Proteomic Analysis of Apple Response to Penicillium expansum Infection Based on Label-Free and Parallel Reaction Monitoring Techniques
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pathogen
2.2. Fruit
2.3. Determination of Sampling Time Points and Preparation of Apple Samples
2.4. Proteome Sample Preparation
2.5. Proteomic Mass Spectrometry
2.6. PRM Verification of DEPs
2.7. Data Analysis
3. Results
3.1. Result Analysis
3.1.1. Selection of Key Time Points for Sampling
3.1.2. Screening for Differentially Expressed Proteins
3.1.3. Subcellular Localization of Differentially Expressed Proteins
3.1.4. GO Enrichment Analysis of Differentially Expressed Proteins
3.1.5. KEGG Enrichment Analysis of Differentially Expressed Proteins
3.1.6. Validation of Selected Candidates by PRM
3.1.7. Expression of Pathogenesis-Related Proteins
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, H.K.; Wan, Y.Z.; Wang, M.; Huo, X.X. History, status, and prospects of the apple industry in China. J. Am. Pomol. Soc. 2015, 69, 174–185. [Google Scholar]
- Patriarca, A. Fungi and mycotoxin problems in the apple industry. Curr. Opin. Food Sci. 2019, 29, 42–47. [Google Scholar] [CrossRef]
- Gebrie, S.A. Biotrophic fungi infection and plant defense mechanism. J. Plant Pathol. Microbiol. 2016, 7, 2. [Google Scholar] [CrossRef] [Green Version]
- Tannous, J.; Kumar, D.; Sela, N.; Sionov, E.; Prusky, D.; Keller, N.P. Fungal attack and host defence pathways unveiled in near-avirulent interactions of Penicillium expansum creA mutants on apples. Mol. Plant Pathol. 2018, 19, 2635–2650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Li, B.Q.; Xu, X.D.; Zhang, Z.Q.; Tian, S.P. The pH-responsive PacC transcription factor plays pivotal roles in virulence and patulin biosynthesis in Penicillium expansum. Environ. Microbiol. 2018, 20, 4063–4078. [Google Scholar] [CrossRef] [PubMed]
- Prusky, D.; McEvoy, J.L.; Saftner, R.; Conway, W.S.; Jones, R. Relationship between host acidification and virulence of Penicillium spp. on apple and citrus fruit. Phytopathology 2004, 94, 44–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, E.; Ballester, A.R.; Raphael, G.; Feigenberg, O.; Liu, Y.S.; Norelli, J.; Droby, S. Identification and characterization of LysM effectors in Penicillium expansum. PLoS ONE 2017, 12, e0186023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.H.; Yu, Y.; Bi, C.W.; Kang, Z.S. Quantitative proteomics reveals the defense response of wheat against Puccinia striiformis f. sp. tritici. Sci. Rep. 2016, 6, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Ravi, G.; Woo, M.C.; Seungmin, S.; Hyun, L.G.; Woo, J.J.; Wook, K.S.; Tae, K.S. Comparative proteome profiling of susceptible and resistant rice cultivars identified an arginase involved in rice defense against Xanthomonas oryzae pv. oryzae. Plant Physiol. Biochem. 2021, 171, 105–114. [Google Scholar] [CrossRef]
- Lu, B.W.; An, F.X.; Cao, L.J.; Yang, Y.J.; Liu, P.M.; Wang, X.; Liu, J. Proteomic profiling uncovered the cytosolic superoxide dismutase BsSOD1 associated with plant defense in the herbal orchid Bletilla striata. Funct. Plant Biol. 2020, 47, 937–944. [Google Scholar] [CrossRef]
- Grandellis, C.; Vranych, C.V.; Piazza, A.; Garavaglia, B.S.; Gottig, N.; Ottado, J. An overview of proteomics tools for understanding plant defense against pathogens. Curr. Issues Mol. Biol. 2016, 19, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.Y.; Misran, A.; Daim, L.D.J.; Lau, B.Y.C. Optimization of protein extraction for proteomic analyses of fresh and frozen “Musang King” durian pulps. Food Chem. 2021, 343, 128471. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, Y.; Barkallah, M.; Bouazizi, E.; Hibar, K.; Gdoura, R.; Triki, M.A. Lignification, phenol accumulation, induction of PR proteins, and antioxidant-related enzymes are key factors in the resistance of Olea europaea to verticillium wilt of olive. Acta Physiol. Plant. 2017, 39, 1–15. [Google Scholar] [CrossRef]
- Dongus, J.A.; Parker, J.E. EDS1 signaling: At the nexus of intracellular and surface receptor immunity. Curr. Opin. Plant Biol. 2021, 62, 102039. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; Lott, A.A.; Zhu, W.; Dufresne, C.P.; Chen, S.X. Mitogen-activated protein kinase 4-regulated metabolic networks. Int. J. Mol. Sci. 2022, 23, 880. [Google Scholar] [CrossRef]
- Zhang, T.; Schneider, J.D.; Lin, C.W.; Geng, S.S.; Ma, T.Y.; Lawrence, S.R.; Chen, S.X. MPK4 phosphorylation dynamics and interacting proteins in plant immunity. J. Proteome Res. 2019, 18, 826–840. [Google Scholar] [CrossRef]
- Tereza, T.; Despina, S.; Anna, K.; Tereza, V.; Jozef, Š. Multifaceted roles of heat shock protein 90 molecular chaperones in plant development. J. Exp. Bot. 2020, 71, 3966–3985. [Google Scholar] [CrossRef]
- Liu, J.Z.; Horstman, H.D.; Braun, E.; Graham, M.A.; Zhang, C.Q.; Navarre, D.; Whitham, S.A. Soybean homologs of MPK4 negatively regulate defense responses and positively regulate growth and development. Plant Physiol. 2011, 157, 1363–1378. [Google Scholar] [CrossRef] [Green Version]
- Philipp, W.; Britta, E.; Tina, R. ZmCPK1, a calcium-independent kinase member of the Zea mays CDPK gene family, functions as a negative regulator in cold stress signalling. Plant Cell Environ. 2015, 38, 544–558. [Google Scholar] [CrossRef]
- Yu, H.X.; Xiao, A.F.; Dong, R.; Fan, Y.Q.; Zhang, X.P.; Liu, C.; Zhang, Z.M. Suppression of innate immunity mediated by the CDPK-Rboh complex is required for rhizobial colonization in medicago truncatula nodules. New Phytol. 2018, 220, 425–434. [Google Scholar] [CrossRef] [Green Version]
- Tamara, V.; Jorge, V.; Satish, K.; Mats, H.; Carmen, C. Emerging complexity in reactive oxygen species production and signaling during the response of plants to pathogens. Plant Physiol. 2010, 154, 444–448. [Google Scholar] [CrossRef]
- Cejudo, F.J.; Sandalio, L.M.; Van Breusegem, F. Understanding plant responses to stress conditions: Redox-based strategies. J. Exp. Bot. 2021, 72, 5785–5788. [Google Scholar] [CrossRef]
- Chang, C.C.C.; Slesak, I.; Jorda, L.; Sotnikov, A.; Melzer, M.; Miszalski, Z.; Karpinski, S. Arabidopsis chloroplastic glutathione peroxidases play a role in cross talk between photooxidative stress and immune responses. Plant Physiol. 2009, 150, 670–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarah, K.N. A review on plant peroxidases. Nova Biol. Reper. 2019, 5, 428–437. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Tian, Y.S.; Xing, X.J.; Xu, Z.S.; Zhu, B.; Fu, X.Y.; Yao, Q.H. Enhancement of phenol stress tolerance in transgenic Arabidopsis plants overexpressing glutathione S-transferase. Plant Growth Regul. 2017, 82, 37–45. [Google Scholar] [CrossRef]
- Wang, J.M.; Liu, H.Y.; Xu, H.M.; Li, M.; Kang, Z.S. Analysis of differential transcriptional profiling in wheat infected by Blumeria graminis f. sp. Tritici using GeneChip. Mol. Biol. Rep. 2012, 39, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Dipali, S.; Giti, V.; Singh, C.A.; Veena, P.; Debasis, C. Rice (Oryza sativa L.) tau class glutathione S-transferase (OsGSTU30) overexpression in Arabidopsis thaliana modulates a regulatory network leading to heavy metal and drought stress tolerance. Metallomics 2019, 11, 375–389. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, Y.J.; Zeng, Q.Y. Overexpression of three orthologous glutathione s-transferases from populus increased salt and drought resistance in Arabidopsis. Biochem. Syst. Ecol. 2019, 83, 57–61. [Google Scholar] [CrossRef]
- Svoboda, T.; Thon, M.R.; Strauss, J. The role of plant hormones in the interaction of Colletotrichum species with their host plants. Int. J. Mol. Sci. 2021, 22, 12454. [Google Scholar] [CrossRef]
- Zhu, F.Y.; Wang, Z.W.; Fang, Y.; Tong, J.H.; Xiang, J.; Yang, K.K.; Wang, R.Z. Study on the role of phytohormones in resistance to watermelon Fusarium wilt. Plants 2022, 11, 156. [Google Scholar] [CrossRef]
- Song, K.; Chen, B.; Cui, Y.; Zhou, L.; Chan, K.G.; Zhang, H.Y.; He, Y.W. The plant defense signal salicylic acid activates the RpfB-dependent quorum sensing signal turnover via altering the culture and cytoplasmic pH in the phytopathogen Xanthomonas campestris. Mbio 2022, 13, e03644-21. [Google Scholar] [CrossRef] [PubMed]
- Czekus, Z.; Kukri, A.; Hamow, K.A.; Szalai, G.; Tari, I.; Ordog, A.; Poor, P. Activation of local and systemic defence responses by flg22 is dependent on daytime and ethylene in intact tomato plants. Int. J. Mol. Sci. 2021, 22, 8354. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.P.; Wang, Y.A.; Xian, Q.Q.; Chen, X.H.; Xu, J. Transcriptome analysis reveals ethylene-mediated defense responses to Fusarium oxysporum f. sp. Cucumerinum infection in Cucumis sativus L. BMC Plant Biol. 2020, 20, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Jiang, T.; Du, K.T.; Chen, H.; Cao, Y.Y.; Xie, J.P.; Zhou, T. Maize phenylalanine ammonia-lyases contribute to resistance to sugarcane mosaic virus infection, most likely through positive regulation of salicylic acid accumulation. Mol. Plant Pathol. 2019, 20, 1365–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, G.X.; Sun, J.T.; Shang, J.P.; Gong, Z.H. Virus-induced gene silencing for phenylalanine ammonia-lyase affects pepper adaption to low temperature. Biol. Plant. 2019, 63, 601–609. [Google Scholar] [CrossRef]
- Christensen, S.A.; Nemchenko, A.; Borrego, E.; Murray, I.; Sobhy, I.S.; Bosak, L.; Kolomiets, M.V. The maize lipoxygenase, ZmLOX10, mediates green leaf volatile, jasmonate, and herbivore-induced plant volatile production for defense against insect attack. Plant J. 2013, 74, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Ruth, M.; Yovanny, I.; Tamara, V.; Satish, K.; Tomás, C.; Mats, H.; Carmen, C. 9-lipoxygenase-derived oxylipins activate brassinosteroid signaling to promote cell wall-based defense and limit pathogen infection. Plant Physiol. 2015, 169, 2324–2334. [Google Scholar] [CrossRef] [Green Version]
- Khodadadi, F.; Tohidfar, M.; Vahdati, K.; Dandekar, A.M.; Leslie, C.A. Functional analysis of walnut polyphenol oxidase gene (JrPPO1) in transgenic tobacco plants and PPO induction in response to walnut bacterial blight. Plant Pathol. 2020, 69, 756–764. [Google Scholar] [CrossRef]
- Niranjan-Raj, S.; Nagaraju, L.S.; Chandra, N.S. Molecular cloning and characterization of pearl millet polyphenol oxidase and its role in defense against downy mildew. J. Plant Prot. Res. 2019, 59, 423–427. [Google Scholar] [CrossRef]
- Yuldashov, U.X.; Matniyazova, H.X.; Azimov, A.A.; Sherimbetov, A.G.; Khamdullaev, S.A.; Rasulova, O.O.; Shavkiev, J.S. Dependence of peroxidase (PO) and polyphenol oxidase (PPO) enzymes activity on plant productivity under the influence of phytopathogen micromycetes in soybean plant (Glycine max (L.) Merr.). Plant Cell Biotechnol. Mol. Biol. 2021, 22, 293–303. Available online: https://www.ikprress.org/index.php/PCBMB/article/view/6383 (accessed on 4 May 2021).
- Renault, H.; Bassard, J.E.; Hamberger, B.; Werck-Reichhart, D. Cytochrome P450-mediated metabolic engineering: Current progress and future challenges. Curr. Opin. Plant Biol. 2014, 19, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Jensen, K.; Møller, B.L. Plant NADPH-cytochrome P450 oxidoreductases. Phytochemistry 2009, 71, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Xia, S.; Wang, X.; Zhang, J.; Qin, H.; Zhang, Y.; Bie, S. Cloning and expression analysis of chalcone synthase and chalcone isomerase encoding genes in Gossypium hirsutum. Agric. Biotechnol. 2018, 7, 15–21+26. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, J.L.; Zhang, X.L.; Zhu, L.M.; Wang, X.F.; Guo, N.; Xing, H. Overexpression of chalcone isomerase (CHI) increases resistance against Phytophthora sojae in soybean. J. Plant Biol. 2018, 61, 309–319. [Google Scholar] [CrossRef]
- Zhang, Q.L.; Xu, C.R.; Wei, H.Y.; Fan, W.Q.; Li, T.Z. Two pathogenesis-related proteins interact with leucine-rich repeat proteins to promote Alternaria leaf spot resistance in apple. Hortic. Res. 2021, 8, 219. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.D.; Nie, J.J.; Lv, L.Q.; Gong, W.; Wang, S.L.; Yang, M.M.; Huang, L.L. A valsa mali effector protein 1 targets apple (Malus domestica) pathogenesis-related 10 protein to promote virulence. Front. Plant Sci. 2021, 12, 741342. [Google Scholar] [CrossRef]
- Pere, M.; Gautier, A.; Marie-Christine, P.; Camille, R.; Christophe, R.; Didier, M.; Jean-François, C. Identification of a Vitis vinifera endo-β-1,3-glucanase with antimicrobial activity against Plasmopara viticola. Mol. Plant Pathol. 2017, 18, 708–719. [Google Scholar] [CrossRef]
- Farahani, A.S.; Taghavi, S.M.; Afsharifar, A.; Niazi, A. Changes in expression of pathogenesis-related gene 1, pathogenesis-related gene 2, phenylalanine ammonia-lyase, and catalase in tomato in response to Pectobacterium carotovorum subsp. carotovorum. J. Plant Pathol. 2016, 98, 525–530. [Google Scholar] [CrossRef]





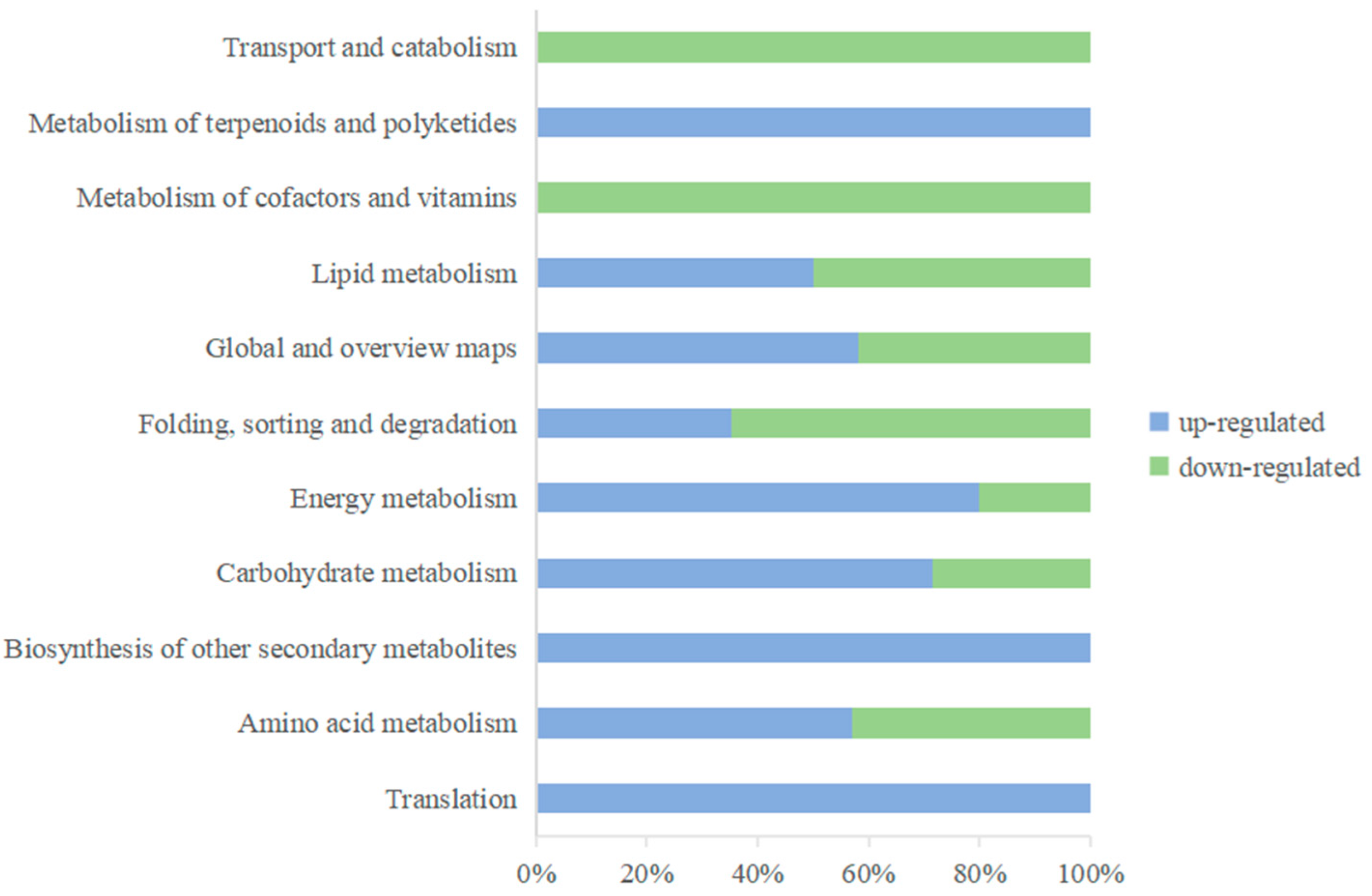

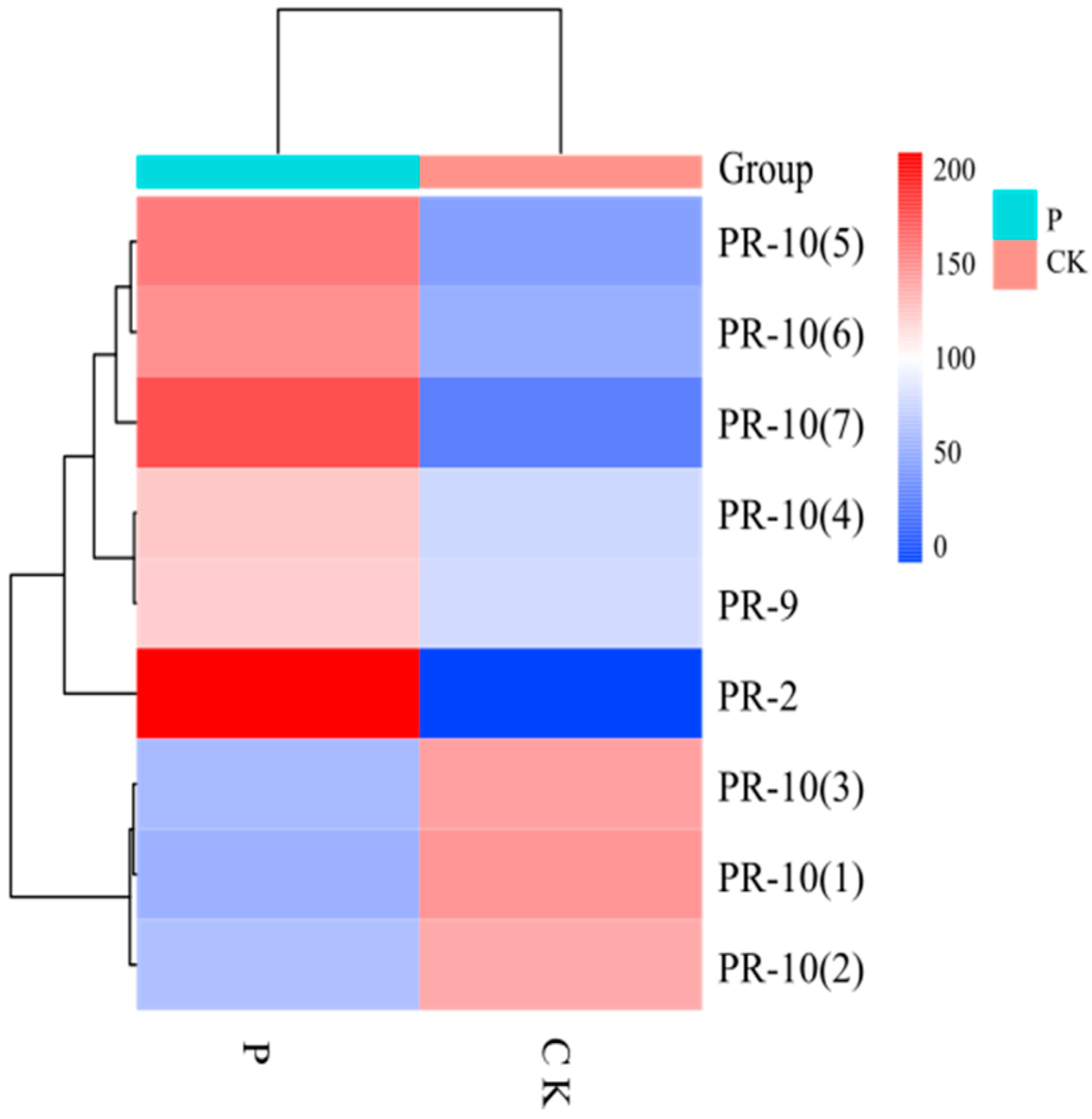
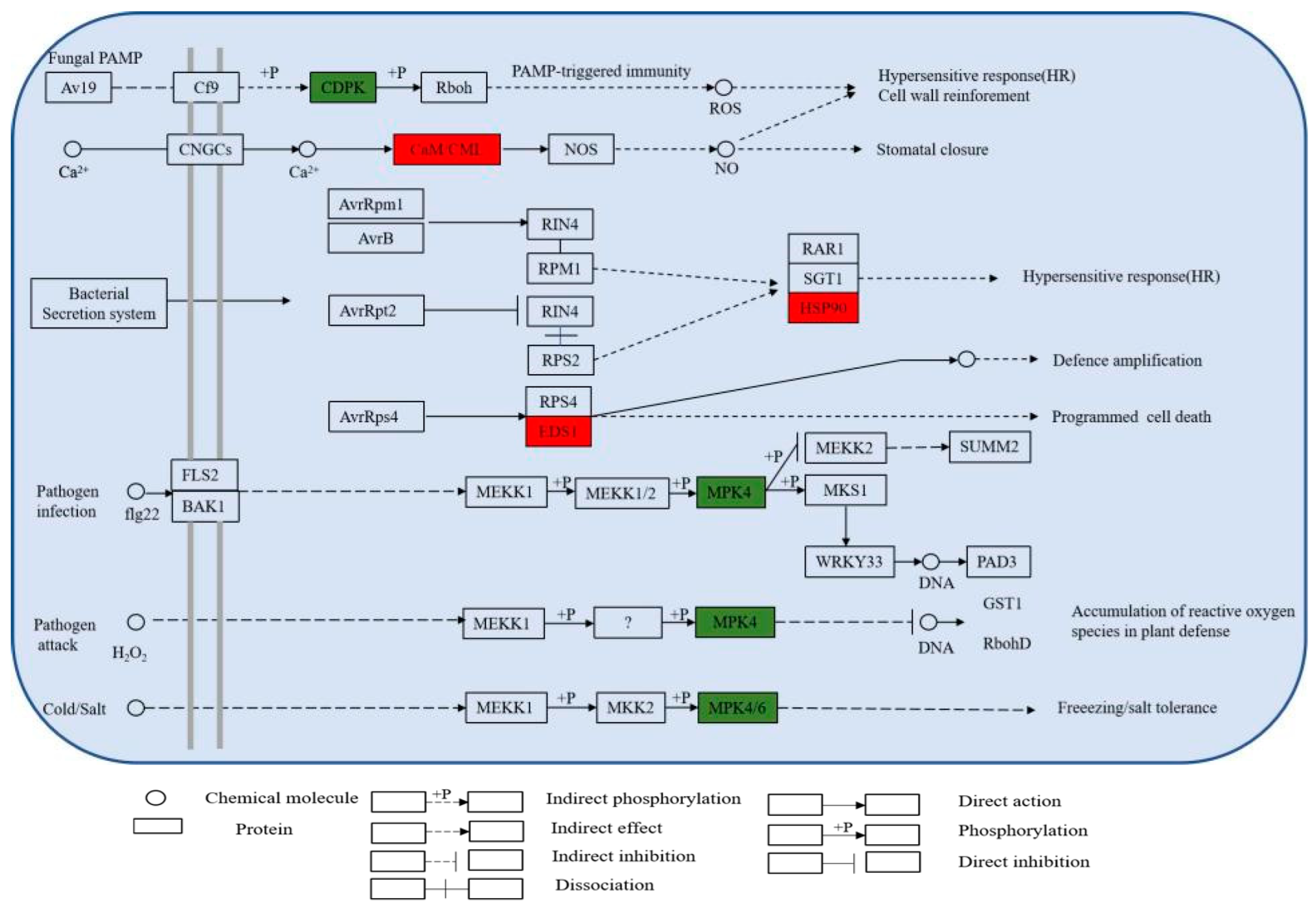

| Pathways | The Number of Upregulated DEPs | The Number of Downregulated DEPs |
|---|---|---|
| Plant-pathogen interaction | 3 | 2 |
| Pyruvate metabolism | 4 | 2 |
| Oxidative phosphorylation | 4 | 0 |
| Alpha-linolenic acid metabolism | 1 | 1 |
| Phenylpropanoid biosynthesis | 5 | 0 |
| Flavonoid biosynthesis | 1 | 0 |
| Glutathione metabolism | 1 | 2 |
| MAPK signaling pathway—plant | 0 | 1 |
| Ubiquitin-mediated proteolysis | 0 | 1 |
| Proteins | Description | FC | p-Value |
|---|---|---|---|
| A0A498K3F2 | Superoxide dismutase [Cu-Zn] | 0.609 | 0.00244296 |
| A0A498K8S2 | Superoxide dismutase [Cu-Zn] | 0.612 | 0.00281705 |
| A0A498INN8 | Superoxide dismutase | 0.645 | 0.01964518 |
| A0A498JNW6 | Glutathione peroxidase | 0.292 | 8.84 × 10−10 |
| S4UL39 | Lipoxygenas | 2.941 | 1.54 × 10−7 |
| A0A498HL71 | Methyltransferase | 2.158 | 0.00218763 |
| A0A498JZC5 | Phenylalanine ammonia-lyase | 2.949 | 9.91 × 10−5 |
| A0A498K3I5 | Chalcone isomerase | 100 | 1.25 × 10−16 |
| A0A498HJB2 | NADPH–cytochrome P450 reductase | 100 | 1.25 × 10−16 |
| A0A498IC26 | NADPH–cytochrome P450 reductase | 2.202 | 0.00085584 |
| Q93XM8 | Polyphenol oxidase 2 | 100 | 1.25 × 10−16 |
| A0A540LHP9 | Peroxidase | 1.855 | 6.51 × 10−5 |
| A0A498HW70 | Glutathione transferase | 1.664 | 0.00087862 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, M.; Wang, K.; Li, J.; Tan, Z.; Godana, E.A.; Zhang, H. Proteomic Analysis of Apple Response to Penicillium expansum Infection Based on Label-Free and Parallel Reaction Monitoring Techniques. J. Fungi 2022, 8, 1273. https://doi.org/10.3390/jof8121273
Xu M, Wang K, Li J, Tan Z, Godana EA, Zhang H. Proteomic Analysis of Apple Response to Penicillium expansum Infection Based on Label-Free and Parallel Reaction Monitoring Techniques. Journal of Fungi. 2022; 8(12):1273. https://doi.org/10.3390/jof8121273
Chicago/Turabian StyleXu, Meng, Kaili Wang, Jun Li, Zhuqing Tan, Esa Abiso Godana, and Hongyin Zhang. 2022. "Proteomic Analysis of Apple Response to Penicillium expansum Infection Based on Label-Free and Parallel Reaction Monitoring Techniques" Journal of Fungi 8, no. 12: 1273. https://doi.org/10.3390/jof8121273
APA StyleXu, M., Wang, K., Li, J., Tan, Z., Godana, E. A., & Zhang, H. (2022). Proteomic Analysis of Apple Response to Penicillium expansum Infection Based on Label-Free and Parallel Reaction Monitoring Techniques. Journal of Fungi, 8(12), 1273. https://doi.org/10.3390/jof8121273







