Genomic Analysis of Stropharia rugosoannulata Reveals Its Nutritional Strategy and Application Potential in Bioremediation
Abstract
:1. Introduction
2. Materials and Methods
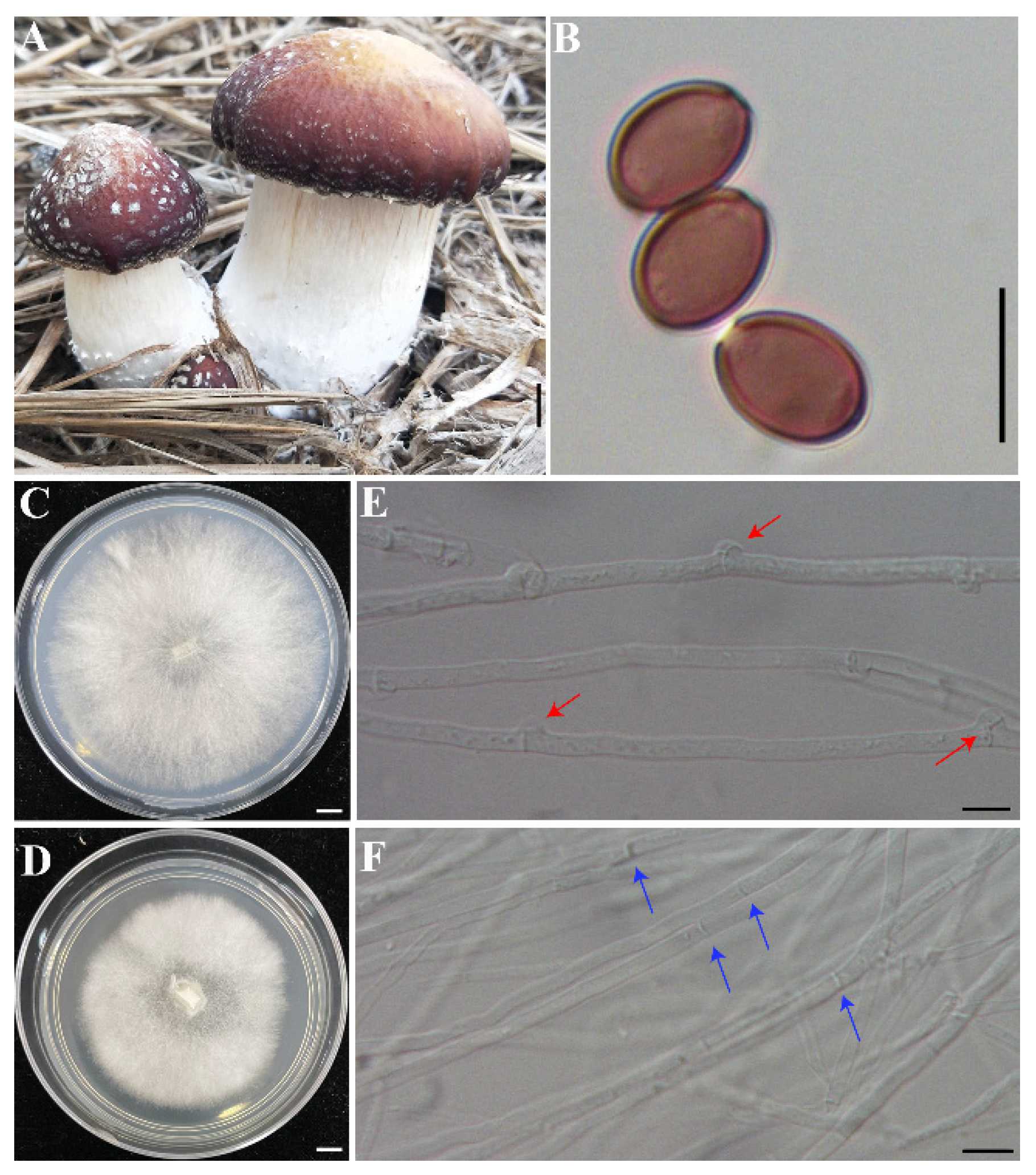
2.1. Origin of the Strain, Culture Conditions, and DNA/RNA Preparation
2.2. Library Construction, Genome/Transcriptome Sequencing, and Assembly
2.3. Repeat Annotation, Gene Prediction, Gene Function, and Noncoding RNA Annotation
2.4. Phylogenomic Analyses
2.5. Putative Peroxidase- and Carbohydrate-Active Enzyme (CAZyme)-Encoding Genes in the Genome of S. rugosoannulata
2.6. Classification of the Nutritional Strategy of S. rugosoannulata by Linear Discriminant Analysis (LDA) Based on the PCWD Gene Families
2.7. Prediction of the Secondary Metabolite Gene Clusters and Cytochrome P450- Encoding Genes
3. Results
3.1. Sequencing Output Processing and De Novo Genome Assembly
3.2. Gene Prediction and Genome-Wide Functional Annotation
3.3. Phylogenomic Analyses
3.4. Putative Peroxidase-Encoding Genes in the Genome of S. rugosoannulata
3.5. The Nutritional Strategy of S. rugosoannulata
3.6. Detection of Secondary Metabolite Clusters
3.7. No Genes Encoding Psilocybin Biosynthesis Enzymes Were Predicted in the Genome of S. rugosoannulata
3.8. Gene Encoding Cytochrome P450 in the Genome of S. rugosoannulata
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Szudyga, K. Stropharia rugosoannulata. In The Biology and Cultivation of Edible Mushrooms; Chang, S.T., Hayes, W.A., Eds.; Academic Press: Cambridge, MA, USA, 1978; pp. 559–571. [Google Scholar]
- De Oliveira, J.B.H.; Pereira, P.R.C.; dos Santos, V.S.; Ferreira, J.M.; Dutra, J.C.V. Stropharia. In Beneficial Microbes in Agro-Ecology: Bacteria and Fungi; Amaresan, N., Kumar, S.M., Annapurna, K., Kumar, K., Sankranarayanan, A., Eds.; Academic Press: Cambridge, MA, USA, 2020; p. 752. [Google Scholar]
- Domondon, D.L.; Poppe, J. Fruit optimization with wastes used for outdoor cultivation of king Stropharia. In Science and Cultivation of Edible Fungi; van Griensven, L.J.L.D., Ed.; Balkema: Rotterdam, The Netherlands, 2000; Volume 2, pp. 909–918. [Google Scholar]
- Wu, J.; Tokuyama, S.; Nagai, K.; Yasuda, N.; Noguchi, K.; Matsumoto, T.; Hirai, H.; Kawagishi, H. Strophasterols A to D with an unprecedented steroid skeleton: From the mushroom Stropharia rugosoannulata. Angew. Chem. Int. Ed. 2012, 51, 10820–10822. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Chen, Y.-Q.; Xiao, X.-W.; Zhong, R.-T.; Yang, C.-F.; Liu, B.; Zhao, C. Nutrient properties and nuclear magnetic resonance-based metabonomic analysis of macrofungi. Foods 2019, 8, 397. [Google Scholar] [CrossRef] [Green Version]
- Steffen, K.T.; Schubert, S.; Tuomela, M.; Hatakka, A.; Hofrichter, M. Enhancement of bioconversion of high-molecular mass polycyclic aromatic hydrocarbons in contaminated non-sterile soil by litter-decomposing fungi. Biodegradation 2007, 18, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Jarosz-Wilkołazka, A.; Kochmańska-Rdest, J.; Malarcz̄yk, E.; Wardas, W.; Leonowicz, A. Fungi and their ability to decolourize azo and anthraquinonic dyes. Enzym. Microb. Technol. 2002, 30, 566–572. [Google Scholar] [CrossRef]
- Weis, M.; Geyer, R.; Günther, T.; Kaestner, M. Fate and stability of 14C-labeled 2,4,6-trinitrotoluene in contaminated soil following microbial bioremediation processes. Environ. Toxicol. Chem. 2004, 23, 2049–2060. [Google Scholar] [CrossRef]
- Kabiersch, G.; Rajasärkkä, J.; Ullrich, R.; Tuomela, M.; Hofrichter, M.; Virta, M.; Hatakka, A.; Steffen, K. Fate of bisphenol A during treatment with the litter-decomposing fungi Stropharia rugosoannulata and Stropharia coronilla. Chemosphere 2011, 83, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Anasonye, F.; Winquist, E.; Kluczek-Turpeinen, B.; Räsänen, M.; Salonen, K.; Steffen, K.T.; Tuomela, M. Fungal enzyme production and biodegradation of polychlorinated dibenzo-p-dioxins and dibenzofurans in contaminated sawmill soil. Chemosphere 2014, 110, 85–90. [Google Scholar] [CrossRef] [Green Version]
- Castellet-Rovira, F.; Lucas, D.; Villagrasa, M.; Rodríguez-Mozaz, S.; Barceló, D.; Sarrà, M. Stropharia rugosoannulata and Gymnopilus luteofolius: Promising fungal species for pharmaceutical biodegradation in contaminated water. J. Environ. Manag. 2018, 207, 396–404. [Google Scholar] [CrossRef]
- Hu, K.; Torán, J.; López-García, E.; Barbieri, M.V.; Postigo, C.; de Alda, M.L.; Caminal, G.; Sarrà, M.; Blánquez, P. Fungal bioremediation of diuron-contaminated waters: Evaluation of its degradation and the effect of amendable factors on its removal in a trickle-bed reactor under non-sterile conditions. Sci. Total Environ. 2020, 743, 140628. [Google Scholar] [CrossRef]
- Bruhn, J.N.; Abright, N.; Mihail, J.D. Forest farming of wine-cap Stropharia mushrooms. Agrofor. Syst. 2010, 79, 267–275. [Google Scholar] [CrossRef]
- Li, D.Y.; Wei, G.J.; Qan, Z.S.; Huang, Y.D.; Hou, Q.G.; Huang, G.J.; Lan, X.D. Stropharia rugoso-annulata cultivation technique in small arch shed with mulberry sawdust. South. Hortic. 2012, 23, 48–49. [Google Scholar]
- Gong, S.; Chen, C.; Zhu, J.; Qi, G.; Jiang, S. Effects of wine-cap Stropharia cultivation on soil nutrients and bacterial communities in forestlands of northern China. PeerJ 2018, 6, e5741. [Google Scholar] [CrossRef] [Green Version]
- Pozdnyakova, N.; Schlosser, D.; Dubrovskaya, E.; Balandina, S.; Sigida, E.; Grinev, V.; Turkovskaya, O. The degradative activity and adaptation potential of the litter-decomposing fungus Stropharia rugosoannulata. World J. Microbiol. Biotechnol. 2018, 34, 133. [Google Scholar] [CrossRef] [PubMed]
- Anasonye, F.; Winquist, E.; Räsänen, M.; Kontroet, J.; Björklöf, K.; Vasilyeva, G.; Jørgensen, K.S.; Steffen, K.T.; Tuomela, M. Bioremediation of TNT contaminated soil with fungi under laboratory and pilot scale conditions. Int. Biodeterior. Biodegrad. 2015, 105, 7–12. [Google Scholar] [CrossRef]
- Gramss, G.; Voigt, K.-D. Basidiospores from wood-decay fungi transform laccase substrates in the absence of glucose and nitrogen supplements. J. Fungi 2020, 6, 62. [Google Scholar] [CrossRef]
- Liers, C.; Arnstadt, T.; Ullrich, R.; Hofrichter, M. Patterns of lignin degradation and oxidative enzyme secretion by different wood- and litter-colonizing basidiomycetes and ascomycetes grown on beech-wood. FEMS Microbiol. Ecol. 2011, 78, 91–102. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wu, S.; Ma, X.; Chen, W.; Zhang, J.; Duan, S.; Gao, Y.; Kui, L.; Huang, W.; Wu, P.; et al. The genome sequences of 90 mushrooms. Sci. Rep. 2018, 8, 9982. [Google Scholar] [CrossRef]
- Suzuki, T.; Ono, A.; Choi, J.-H.; Wu, J.; Kawagishi, H.; Dohra, H. The complete mitochondrial genome sequence of the edible mushroom Stropharia rugosoannulata (Strophariaceae, Basidiomycota). Mitochondrial DNA Part B 2019, 4, 570–572. [Google Scholar] [CrossRef]
- Shang, J.J.; Hou, D.; Li, Y.; Zhou, C.L.; Guo, T.; Tang, L.H.; Mao, W.J.; Chem, Q.; Bao, D.P.; Yang, R.H. Analyses of mating systems in Stropharia rugosoannulata based on genomic data. Mycosystema 2020, 39, 1152–1161. [Google Scholar] [CrossRef]
- Ren, J.; Wang, Q.; Zuo, J.; Jiang, S. Study of thermotolerant mechanism of Stropharia rugosoannulata under high temperature stress based on the transcriptome sequencing. Mycoscience 2021, 62, 95–105. [Google Scholar] [CrossRef]
- Hao, H.B.; Zhang, J.J.; Wu, S.D.; Bai, J.; Zhou, X.Y.; Zhang, J.X.; Kuai, B.K.; Chen, H. Transcriptomic analysis of Stropharia Rugosoannulata reveals potential carbohydrate metabolism and cold resistance mechanisms under low-temperature stress. AMB Express, 2021; in press. [Google Scholar] [CrossRef]
- Floudas, D.; Bentzer, J.; Ahrén, D.; Johansson, T.; Persson, P.; Tunlid, A. Uncovering the hidden diversity of litter-decomposition mechanisms in mushroom-forming fungi. ISME J. 2020, 14, 2046–2059. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L. GMATA: An integrated software package for genome-scale SSR mining, marker development and viewing. Front. Plant Sci. 2016, 7, 1350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abrusán, G.; Grundmann, N.; DeMester, L.; Makalowski, W. TEclass—A tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 2009, 25, 1329–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohler, A.; Kuo, A.; Nagy, L.G.; Morin, E.; Barry, K.W.; Buscot, F.; Canbäck, B.; Choi, C.; Cichocki, N.; Clum, A.; et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat. Genet. 2015, 47, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Fricke, J.; Blei, F.; Hoffmeister, D. Enzymatic synthesis of psilocybin. Angew. Chem. Int. Ed. 2017, 56, 12352–12355. [Google Scholar] [CrossRef] [PubMed]
- Sirén, J.; Valimaki, N.; Mäkinen, V. Indexing graphs for path queries with applications in genome research. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014, 11, 375–388. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Burge, S.W.; Bateman, A.; Daub, J.; Eberhardt, R.; Eddy, S.; Floden, E.; Gardner, P.P.; Jones, T.A.; Tate, J.; et al. Rfam 12.0: Updates to the RNA families database. Nucleic Acids Res. 2015, 43, D130–D137. [Google Scholar] [CrossRef]
- Li, L.; Stoeckert, C.J., Jr.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [Green Version]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [Green Version]
- Vaidya, G.; Lohman, D.J.; Meier, R. Sequence Matrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FigTree, version 1.4.2; 2014. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 3 January 2022).
- Choi, J.; Détry, N.; Kim, K.-T.; Asiegbu, F.O.; Valkonen, J.P.; Lee, Y.-H. fPoxDB: Fungal peroxidase database for comparative genomics. BMC Microbiol. 2014, 14, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merchante, L.F.S.; Grandvalet, Y.; Govaert, G. An efficient approach to sparse linear discriminant analysis. In ICML ’12: Proceedings of the 29th International Conference on Machine Learning; Langford, J., Pineau, J., Eds.; Omnipress: Madison, WI, USA, 2012; pp. 1291–1298. [Google Scholar]
- Nelson, D.R. The cytochrome P450 homepage. Hum. Genom. 2009, 4, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Matheny, P.B.; Curtis, J.M.; Hofstetter, V.; Aime, M.C.; Moncalvo, J.-M.; Ge, Z.-W.; Yang, Z.-L.; Slot, J.C.; Ammirati, J.F.; Baroni, T.J.; et al. Major clades of Agaricales: A multilocus phylogenetic overview. Mycologia 2006, 98, 982–995. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Floudas, D.; Binder, M.; Riley, R.; Barry, K.; Blanchette, R.A.; Henrissat, B.; Martínez, A.T.; Otillar, R.; Spatafora, J.W.; Yadav, J.S.; et al. The paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 2012, 336, 1715–1719. [Google Scholar] [CrossRef] [Green Version]
- Colpa, D.; Fraaije, M.W.; van Bloois, E. DyP-type peroxidases: A promising and versatile class of enzymes. J. Ind. Microbiol. Biotechnol. 2014, 41, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Tsunematsu, Y. Genomics-directed activation of cryptic natural product pathways deciphers codes for biosynthesis and molecular function. J. Nat. Med. 2021, 75, 261–274. [Google Scholar] [CrossRef]
- Garg, U.; Knoblauch, J.; Frazee, C.C., III; Baron, A.; Dudley, M. Accidental death involving psilocin from ingesting “magic mushroom”. In Toxicology Cases for the Clinical and Forensic Laboratory; Elsevier: Amsterdam, The Netherlands, 2020; pp. 379–382. [Google Scholar] [CrossRef]
- Reingardiene, D.; Vilcinskaite, J.; Lazauskas, R. Hallucinogenic mushrooms. Medicine 2005, 41, 1067–1070. [Google Scholar]
- Sticht, G.; Käferstein, H. Detection of psilocin in body fluids. Forensic Sci. Int. 2000, 113, 403–407. [Google Scholar] [CrossRef]
- Siijve, T.; de Meijer, A.A.R. Macromycetes from the state of Paraná, Brazil. 4. The psychoactive species. Arq. Biol. Technol. 1993, 36, 313–329. [Google Scholar]
- Rodríguez-Sáiz, M.; Barredo, J.L.; Moreno, M.A.; Fernández-Cañón, J.M.; Peñalva, M.A.; Díez, B. Reduced function of a phenylacetate-oxidizing cytochrome p450 caused strong genetic improvement in early phylogeny of penicillin-producing strains. J. Bacteriol. 2001, 183, 5465–5471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Peng, J.; Sun, L.; Bonito, G.; Wang, J.; Cui, W.; Fu, Y.; Li, Y. Genome sequencing illustrates the genetic basis of the pharmacological properties of Gloeostereum incarnatum. Genes 2019, 10, 188. [Google Scholar] [CrossRef] [Green Version]
- Gao, W.; Qu, J.; Zhang, J.; Sonnenberg, A.; Chen, Q.; Zhang, Y.; Huang, C. A genetic linkage map of Pleurotus tuoliensis integrated with physical mapping of the de novo sequenced genome and the mating type loci. BMC Genom. 2018, 19, 18. [Google Scholar] [CrossRef]
- Shao, Y.; Guo, H.; Zhang, J.; Liu, H.; Wang, K.; Zuo, S.; Xu, P.; Xia, Z.; Zhou, Q.; Zhang, H.; et al. The Genome of the medicinal macrofungus sanghuang provides insights into the synthesis of diverse secondary metabolites. Front. Microbiol. 2020, 10, 3035. [Google Scholar] [CrossRef] [Green Version]
- Chowdhary, P.; Gargi, S.; Garima, R.; Ferreira, L.F.R.; Bharagava, R.N. Microbial manganese peroxidase: A ligninolytic enzyme and its ample opportunities in research. SN Appl. Sci. 2019, 1, 45. [Google Scholar] [CrossRef] [Green Version]
- Wojaczyńska, E.; Wojaczyński, J. Enantioselective synthesis of sulfoxides: 2000–2009. Chem. Rev. 2010, 110, 4303–4356. [Google Scholar] [CrossRef]
- Cortez, V.G.; Silveira, R.M.B. The agaric genus Stropharia (Strophariaceae) in Rio Grande do Sul state, Brazil. Fungal Divers. 2008, 32, 31–57. [Google Scholar]
- Kabel, M.A.; Jurak, E.; Mäkelä, M.; de Vries, R. Occurrence and function of enzymes for lignocellulose degradation in commercial Agaricus bisporus cultivation. Appl. Microbiol. Biotechnol. 2017, 101, 4363–4369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mydy, L.S.; Bailey, D.C.; Patel, K.D.; Rice, M.R.; Gulick, A.M. The siderophore synthetase IucA of the aerobactin biosynthetic pathway uses an ordered mechanism. Biochemistry 2020, 59, 2143–2153. [Google Scholar] [CrossRef]
- Ahmed, E.; Holmström, S.J.M. Siderophores in environmental research: Roles and applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, H.T.; Vijayakumar, V.; Gluck-Thaler, E.; Korotkin, H.B.; Matheny, P.B.; Slot, J.C. Horizontal gene cluster transfer increased hallucinogenic mushroom diversity. Evol. Lett. 2018, 2, 88–101. [Google Scholar] [CrossRef] [PubMed]








| Genome | Value |
|---|---|
| Length of genome assembly, bp | 48,331,048 |
| No. of scaffolds | 21 |
| Length of the largest scaffold, bp | 4,928,370 |
| Length of the smallest scaffold, bp | 182,917 |
| N50, bp | 2,961,130 |
| N90, bp | 1,348,662 |
| Scaffolds ≥ 5 kb, percentage of assembly, % | 100 |
| GC content, % | 47.35 |
| No. of protein-coding genes | 11,750 |
| Average protein length, aa | 485 |
| Average exon size, bp | 232.84 |
| Average No. of exons per gene | 6.25 |
| Average intron size, bp | 92.77 |
| Classification | Order | Superfamily | Number of Elements | Length of Sequence (bp) | Percentage of Sequence (%) |
|---|---|---|---|---|---|
| Class I (retrotransposons) | 7915 | 6,192,475 | 12.81 | ||
| LTR | 5813 | 5,386,682 | 11.15 | ||
| Gypsy | 2292 | 3,629,891 | 7.51 | ||
| Copia | 987 | 521,946 | 1.08 | ||
| Unknown | 2534 | 1,186,845 | 2.55 | ||
| LINE | 1986 | 790,514 | 1.64 | ||
| SINE | 116 | 15,279 | 0.03 | ||
| Class II (DNA transposons) | 3706 | 2,254,847 | 4.67 | ||
| DNA | 2473 | 1,745,535 | 3.61 | ||
| MITE | 1090 | 417,182 | 0.86 | ||
| RC | Helitron | 143 | 92,130 | 0.19 | |
| Total TEs | 11,621 | 8,447,322 | 17.48 | ||
| Tandem Repeats | 4087 | 227,073 | 0.47 | ||
| Ttandem repeat | 2288 | 204,520 | 0.42 | ||
| SSR | 1799 | 22,553 | 0.05 | ||
| Simple repeats | 203 | 19,120 | 0.04 | ||
| Unknown | 2463 | 927,192 | 1.92 | ||
| Low complexity | 9 | 1419 | 0.00 | ||
| Total repeats | 18,383 | 9,622,126 | 19.91 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Meng, G.; Ni, S.; Zhang, H.; Dong, C. Genomic Analysis of Stropharia rugosoannulata Reveals Its Nutritional Strategy and Application Potential in Bioremediation. J. Fungi 2022, 8, 162. https://doi.org/10.3390/jof8020162
Yang Y, Meng G, Ni S, Zhang H, Dong C. Genomic Analysis of Stropharia rugosoannulata Reveals Its Nutritional Strategy and Application Potential in Bioremediation. Journal of Fungi. 2022; 8(2):162. https://doi.org/10.3390/jof8020162
Chicago/Turabian StyleYang, Ying, Guoliang Meng, Shujun Ni, Haifeng Zhang, and Caihong Dong. 2022. "Genomic Analysis of Stropharia rugosoannulata Reveals Its Nutritional Strategy and Application Potential in Bioremediation" Journal of Fungi 8, no. 2: 162. https://doi.org/10.3390/jof8020162
APA StyleYang, Y., Meng, G., Ni, S., Zhang, H., & Dong, C. (2022). Genomic Analysis of Stropharia rugosoannulata Reveals Its Nutritional Strategy and Application Potential in Bioremediation. Journal of Fungi, 8(2), 162. https://doi.org/10.3390/jof8020162







