Calcineurin Inhibitors Synergize with Manogepix to Kill Diverse Human Fungal Pathogens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains Used
2.2. Culture Media and Conditions
2.3. Dose-Response Assays
2.4. Bioinformatic Analyses
2.5. Chemical Genetic Interaction Screen
2.6. Cell Wall Staining and Quantification
2.7. Flow Cytometry
2.8. Microscopy
3. Results
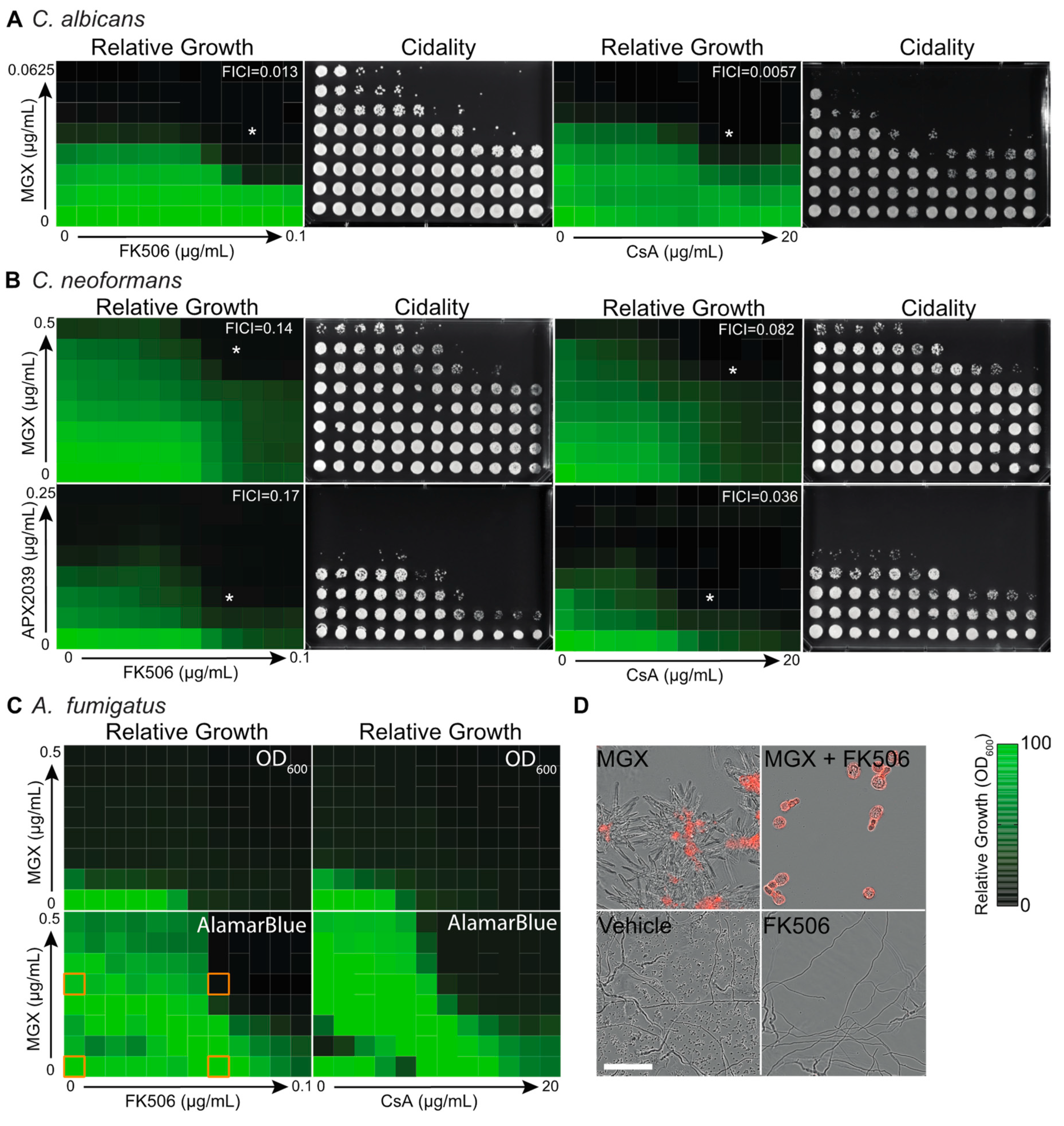
3.1. Gwt1 Inhibitors Synergize with Classical Immunosuppressive Calcineurin Inhibitors
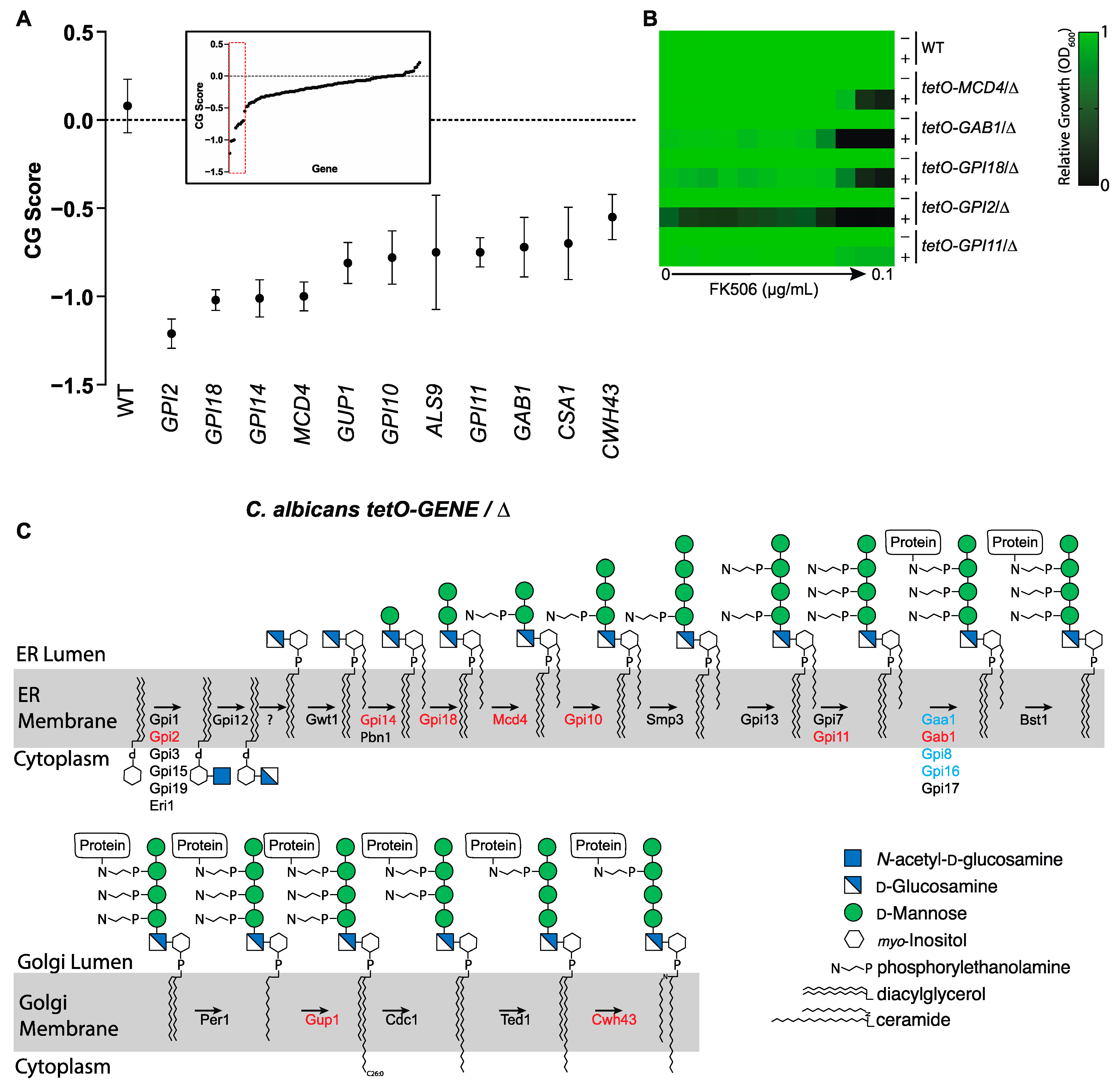
3.2. Chemical-Genetic Interactions Confirm Target-Based Synergy
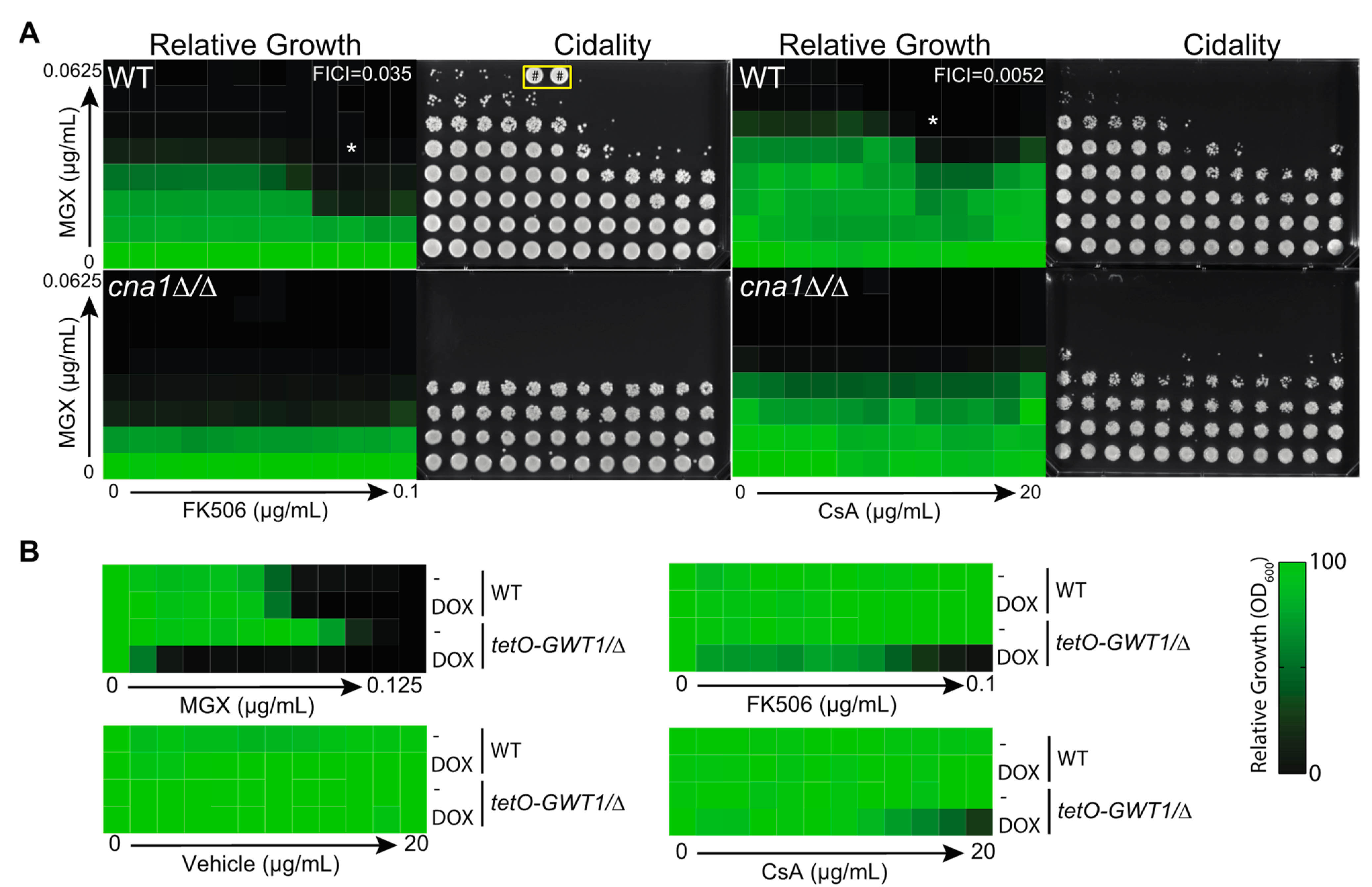
3.3. Calcineurin Mediates Tolerance to Impairment of GPI-Anchor Biosynthesis at Multiple Steps in the Biosynthetic Pathway
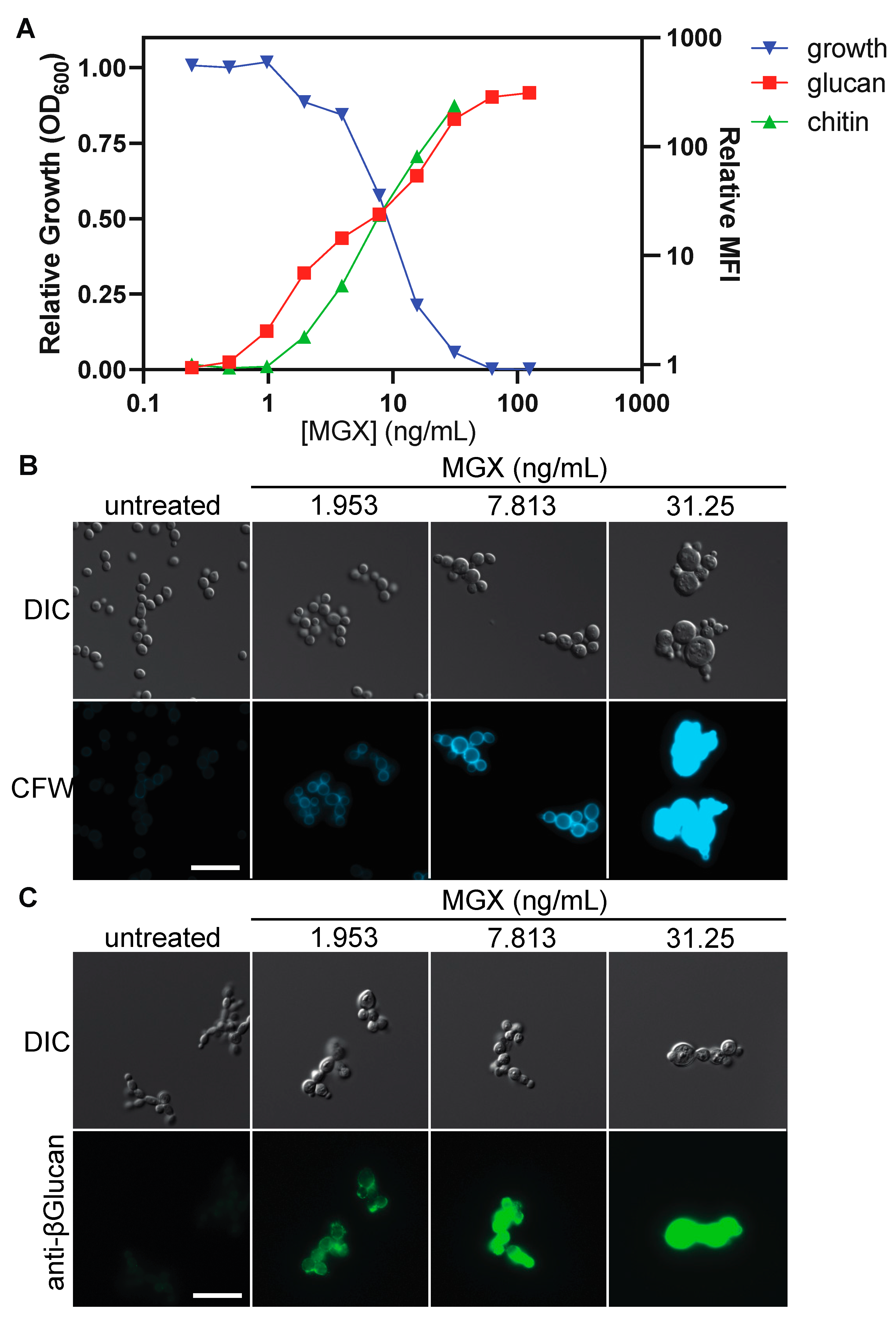
3.4. Impairing GPI Anchor Biosynthesis at Diverse Steps Alters Cell Wall Architecture to Expose (1→3)-β-D-glucan and Increase Chitin Deposition
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CsA | Cyclosporin A |
| CFW | Calcofluor-white |
| DOX | Doxycycline |
| FICI | Fractional inhibitory concentration index |
| GlcN-PI | glucosaminyl-phosphatidyl inositol |
| GPI | glycosylphosphatidylinositol |
| Gwt1 | GPI-anchored wall protein transfer |
| Mcd4 | Morphogenesis checkpoint dependent 4 |
| MGX | manogepix |
| FMGX | fosmanogepix |
References
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef] [Green Version]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Low, C.-Y.; Rotstein, C. Emerging fungal infections in immunocompromised patients. F1000 Med. Rep. 2011, 3, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive mycoses in North America. Antimicrob. Agents Chemother. 2010, 36, 1–53. [Google Scholar] [CrossRef] [PubMed]
- Enoch, D.A.; Ludlam, H.A.; Brown, N.M. Invasive fungal infections: A review of epidemiology and management options. J. Med. Microbiol. 2006, 55, 809–818. [Google Scholar] [CrossRef]
- Wilson, L.S.; Reyes, C.M.; Stolpman, M.; Speckman, J.; Allen, K.; Beney, J. The direct cost and incidence of systemic fungal infections. Value Health 2002, 5, 26–34. [Google Scholar] [CrossRef] [Green Version]
- Robbins, N.; Wright, G.D.; Cowen, L.E. Antifungal Drugs: The Current Armamentarium and Development of New Agents. Microbiol. Spectr. 2016, 4, 903–922. [Google Scholar] [CrossRef]
- McCarthy, M.W.; Kontoyiannis, D.P.; Cornely, O.A.; Perfect, J.R.; Walsh, T.J. Novel Agents and Drug Targets to Meet the Challenges of Resistant Fungi. J. Infect. Dis. 2017, 216, S474–S483. [Google Scholar] [CrossRef] [Green Version]
- Hoenigl, M.; Sprute, R.; Arastehfar, A.; Perfect, J.R.; Lass-Flörl, C.; Bellmann, R.; Prattes, J.; Iii, G.R.T.; Wiederhold, N.P.; Al Obaidi, M.M.; et al. Invasive candidiasis: Investigational drugs in the clinical development pipeline and mechanisms of action. Expert Opin. Investig. Drugs 2022, 31, 795–812. [Google Scholar] [CrossRef]
- Perfect, J.R.; Ghannoum, M. Emerging Issues in Antifungal Resistance. Infect. Dis. Clin. North Am. 2020, 34, 921–943. [Google Scholar] [CrossRef]
- Perfect, J.R. The antifungal pipeline: A reality check. Nat. Rev. Drug Discov. 2017, 16, 603–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mutz, M.; Roemer, T. The GPI anchor pathway: A promising antifungal target? Future Med. Chem. 2016, 8, 1387–1391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, U.; Khan, M.A. Targeting the GPI biosynthetic pathway. Pathog. Glob. Health 2018, 112, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.J.; Ibrahim, A.S. Fosmanogepix: A Review of the First-in-Class Broad Spectrum Agent for the Treatment of Invasive Fungal Infections. J. Fungi 2020, 6, 239. [Google Scholar] [CrossRef] [PubMed]
- Zhen, C.; Lu, H.; Jiang, Y. Novel Promising Antifungal Target Proteins for Conquering Invasive Fungal Infections. Front. Microbiol. 2022, 13, 911322. [Google Scholar] [CrossRef] [PubMed]
- Pittet, M.; Conzelmann, A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 2007, 1771, 405–420. [Google Scholar] [CrossRef] [Green Version]
- Ostrosky-Zeichner, L.; Casadevall, A.; Galgiani, J.N.; Odds, F.C.; Rex, J.H. An insight into the antifungal pipeline: Selected new molecules and beyond. Nat. Rev. Drug Discov. 2010, 9, 719–727. [Google Scholar] [CrossRef]
- Liston, S.D.; Whitesell, L.; McLellan, C.A.; Mazitschek, R.; Petraitis, V.; Petraitiene, R.; Kavaliauskas, P.; Walsh, T.J.; Cowen, L.E. Antifungal Activity of Gepinacin Scaffold Glycosylphosphatidylinositol Anchor Biosynthesis Inhibitors with Improved Metabolic Stability. Antimicrob. Agents Chemother. 2020, 64, e00899-20. [Google Scholar] [CrossRef]
- McLellan, C.A.; Whitesell, L.; King, O.D.; Lancaster, A.; Mazitschek, R.; Lindquist, S. Inhibiting GPI anchor biosynthesis in fungi stresses the endoplasmic reticulum and enhances immunogenicity. ACS Chem. Biol. 2012, 7, 1520–1528. [Google Scholar] [CrossRef]
- Watanabe, N.-A.; Miyazaki, M.; Horii, T.; Sagane, K.; Tsukahara, K.; Hata, K. E1210, a new broad-spectrum antifungal, suppresses Candida albicans hyphal growth through inhibition of glycosylphosphatidylinositol biosynthesis. Antimicrob. Agents Chemother. 2012, 56, 960–971. [Google Scholar] [CrossRef]
- Umemura, M.; Okamoto, M.; Nakayama, K.-I.; Sagane, K.; Tsukahara, K.; Hata, K.; Jigami, Y. GWT1 gene is required for inositol acylation of glycosylphosphatidylinositol anchors in yeast. J. Biol. Chem. 2003, 278, 23639–23647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukahara, K.; Hata, K.; Nakamoto, K.; Sagane, K.; Watanabe, N.-A.; Kuromitsu, J.; Kai, J.; Tsuchiya, M.; Ohba, F.; Jigami, Y.; et al. Medicinal genetics approach towards identifying the molecular target of a novel inhibitor of fungal cell wall assembly. Mol. Microbiol. 2003, 48, 1029–1042. [Google Scholar] [CrossRef] [PubMed]
- Trzoss, M.; Covel, J.A.; Kapoor, M.; Moloney, M.K.; Soltow, Q.A.; Webb, P.J.; Shaw, K.J. Synthesis of analogs of the Gwt1 inhibitor manogepix (APX001A) and in vitro evaluation against Cryptococcus spp. Bioorg. Med. Chem. Lett. 2019, 29, 126713. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.J.; Köhler, J.R.; DiDomenico, B.; Loebenberg, D.; Cacciapuoti, A.; Fink, G.R. Nonfilamentous C. albicans mutants are avirulent. Cell 1997, 90, 939–949. [Google Scholar] [CrossRef] [Green Version]
- Noble, S.M.; French, S.; Kohn, L.A.; Chen, V.; Johnson, A.D. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 2010, 42, 590–598. [Google Scholar] [CrossRef] [Green Version]
- Salgado, P.S.; Yan, R.; Taylor, J.D.; Burchell, L.; Jones, R.; Hoyer, L.L.; Matthews, S.J.; Simpson, P.J.; Cota, E. Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 2011, 108, 15775–15779. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, R.T.; Fink, G.R. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2006, 2, e35. [Google Scholar] [CrossRef]
- Bonilla, M.; Nastase, K.K.; Cunningham, K.W. Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 2002, 21, 2343–2353. [Google Scholar] [CrossRef] [Green Version]
- Iyer, K.R.; Robbins, N.; Cowen, L.E. The role of Candida albicans stress response pathways in antifungal tolerance and resistance. iScience 2022, 25, 103953. [Google Scholar] [CrossRef]
- Mann, P.A.; McLellan, C.A.; Koseoglu, S.; Si, Q.; Kuzmin, E.; Flattery, A.; Harris, G.; Sher, X.; Murgolo, N.; Wang, H.; et al. Chemical Genomics-Based Antifungal Drug Discovery: Targeting Glycosylphosphatidylinositol (GPI) Precursor Biosynthesis. ACS Infect. Dis. 2015, 1, 59–72. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Bale, M.; Buschelman, B.; Lancaster, M.; Espinel-Ingroff, A.; Rex, J.H.; Rinaldi, M.G. Selection of candidate quality control isolates and tentative quality control ranges for in vitro susceptibility testing of yeast isolates by National Committee for Clinical Laboratory Standards proposed standard methods. J. Clin. Microbiol. 1994, 32, 1650–1653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noble, S.M.; Johnson, A.D. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 2005, 4, 298–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.D.; Robbins, N.; Zaas, A.K.; Schell, W.A.; Perfect, J.R.; Cowen, L.E. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 2009, 5, e1000532. [Google Scholar] [CrossRef] [PubMed]
- Roemer, T.; Jiang, B.; Davison, J.; Ketela, T.; Veillette, K.; Breton, A.; Tandia, F.; Linteau, A.; Sillaots, S.; Marta, C.; et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 2003, 50, 167–181. [Google Scholar] [CrossRef]
- Nielsen, K.; Cox, G.M.; Wang, P.; Toffaletti, D.L.; Perfect, J.R.; Heitman, J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect. Immun. 2003, 71, 4831–4841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nierman, W.C.; Pain, A.; Anderson, M.J.; Wortman, J.R.; Kim, H.S.; Arroyo, J.; Berriman, M.; Abe, K.; Archer, D.B.; Bermejo, C.; et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 2005, 438, 1151–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skrzypek, M.S.; Binkley, J.; Binkley, G.; Miyasato, S.R.; Simison, M.; Sherlock, G. The Candida Genome Database (CGD): Incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017, 45, D592–D596. [Google Scholar] [CrossRef] [Green Version]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nature Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Pierleoni, A.; Martelli, P.L.; Casadio, R. PredGPI: A GPI-anchor predictor. BMC Bioinform. 2008, 9, 392. [Google Scholar] [CrossRef] [Green Version]
- Komath, S.S.; Singh, S.L.; Pratyusha, V.A.; Sah, S.K. Generating anchors only to lose them: The unusual story of glycosylphosphatidylinositol anchor biosynthesis and remodeling in yeast and fungi. IUBMB Life 2018, 70, 355–383. [Google Scholar] [CrossRef]
- Fu, C.; Zhang, X.; Veri, A.O.; Iyer, K.R.; Lash, E.; Xue, A.; Yan, H.; Revie, N.M.; Wong, C.; Lin, Z.-Y.; et al. Leveraging machine learning essentiality predictions and chemogenomic interactions to identify antifungal targets. Nat. Commun. 2021, 12, 6497. [Google Scholar] [CrossRef] [PubMed]
- Boucher, B.; Jenna, S. Genetic interaction networks: Better understand to better predict. Front. Genet. 2013, 4, 290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ianevski, A.; Giri, A.K.; Aittokallio, T. SynergyFinder 3.0: An interactive analysis and consensus interpretation of multi-drug synergies across multiple samples. Nucleic. Acids Res. 2022, 50, W739–W743. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jiang, B.; Ketela, T.; Lemieux, S.; Veillette, K.; Martel, N.; Davison, J.; Sillaots, S.; Trosok, S.; Bachewich, C.; et al. Genome-Wide Fitness Test and Mechanism-of-Action Studies of Inhibitory Compounds in Candida albicans. PLoS Pathog. 2007, 3, e92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Meara, T.R.; Veri, A.O.; Ketela, T.; Jiang, B.; Roemer, T.; Cowen, L.E. Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat. Commun. 2015, 6, 6741. [Google Scholar] [CrossRef] [Green Version]
- Eisenhaber, B.; Schneider, G.; Wildpaner, M.; Eisenhaber, F. A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe. J. Mol. Biol. 2004, 337, 243–253. [Google Scholar] [CrossRef]
- Neelamegham, S.; Aoki-Kinoshita, K.; Bolton, E.; Frank, M.; Lisacek, F.; Lütteke, T.; O’Boyle, N.; Packer, N.; Stanley, P.; Toukach, P.; et al. Updates to the Symbol Nomenclature for Glycans guidelines. Glycobiology 2019, 29, 620–624. [Google Scholar] [CrossRef]
- Gow, N.A.R.; Latge, J.-P.; Munro, C.A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5, 28513415. [Google Scholar] [CrossRef] [Green Version]
- Tyers, M.; Wright, G.D. Drug combinations: A strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. 2019, 17, 141–155. [Google Scholar] [CrossRef]
- Johnson, M.D.; Perfect, J.R. Combination antifungal therapy: What can and should we expect? Bone Marrow Transpl. 2007, 40, 297–306. [Google Scholar] [CrossRef]
- Roemer, T.; Boone, C. Systems-level antimicrobial drug and drug synergy discovery. Nat. Chem. Biol. 2013, 9, 222–231. [Google Scholar] [CrossRef]
- Spitzer, M.; Robbins, N.; Wright, G.D. Combinatorial strategies for combating invasive fungal infections. Virulence 2017, 8, 169–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapoor, M.; Moloney, M.; Soltow, Q.A.; Pillar, C.M.; Shaw, K.J. Evaluation of Resistance Development to the Gwt1 Inhibitor Manogepix (APX001A) in Candida Species. Antimicrob. Agents Chemother. 2019, 64, e01387-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locke, J.B.; Almaguer, A.L.; Zuill, D.E.; Bartizal, K. Characterization of In Vitro Resistance Development to the Novel Echinocandin CD101 in Candida Species. Antimicrob. Agents Chemother. 2016, 60, 6100–6107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liston, S.D.; Whitesell, L.; Kapoor, M.; Shaw, K.J.; Cowen, L.E. Enhanced Efflux Pump Expression in Candida Mutants Results in Decreased Manogepix Susceptibility. Antimicrob. Agents Chemother. 2020, 64, e00261-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niimi, M.; Niimi, K.; Tanabe, K.; Cannon, R.D.; Lamping, E. Inhibitor-Resistant Mutants Give Important Insights into Candida albicans ABC Transporter Cdr1 Substrate Specificity and Help Elucidate Efflux Pump Inhibition. Antimicrob. Agents Chemother. 2022, 66, e0174821. [Google Scholar] [CrossRef]
- Déri, M.; Szakál-Tóth, Z.; Fekete, F.; Mangó, K.; Incze, E.; Minus, A.; Merkely, B.; Sax, B.; Monostory, K. CYP3A-status is associated with blood concentration and dose-requirement of tacrolimus in heart transplant recipients. Sci. Rep. 2021, 11, 21389. [Google Scholar] [CrossRef]
- Powell, J.D.; Zheng, Y. Dissecting the mechanism of T-cell anergy with immunophilin ligands. Curr. Opin. Investig. Drugs 2006, 7, 1002–1007. [Google Scholar] [PubMed]
- Hoy, M.J.; Park, E.; Lee, H.; Lim, W.Y.; Cole, D.C.; DeBouver, N.D.; Bobay, B.G.; Pierce, P.G.; Fox, D.; Ciofani, M.; et al. Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus. MBio 2022, 13, e0104922. [Google Scholar] [CrossRef] [PubMed]
- Gobeil, S.M.-C.; Bobay, B.G.; Juvvadi, P.R.; Cole, D.C.; Heitman, J.; Steinbach, W.J.; Venters, R.A.; Spicer, L.D. Leveraging Fungal and Human Calcineurin-Inhibitor Structures, Biophysical Data, and Dynamics To Design Selective and Nonimmunosuppressive FK506 Analogs. MBio 2021, 12, e0300021. [Google Scholar] [CrossRef]
- Gaynor, E.C.; Mondésert, G.; Grimme, S.J.; Reed, S.I.; Orlean, P.; Emr, S.D. MCD4 encodes a conserved endoplasmic reticulum membrane protein essential for glycosylphosphatidylinositol anchor synthesis in yeast. Mol. Biol. Cell 1999, 10, 627–648. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liston, S.D.; Whitesell, L.; Kapoor, M.; Shaw, K.J.; Cowen, L.E. Calcineurin Inhibitors Synergize with Manogepix to Kill Diverse Human Fungal Pathogens. J. Fungi 2022, 8, 1102. https://doi.org/10.3390/jof8101102
Liston SD, Whitesell L, Kapoor M, Shaw KJ, Cowen LE. Calcineurin Inhibitors Synergize with Manogepix to Kill Diverse Human Fungal Pathogens. Journal of Fungi. 2022; 8(10):1102. https://doi.org/10.3390/jof8101102
Chicago/Turabian StyleListon, Sean D., Luke Whitesell, Mili Kapoor, Karen J. Shaw, and Leah E. Cowen. 2022. "Calcineurin Inhibitors Synergize with Manogepix to Kill Diverse Human Fungal Pathogens" Journal of Fungi 8, no. 10: 1102. https://doi.org/10.3390/jof8101102





