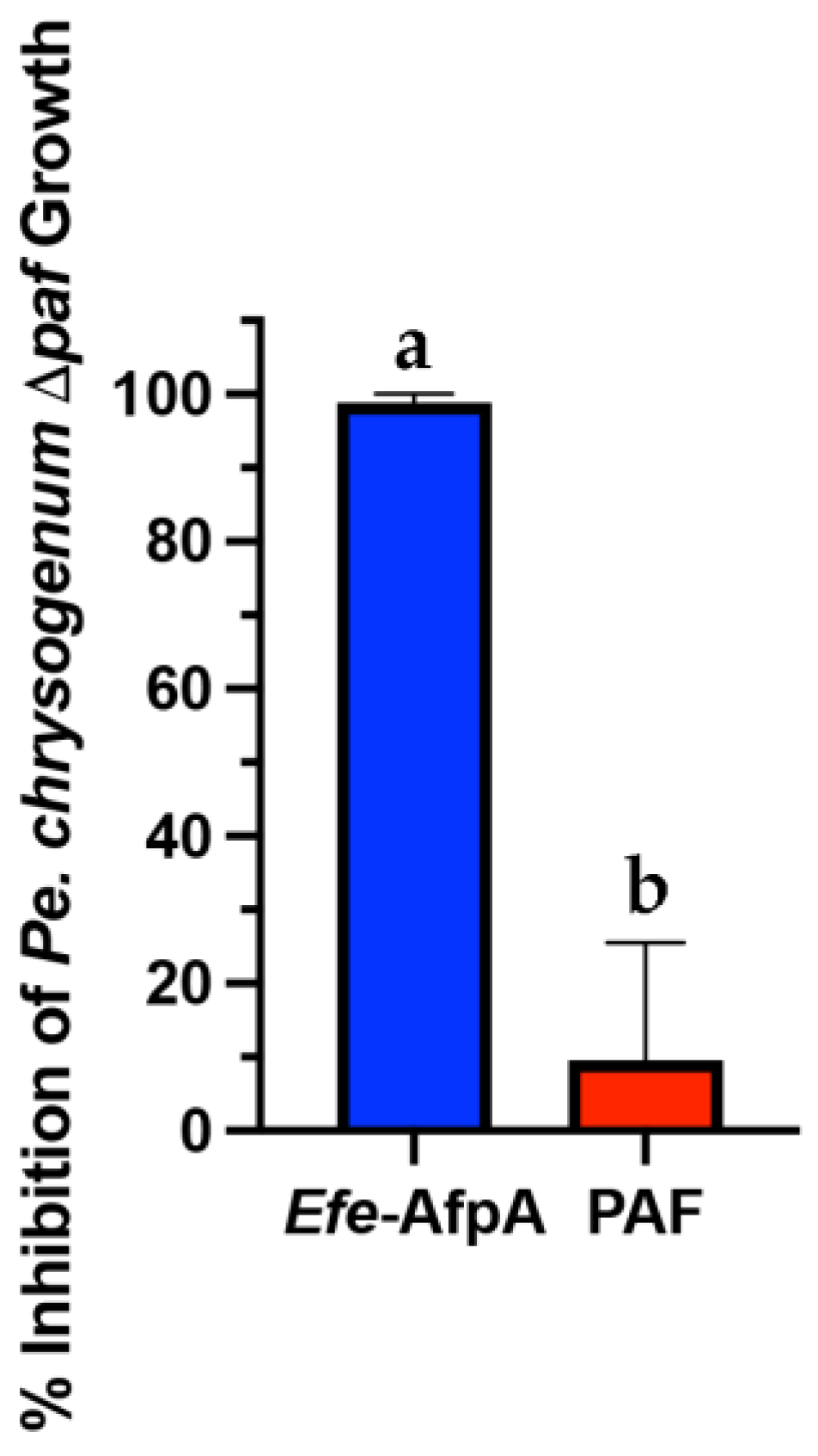
2.8. Penicillium chrysogenum Δpaf Sensitivity to Efe-AfpA
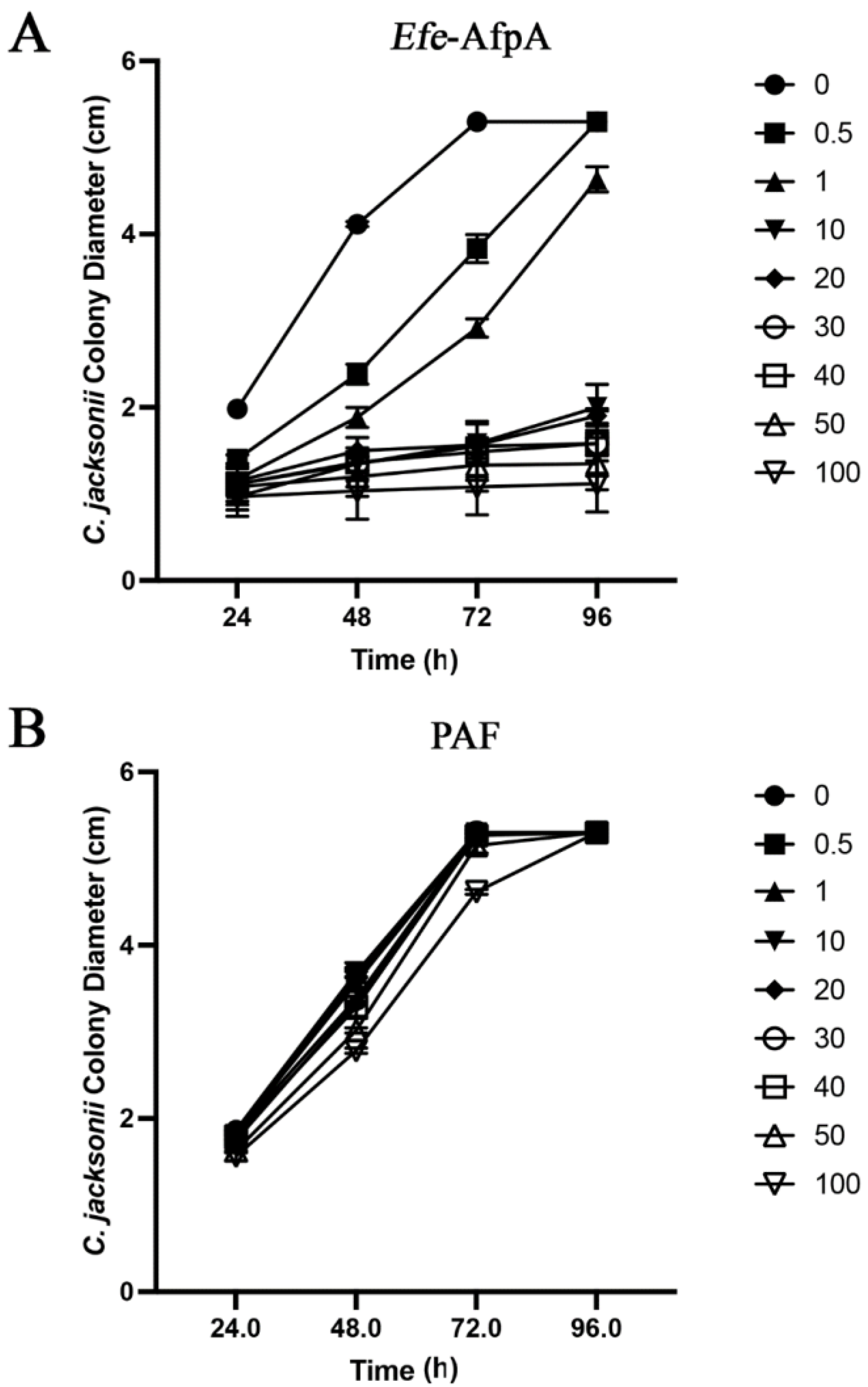
2.9. Clarireedia jacksonii Inhibition Assays
2.10. Strong Creeping Red Fescue and Creeping Bentgrass Greenhouse Infection Assay
2.11. Evaluation of the Effect of Neurospora crassa Glucosylceramides on Efe-AfpA Activity
2.12. Phylogenetic Analysis
2.13. Protein Gel Electrophoresis
3. Results
3.1. Production of Efe-AfpA in E. coli
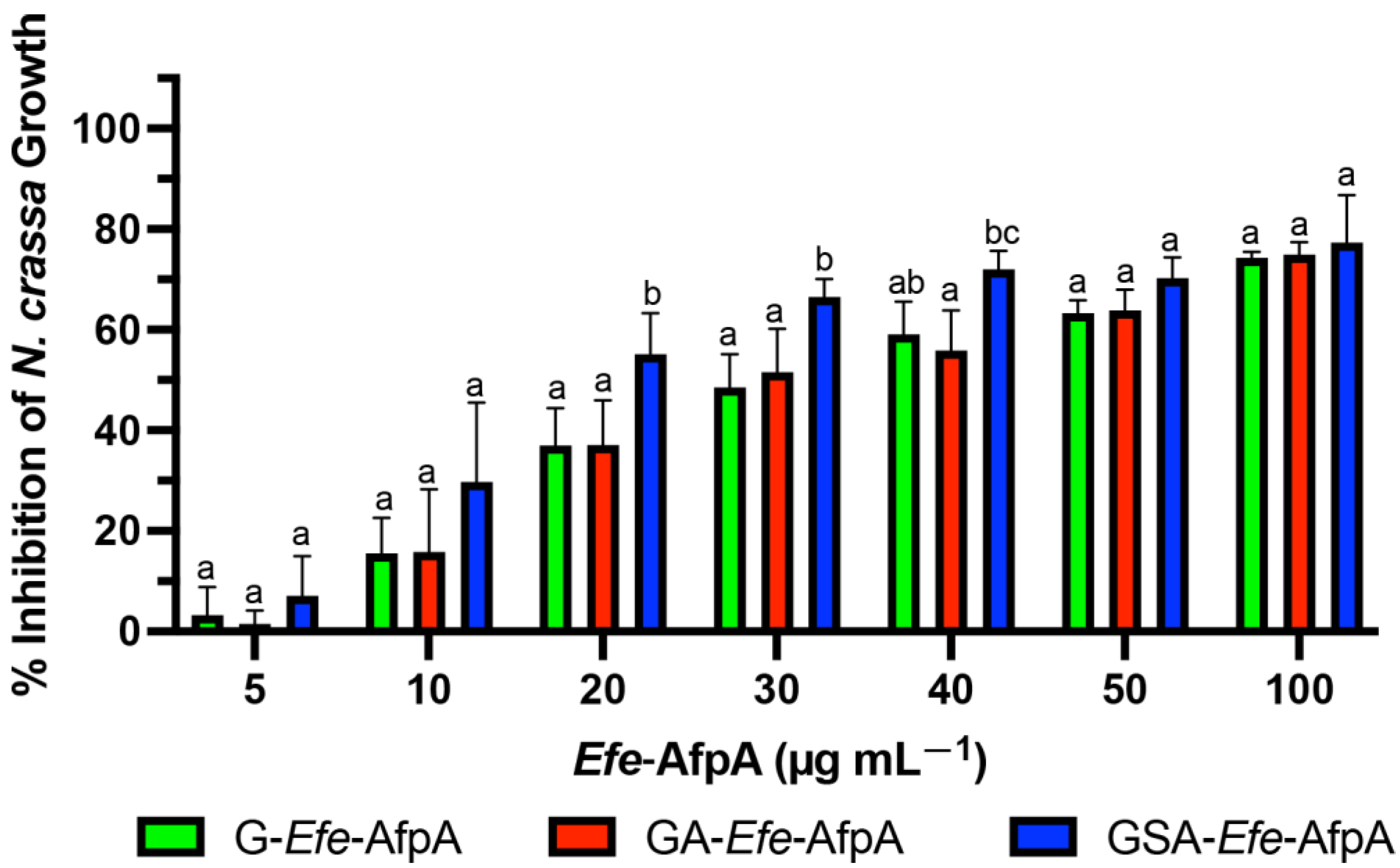
3.2. Activity of the Modified Efe-AfpA Proteins
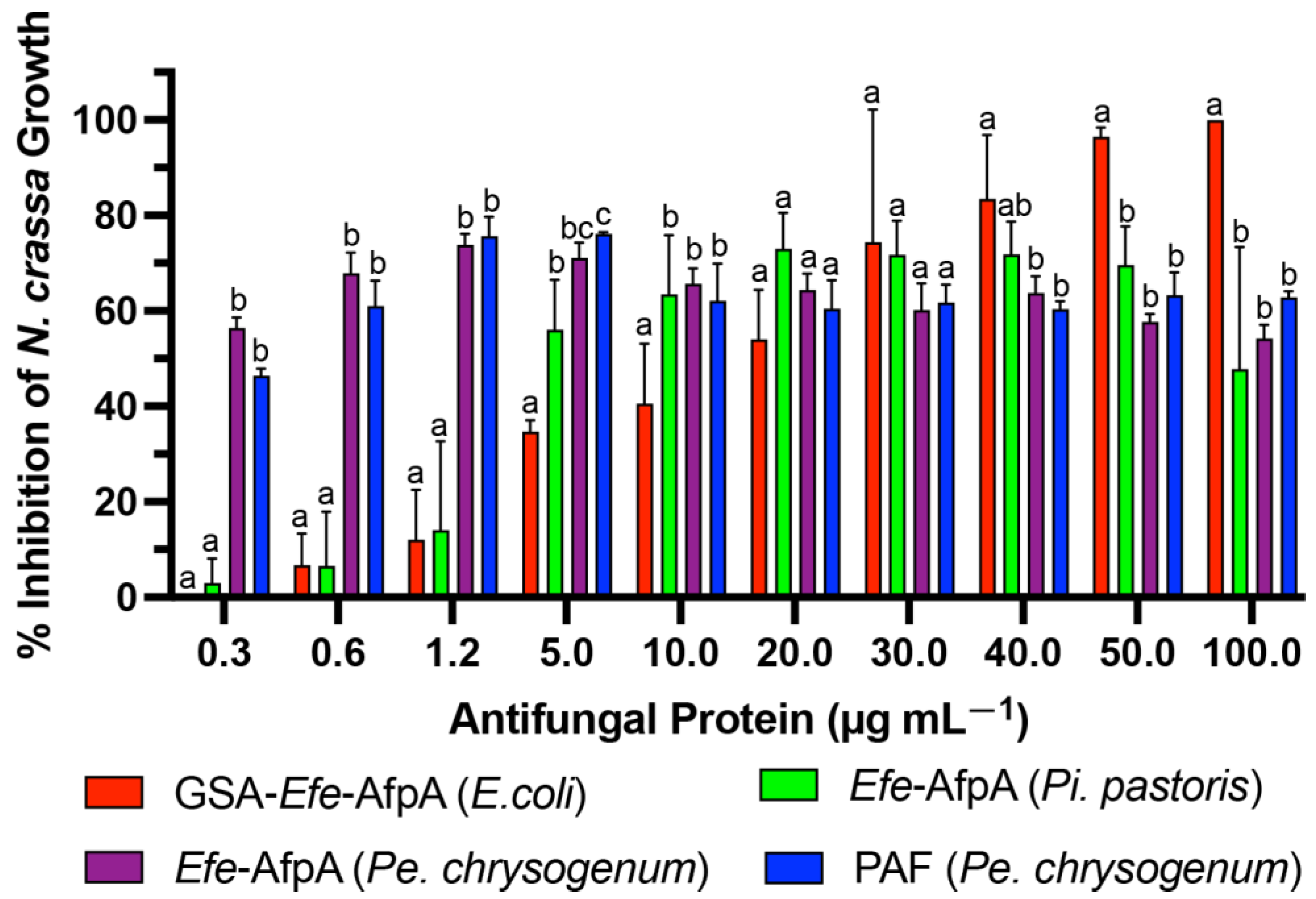
3.3. Expression of Efe-AfpA in Penicillium chrysogenum
3.4. Activity of Efe-AfpA and PAF against Clarireedia jacksonii in Culture
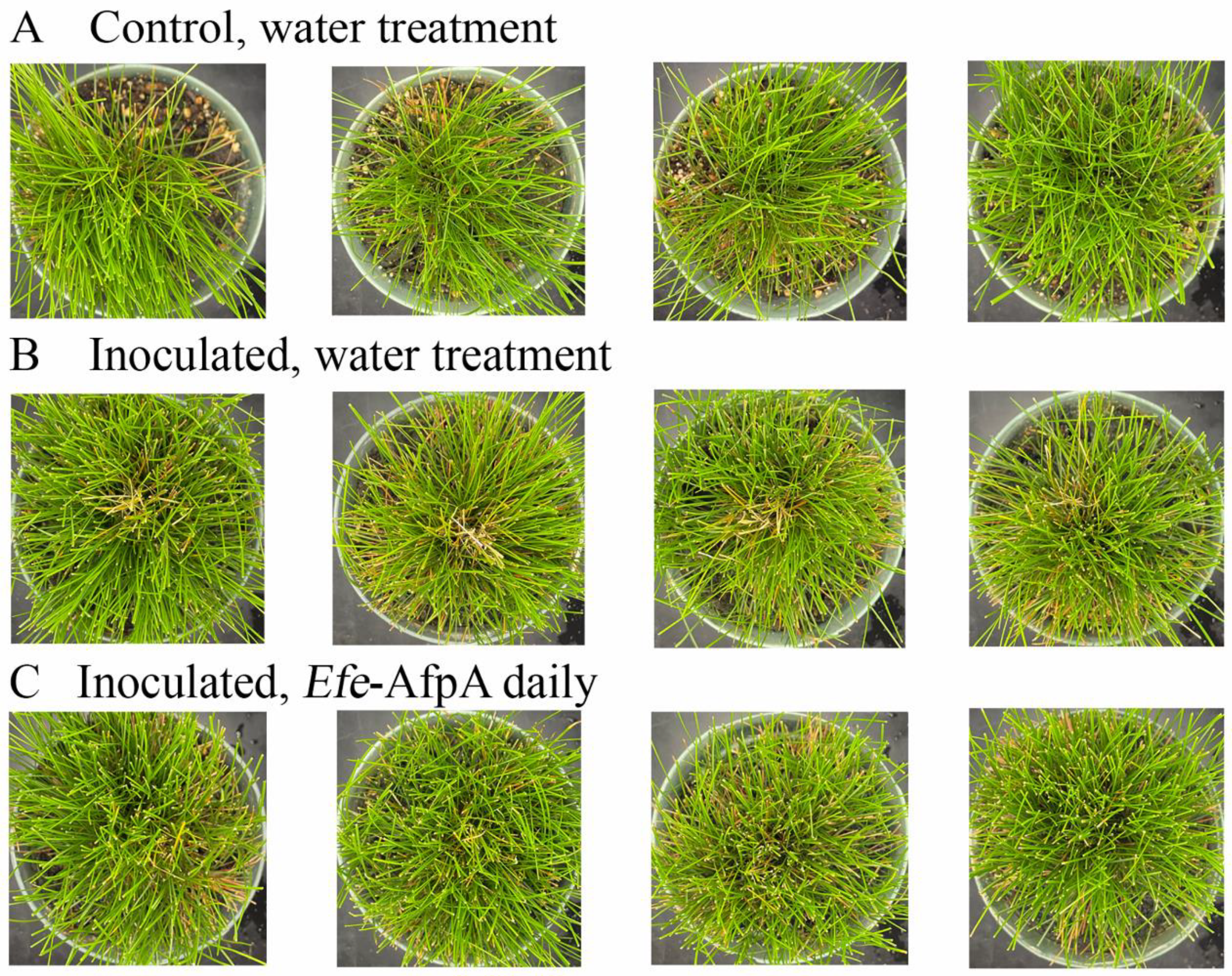
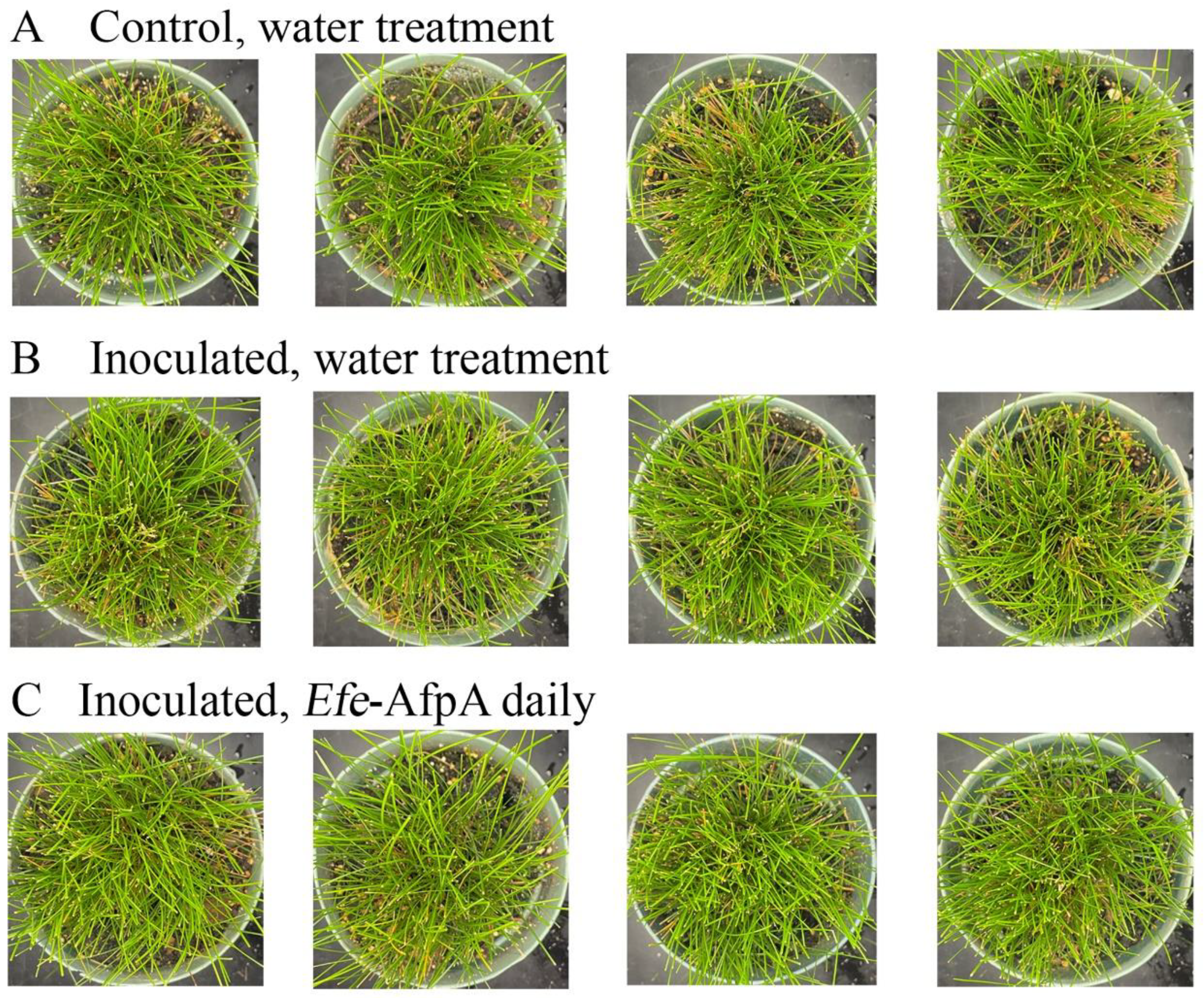
3.5. Activity of Applied Efe-AfpA on Expression of Dollar Spot Symptoms When Strong Creeping Red Fescue and Creeping Bentgrass Plants Were Inoculated with Clarireedia jacksonii in a Greenhouse Assay
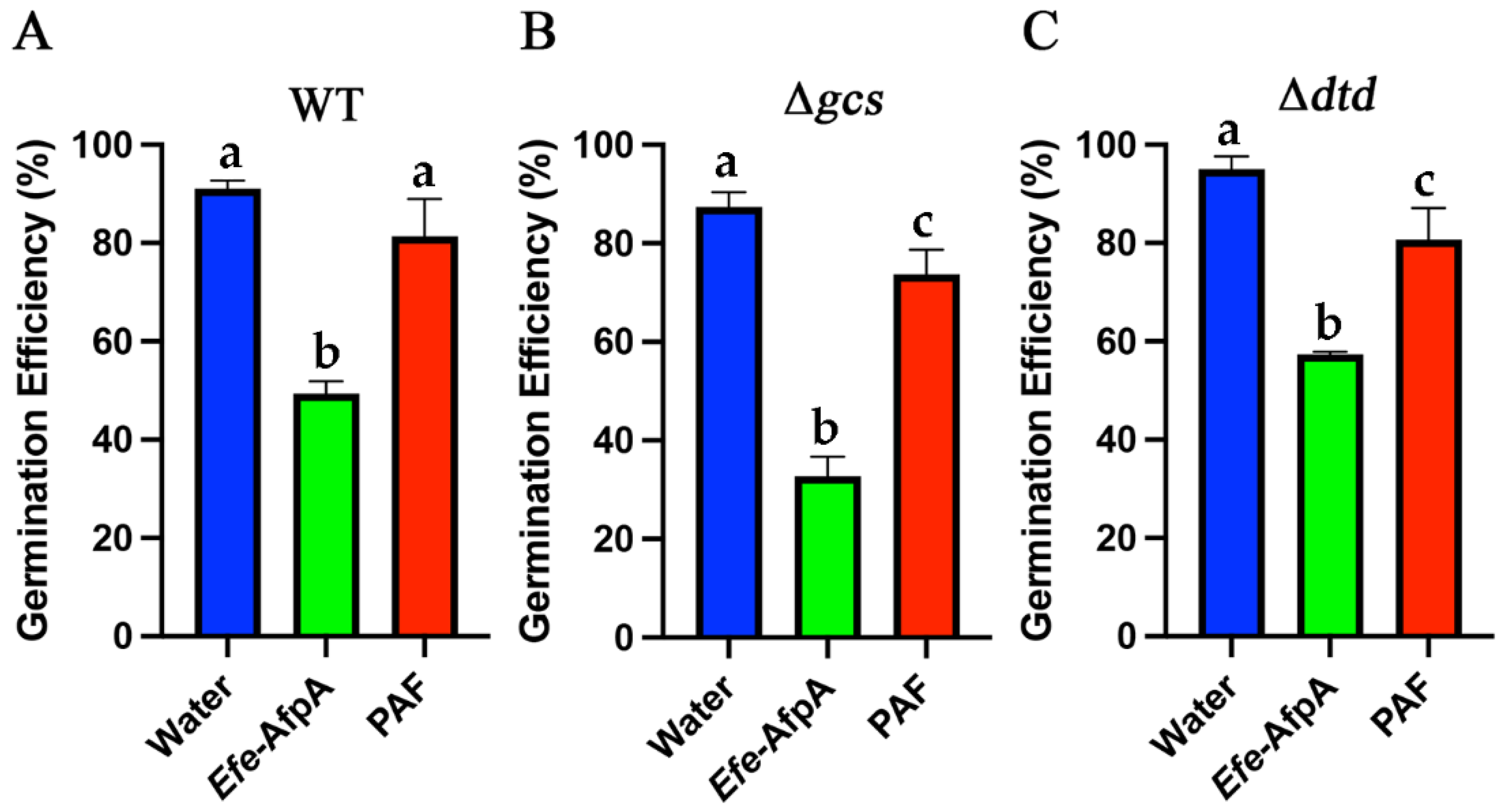
3.6. Efe-AfpA Activity against Neurospora crassa Glucosylceramide Mutant Strains
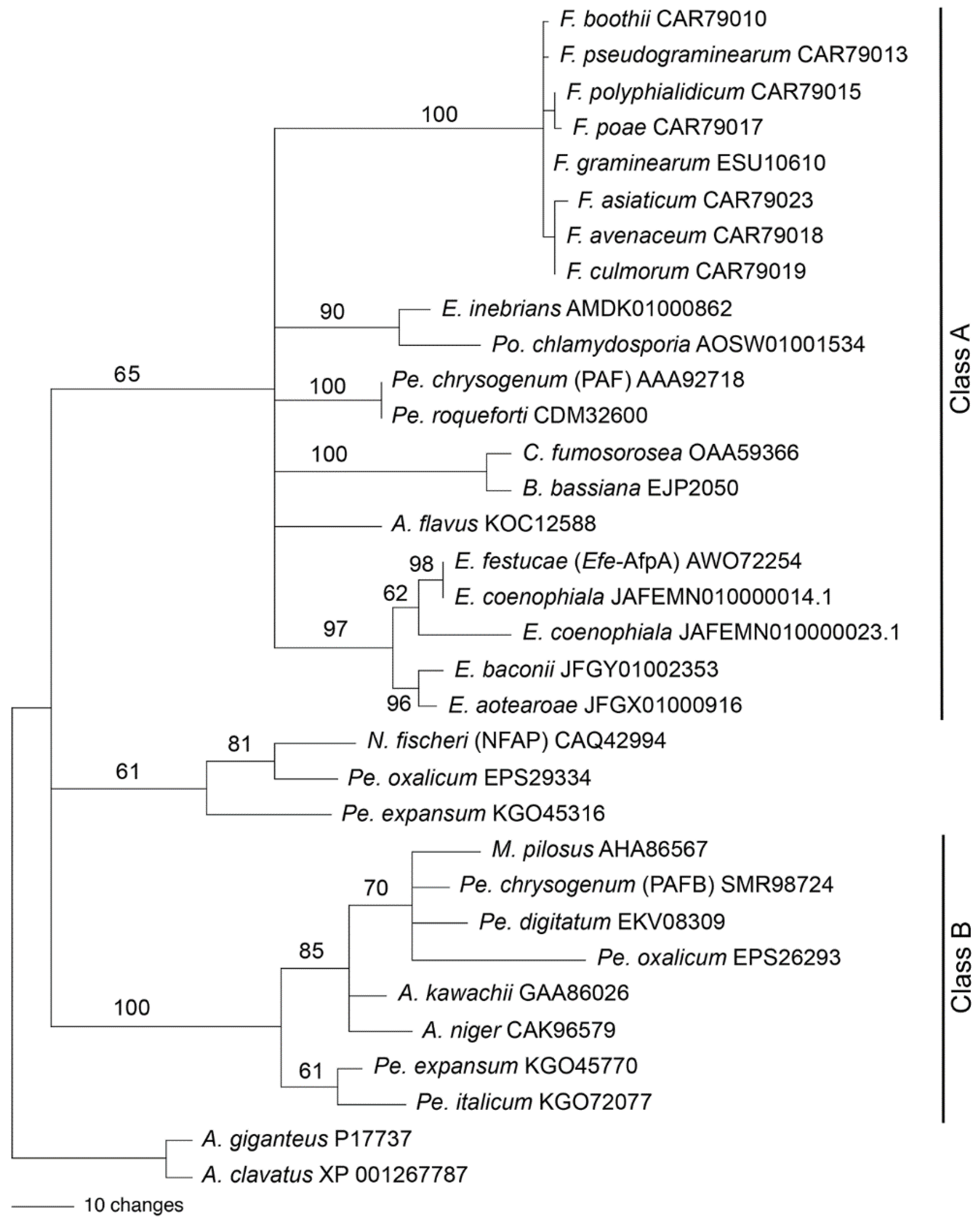
3.7. Relationship of Efe-AfpA to Other Antifungal Proteins from Filamentous Fungi
3.8. Afp-A Genes in Epichloë spp.
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marx, F. Small, basic antifungal proteins secreted from filamentous ascomycetes: A comparative study regarding expression, structure, function and potential application. App. Microbiol. Biotechnol. 2004, 65, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Hegedus, N.; Marx, F. Antifungal proteins: More than antimicrobials? Fungal Biol. Rev. 2013, 26, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Marx, F.; Haas, H.; Reindl, M.; Stoffler, G.; Lottspeich, F.; Redl, B. Cloning, structural organization and regulation of expression of the Penicillium chrysogenum paf gene encoding an abundantly secreted protein with antifungal activity. Gene 1995, 167, 167–171. [Google Scholar] [CrossRef]
- Hegedus, N.; Leiter, E.; Kovacs, B.; Tomori, V.; Kwon, N.-J.; Emri, T.; Marx, F.; Batta, G.; Csernoch, L.; Haas, H. The small molecular mass antifungal protein of Penicillium chrysogenum—A mechanism of action oriented review. J. Basic Microbiol. 2011, 51, 561–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huber, A.; Galgoczy, L.; Varadi, G.; Holzknecht, J.; Kakar, A.; Malanovic, N.; Leber, R.; Koch, J.; Keller, M.A.; Batta, G.; et al. Two small, cysteine-rich and cationic antifungal proteins from Penicillium chrysogenum: A comparative study of PAF and PAFB. Biochim. Biophys. Acta—Biomembr. 2020, 1862, 183246. [Google Scholar] [CrossRef]
- Whendt, S.; Ulbrich, N.; Stahl, U. Molecular cloning, sequence analysis and expression of the gene encoding an antifungal-protein from Aspergillus giganteus. Curr. Genet. 1994, 25, 519–523. [Google Scholar] [CrossRef]
- Meyer, V. A small protein that fights fungi: AFP as a new promising antifungal agent of biotechnological value. Appl. Microbiol. Biotechnol. 2008, 78, 17–28. [Google Scholar] [CrossRef]
- Kovacs, L.; Viragh, M.; Tako, M.; Papp, T.; Vagvolgyi, C.; Galgoczy, L. Isolation and characterization of Neosartorya fischeri antifungal protein (NFAP). Peptides 2011, 32, 1724–1731. [Google Scholar] [CrossRef]
- Delgado, J.; Owens, R.A.; Doyle, S.; Asensio, M.A.; Nunez, F. Antifungal proteins from moulds: Analytical tools and potential application to dry-ripened foods. Appl. Microbiol. Biotechnol. 2016, 100, 6991–7000. [Google Scholar] [CrossRef] [Green Version]
- Leiter, E.; Gall, T.; Csernoch, L.; Pocsi, I. Biofungicide utilizations of antifungal proteins of filamentous ascomycetes: Current and foreseeable future developments. BioControl 2017, 62, 125–138. [Google Scholar] [CrossRef]
- Palicz, Z.; Gall, T.; Leiter, E.; Kollar, S.; Kovacs, I.; Miszti-Blasius, K.; Pocsi, I.; Csernoch, L.; Szentesi, P. Application of a low molecular weight antifungal protein from Penicillium chrysogenum (PAF) to treat pulmonary aspergillosis in mice. Emerg. Microbes Infect. 2016, 5, e114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Culebras, P.V.; Gandia, M.; Boronat, A.; Marcos, J.F.; Manzanares, P. Differential susceptibility of mycotoxin-producing fungi to distinct antifungal proteins (AFPs). Food Microbiol. 2021, 97, 103760. [Google Scholar] [CrossRef] [PubMed]
- Ruemmele, B.A.; Wipff, J.K.; Brilman, L.; Hignight, K.W. Fine-leaved Festuca species. In Turfgrass Biology, Genetics, and Breeding; Cassler, M.D., Duncan, R.R., Eds.; Wiley: Hoboken, NJ, USA, 2003; pp. 129–174. [Google Scholar]
- Braun, R.C.; Patton, A.J.; Watkins, E.; Koch, P.L.; Anderson, N.P.; Bonos, S.A.; Brilman, L.A. Fine fescues: A review of the species, their improvement, production, establishment, and management. Crop Sci. 2020, 60, 1142–1187. [Google Scholar] [CrossRef] [Green Version]
- Schardl, C.L.; Young, C.A.; Hesse, U.; Amyotte, S.G.; Andreeva, K.; Calie, P.J.; Fleetwood, D.J.; Haws, D.C.; Moore, N.; Oeser, B.; et al. Plant-symbiotic fungi as chemical engineers: Multi-genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 2013, 9, e1003323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caradus, J.R.; Johnson, L.J. Epichloë fungal endophytes—From a biological curiosity in wild grasses to an essential component of resilient high performing ryegrass and fescue pastures. J. Fungi 2020, 6, 322. [Google Scholar] [CrossRef]
- Funk, C.R.; White, R.H.; Breen, J.P. Importance of Acremonium endophytes in turfgrass breeding and management. Agric. Ecosyst. Environ. 1993, 44, 215–232. [Google Scholar] [CrossRef]
- Bonos, S.A.; Wilson, M.M.; Meyer, W.A.; Funk, C.R. Suppression of red thread in fine fescues through endophyte-mediated resistance. Appl. Turfgrass Sci. 2005, 10, 1094. [Google Scholar] [CrossRef]
- Clarke, B.B.; White, J.F., Jr.; Hurley, R.H.; Torres, M.S.; Sun, S.; Huff, D.R. Endophyte-mediated suppression of dollar spot disease in fine fescues. Plant Dis. 2006, 90, 994–998. [Google Scholar] [CrossRef] [Green Version]
- Tian, Z.; Wang, R.; Ambrose, K.V.; Clarke, B.B.; Belanger, F.C. The Epichloë festucae antifungal protein has activity against the plant pathogen Sclerotinia homoeocarpa, the causal agent of dollar spot disease. Sci. Rep. 2017, 7, 5643. [Google Scholar] [CrossRef] [Green Version]
- Heineck, G.C.; Qiu, Y.; Ehlke, N.J.; Watkins, E. The fungal endophyte Epichloë festucae var. lolii plays a limited role in mediating crown rust severity in perennial ryegrass. Crop Sci. 2020, 60, 1090–1104. [Google Scholar]
- Ambrose, K.V.; Belanger, F.C. SOLiD-SAGE of endophyte-infected red fescue reveals numerous effects on host transcriptome and an abundance of highly expressed fungal secreted proteins. PLoS ONE 2012, 7, e53214. [Google Scholar] [CrossRef]
- Card, S.D.; Bastias, D.A.; Caradus, J.R. Antagonism to plant pathogens by Epichloë fungal endophytes—A review. Plants 2021, 10, 1997. [Google Scholar] [CrossRef]
- Fernando, K.; Reddy, P.; Spangenberg, G.C.; Rochfort, S.J.; Guthridge, K.M. Metabolic potential of Epichloë endophytes for host grass fungal disease resistance. Microorganisms 2022, 10, 64. [Google Scholar] [CrossRef]
- Salgado-Salazar, C.; Beirn, L.A.; Ismaiel, A.; Boehm, M.J.; Carbone, I.; Putman, A.I.; Tredway, L.P.; Clarke, B.B.; Crouch, J.A. Clarireedia: A new fungal genus comprising four pathogenic species responsible for dollar spot disease of turfgrass. Fungal Biol. 2018, 122, 761–773. [Google Scholar] [CrossRef]
- Sapkota, S.; Catching, K.E.; Raymer, P.L.; Martinez-Espinoza, A.D.; Bahri, B.A. New approaches to an old problem: Dollar spot of turfgrass. Phytopathology 2022, 112, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Walsh, B.; Ikeda, S.S.; Boland, G.J. Biology and management of dollar spot (Sclerotinia homoeocarpa); an important disease of turfgrass. HortScience 1999, 34, 13–21. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Luo, S.; Clarke, B.B.; Belanger, F.C. The Epichloë festucae antifungal protein Efe-AfpA is also a possible effector protein required for the interaction of the fungus with its host grass Festuca rubra subsp. rubra. Microorganisms 2021, 9, 140. [Google Scholar] [CrossRef] [PubMed]
- Schardl, C.L.; Scott, B. Recommendations for gene nomenclature for Epichloë species and related Clavicipitaceae. In Epichloae, Endophytes of Cool Season Grasses: Implications, Utilization and Biology; Young, C.A., Aiken, G.E., McCulley, R.L., Strickland, J.R., Schardl, C.L., Eds.; The Samuel Roberts Noble Foundation: Ardmore, OK, USA, 2012; pp. 84–87. [Google Scholar]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef] [Green Version]
- Sonderegger, C.; Galgoczy, L.; Garrigues, S.; Fizil, A.; Borics, A.; Manzanares, P.; Hededus, N.; Huber, A.; Marcos, J.F.; Batta, G.; et al. A Penicillium chrysogenum-based expression system for the production of small, cysteine-rich antifungal proteins for structural and functional analyses. Microb. Cell Factories 2016, 15, 192. [Google Scholar] [CrossRef] [Green Version]
- Marx, F.; Binder, U.; Leiter, E.; Pocsi, I. The Penicillium chrysogenum antifungal protein PAF, a promising tool for the development of new antifungal therapies and fungal cell biology studies. Cell. Mol. Life Sci. 2008, 65, 445–454. [Google Scholar] [CrossRef]
- Varadi, G.; Toth, G.; Kele, Z.; Galgoczy, L.; Fizil, A.; Batta, G. Synthesis of PAF, an antifungal protein from P. chrysogenum, by native chemical ligation: Native disulfide pattern and fold obtained upon oxidative refolding. Chemistry 2013, 19, 12684–12692. [Google Scholar] [CrossRef] [PubMed]
- Shehata, H.R.; Lyons, E.M.; Jordan, K.S.; Raizada, M.N. Bacterial endophytes from wild and ancient maize are able to suppress the fungal pathogen Sclerotinia homoeocarpa. J. Appl. Microbiol. 2016, 120, 756–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson-Cicalese, J.; Secks, M.E.; Lam, C.K.; Meyer, W.A.; Murphy, J.A.; Belanger, F.C. Cross species inoculation of Chewings and strong creeping red fescues with fungal endophytes. Crop Sci. 2000, 40, 1485–1489. [Google Scholar] [CrossRef]
- Huber, A.; Oemer, G.; Malanovic, N.; Lohner, K.; Kovacs, L.; Salvenmoser, W.; Zschocke, J.; Keller, M.A.; Marx, F. Membrane sphingolipids regulate the fitness and antifungal protein susceptibility of Neurospora crassa. Front. Microbiol. 2019, 10, 605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zauner, S.; Zahringer, U.; Lindner, B.; Warnecke, D.; Sperling, P. Identification and functional characterization of the 2-hydroxy fatty N-acyl-Δ3(E)-desaturase from Fusarium graminearum. J. Biol. Chem. 2008, 283, 36734–36742. [Google Scholar] [CrossRef] [Green Version]
- Colot, H.V.; Park, G.; Turner, G.E.; Ringelberg, C.; Crew, C.M.; Litvinkova, L.; Weiss, R.L.; Borkovich, K.A.; Dunlap, J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 2006, 103, 10352–10357. [Google Scholar] [CrossRef] [Green Version]
- Dunlap, J.C.; Borkovich, K.A.; Henn, M.R.; Turner, G.E.; Sachs, M.S.; Glass, N.L.; McCluskey, K.; Plamann, M.; Galagan, J.E.; Birren, B.W.; et al. Enabling a community to dissect an organism: Overview of the Neurospora functional genomics project. Adv. Genet. 2007, 57, 49–96. [Google Scholar]
- Sonderegger, C.; Fizil, A.; Burtscher, L.B.; Hajdu, D.; Munoz, A.; Gaspari, Z.; Read, N.D.; Batta, G.; Marx, F. D19S mutation of the cationic, cysteine-rich protein PAF: Novel insights into its structral dynamics, thermal unfolding and antifungal function. PLoS ONE 2017, 12, e0169920. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL-X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nuc. Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Ambrose, K.V.; Tian, Z.; Wang, Y.; Smith, J.; Zylstra, G.; Huang, B.; Belanger, F.C. Functional characterization of salicylate hydroxylase from the fungal endophyte Epichloë festucae. Sci. Rep. 2015, 5, 10939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malakhov, M.P.; Mattern, M.R.; Malakhova, O.Z.; Drinker, M.; Weeks, S.D.; Butt, T.R. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genom. 2004, 5, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Lobstein, J.; Emrich, C.A.; Jeans, C.; Faulkner, M.; Riggs, P.; Berkmen, M. Shuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Fact. 2012, 11, 56. [Google Scholar] [CrossRef] [Green Version]
- Binder, U.; Chu, M.; Read, N.D.; Marx, F. The antifungal activity of the Penicillium chrysogenum protein PAF disrupts calcium homeostasis in Neurospora crassa. Eukaryot. Cell 2010, 9, 1374–1382. [Google Scholar] [CrossRef] [Green Version]
- Gaff, D.F.; Okong’O-Ogola, O. The use of non-permeating pigments for testing the survival of cells. J. Exp. Bot. 1971, 22, 756–758. [Google Scholar] [CrossRef]
- Paege, N.; Warnecke, D.; Zauner, S.; Hagen, S.; Rodrigues, A.; Baumann, B.; Thiess, M.; Jung, S.; Meyer, V. Species-specific differences in the susceptibility of fungi to the antifungal protein AFP depend on C-3 saturation of glycosylceramides. MSphere 2019, 4, e00741-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrigues, S.; Gandia, M.; Marcos, J.F. Occurrence and function of fungal antifungal proteins: A case study of the citrus postharvest pathogen Penicillium digitatum. Appl. Microbiol. Biotechnol. 2016, 100, 2243–2256. [Google Scholar] [CrossRef]
- Cheeseman, K.; Ropars, J.; Renault, P.; Dupont, J.; Gouzy, J.; Branca, A.; Abraham, A.-L.; Ceppi, M.; Conseiller, E.; Debuchy, R.; et al. Multiple recent horizontal transfers of a large genomic region in cheese making fungi. Nat. Commun. 2013, 5, 2876. [Google Scholar] [CrossRef] [Green Version]
- Ropars, J.; Rodriguez de la Vega, R.; Lopez-Villavicencio, M.; Gouzy, J.; Sallet, E.; Dumas, E.; Lacoste, S.; Debuchy, R.; Dupont, J.; Branca, A.; et al. Adaptive horizontal gene transfers between multiple cheese-associated fungi. Curr. Biol. 2015, 25, 2562–2569. [Google Scholar] [CrossRef] [Green Version]
- Campos-Olivas, R.; Bruix, M.; Santoro, J.; Lacadena, J.; del Pozo, A.M.; Gavilanes, J.G.; Rico, M. NMR solution structure of the antifungal protein from Aspergillus giganteus: Evidence for cysteine pairing isomerism. Biochemistry 1995, 34, 3009–3021. [Google Scholar] [CrossRef]
- Skouri-Gargouri, H.; Ali, M.B.; Gargouri, A. Molecular cloning, structural analysis and modeling of the AcAFP antifungal peptide from Aspergillus clavatus. Peptides 2009, 30, 1798–1804. [Google Scholar] [CrossRef] [PubMed]
- Hajdu, D.; Huber, A.; Czajlik, A.; Toth, L.; Kele, Z.; Kocsube, S.; Fizil, A.; Marx, F.; Galgloczy, L.; Batta, G. Solution structure and novel insights into phylogeny and mode of action of the Neosartorya (Aspergillus) fischeri antifungal protein (NFAP). Int. J. Biol. Macromol. 2019, 129, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.A.; Tapper, B.A.; Simpson, W.R.; Johnson, R.D.; Mace, W.; Ram, A.; Lukito, Y.; Dupont, P.-Y.; Johnson, L.J.; Scott, D.B.; et al. Epichloë hybrida, sp. nov., an emerging model system for investigating fungal allopolyploidy. Mycologia 2017, 109, 715–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Florea, S.; Jaromczyk, J.; Schardl, C.L. Non-transgenic CRISPR-mediated knockout of entire ergot alkaloid gene clusters in slow-growing asexual polyploid fungi. Toxins 2021, 13, 153. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.-F.; Liu, J.-S.; Staben, C.; Christensen, M.J.; Latch, G.C.M.; Siegel, M.R.; Schardl, C.L. Evolutionary diversification of fungal endophytes of tall fescue grass by hybridization with Epichloë species. Proc. Natl. Acad. Sci. USA 1994, 91, 2542–2546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, C.D.; Craven, K.D.; Leuchtmann, A.; Clement, S.L.; Schardl, C.L. Prevalence of interspecific hybrids amongst asexual fungal endophytes of grasses. Mol. Ecol. 2004, 13, 1455–1467. [Google Scholar] [CrossRef]
- Dinkins, R.D.; Nagabhyru, P.; Graham, M.A.; Boykin, D.; Schardl, C.L. Transcriptome response of Lolium arundinaceum to its fungal endophyte Epichloë coenophiala. New Phytol. 2016, 213, 324–337. [Google Scholar] [CrossRef]
- Wang, R.; Clarke, B.B.; Belanger, F.C. Transcriptome analysis of choke stroma and asymptomatic inflorescence tissues reveals changes in gene expression in both Epichloë festucae and its host plant Festuca rubra subsp. rubra. Microorganisms 2019, 7, 567. [Google Scholar] [CrossRef] [Green Version]
- Hegedus, N.; Sigl, C.; Zadra, I.; Pocsi, I.; Marx, F. The paf gene product modulates asexual development in Penicillium chrysogenum. J. Basic Microbiol. 2011, 51, 253–262. [Google Scholar] [CrossRef] [Green Version]
- Kovacs, B.; Hegedus, N.; Balint, M.; Szabo, Z.; Emri, T.; Kiss, G.; Antal, M.; Pocsi, I.; Leiter, E. Penicillium antifungal protein (PAF) is involved in the apoptotic and autophagic processes of the producer Penicillium chrysogenum. Acta Microbiol. Immunol. Hung. 2014, 61, 379–388. [Google Scholar] [CrossRef]
- Marx, F.; Salvenmoser, W.; Kaiserer, L.; Graessle, S.; Weiler-Gorz, R.; Zadra, I.; Oberparleiter, C. Proper folding of the antifungal protein PAF is required for optimal activity. Res. Microbiol. 2005, 156, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.-Y.; Chen, Y.-P.; Yu, M.-C.; Hwang, I.-E.; Wu, D.-Y.; Liaw, L.-L. Characterization and expression of the antifungal protein from Monascus pilosus and its distribution among various Monascus species. J. Biosci. Bioeng. 2016, 122, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Nguyen, N.; Breen, S.; Outram, M.A.; Dodds, P.N.; Kobe, B.; Solomon, P.S.; Williams, S.J. Production of small cysteine-rich effector proteins in Escherichia coli for structural and functional studies. Mol. Plant Pathol. 2017, 18, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Ruiz, A.; Martinez del Pozo, A.; Lacadena, J.; Mancheno, J.M.; Onaderra, M.; Gavilanes, J.G. Characterization of a natural larger form of the antifungal protein (AFP) from Aspergillus giganteus. Biochim. Biophys. Acta 1997, 1340, 81–87. [Google Scholar] [CrossRef]
- Garrigues, S.; Gandia, M.; Castillo, L.; Coca, M.; Marx, F.; Marcos, J.F.; Manzanares, P. Three antifungal proteins from Penicillium expansum: Different patterns of production an antifungal activity. Front. Microbiol. 2018, 9, 2370. [Google Scholar] [CrossRef] [Green Version]
- Toth, L.; Boros, E.; Poor, P.; Ordog, A.; Kele, Z.; Varadi, G.; Holzknecht, J.; Bratschun-Khan, D.; Nagy, I.; Toth, G.K.; et al. The potential use of the Penicillium chrysogenum antifungal protein PAF, the designed variant PAFopt and its γ-core peptide Pγopt in plant protection. Microb. Biotechnol. 2020, 13, 1403–1414. [Google Scholar] [CrossRef] [Green Version]
- Gandia, M.; Kakar, A.; Giner-Llorca, M.; Holzknecht, J.; Martinez-Culebras, P.; Galgoczy, L.; Marx, F.; Marcos, J.F.; Manzanares, P. Potential of antifungal proteins (AFPs) to control Penicillium postharvest fruit decay. J. Fungi 2021, 7, 449. [Google Scholar] [CrossRef]
- Shi, X.; Cordero, T.; Garrigues, S.; Marcos, J.F.; Daros, J.-A.; Coca, M. Efficient production of antifungal proteins in plants using a new transient expression vector derived from tobacco mosaic virus. Plant Biotechnol. J. 2019, 17, 1069–1080. [Google Scholar] [CrossRef]





| Modification | Protein Designation | Protein Sequence |
|---|---|---|
| None | Efe-AfpA | ITYEGTCSRAKNECKYKNQNNKDTFVKCPSFANKKCTKDNAKCSFDSYSRAVTCH |
| I to A | A-Efe-AfpA | ATYEGTCSRAKNECKYKNQNNKDTFVKCPSFANKKCTKDNAKCSFDSYSRAVTCH |
| A to G | G-Efe-AfpA | GTYEGTCSRAKNECKYKNQNNKDTFVKCPSFANKKCTKDNAKCSFDSYSRAVTCH |
| A + G | GA-Efe-AfpA | GATYEGTCSRAKNECKYKNQNNKDTFVKCPSFANKKCTKDNAKCSFDSYSRAVTCH |
| A + GS | GSA-Efe-AfpA | GSATYEGTCSRAKNECKYKNQNNKDTFVKCPSFANKKCTKDNAKCSFDSYSRAVTCH |
| Expression System | Protein Yield (mg L−1 Culture) | Cultivation Time (Days) * |
|---|---|---|
| Escherichia coli | 1 | 5 |
| Pichia pastoris | 3 | 9 |
| Penicillium chrysogenum | 12 | 9 |
| MIC (μg mL−1) | ||
|---|---|---|
| N. crassa Strain | Efe-AfpA | PAF |
| WT | 0.3 | 0.6 |
| Δgcs | 0.6 | 40 |
| Δdtd | 0.3 | 0.6 |
| Protein (Accession) | pI 1 | Net Charge (pH 7.0) 1 | Arg | Lys | His | Asp | Glu |
|---|---|---|---|---|---|---|---|
| PAF (AAA92718) | 8.64 | 4.7 | 0 | 13 | 0 | 7 | 1 |
| PAFB (SMR98724) | 8.5 | 5.2 | 2 | 8 | 6 | 3 | 3 |
| Efe-AfpA (AWO72254) | 8.89 | 6.0 | 2 | 9 | 1 | 3 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fardella, P.A.; Tian, Z.; Clarke, B.B.; Belanger, F.C. The Epichloë festucae Antifungal Protein Efe-AfpA Protects Creeping Bentgrass (Agrostis stolonifera) from the Plant Pathogen Clarireedia jacksonii, the Causal Agent of Dollar Spot Disease. J. Fungi 2022, 8, 1097. https://doi.org/10.3390/jof8101097
Fardella PA, Tian Z, Clarke BB, Belanger FC. The Epichloë festucae Antifungal Protein Efe-AfpA Protects Creeping Bentgrass (Agrostis stolonifera) from the Plant Pathogen Clarireedia jacksonii, the Causal Agent of Dollar Spot Disease. Journal of Fungi. 2022; 8(10):1097. https://doi.org/10.3390/jof8101097
Chicago/Turabian StyleFardella, Patrick A., Zipeng Tian, Bruce B. Clarke, and Faith C. Belanger. 2022. "The Epichloë festucae Antifungal Protein Efe-AfpA Protects Creeping Bentgrass (Agrostis stolonifera) from the Plant Pathogen Clarireedia jacksonii, the Causal Agent of Dollar Spot Disease" Journal of Fungi 8, no. 10: 1097. https://doi.org/10.3390/jof8101097







