Diversity of Ophiostomatoid Fungi Associated with Dendroctonus armandi Infesting Pinus armandii in Western China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Fungi Isolation
2.2. Morphological and Physiological Characteristics
2.3. DNA Extraction and Sequencing
2.4. Phylogenetic Analyses
3. Results
3.1. Fungal Isolation
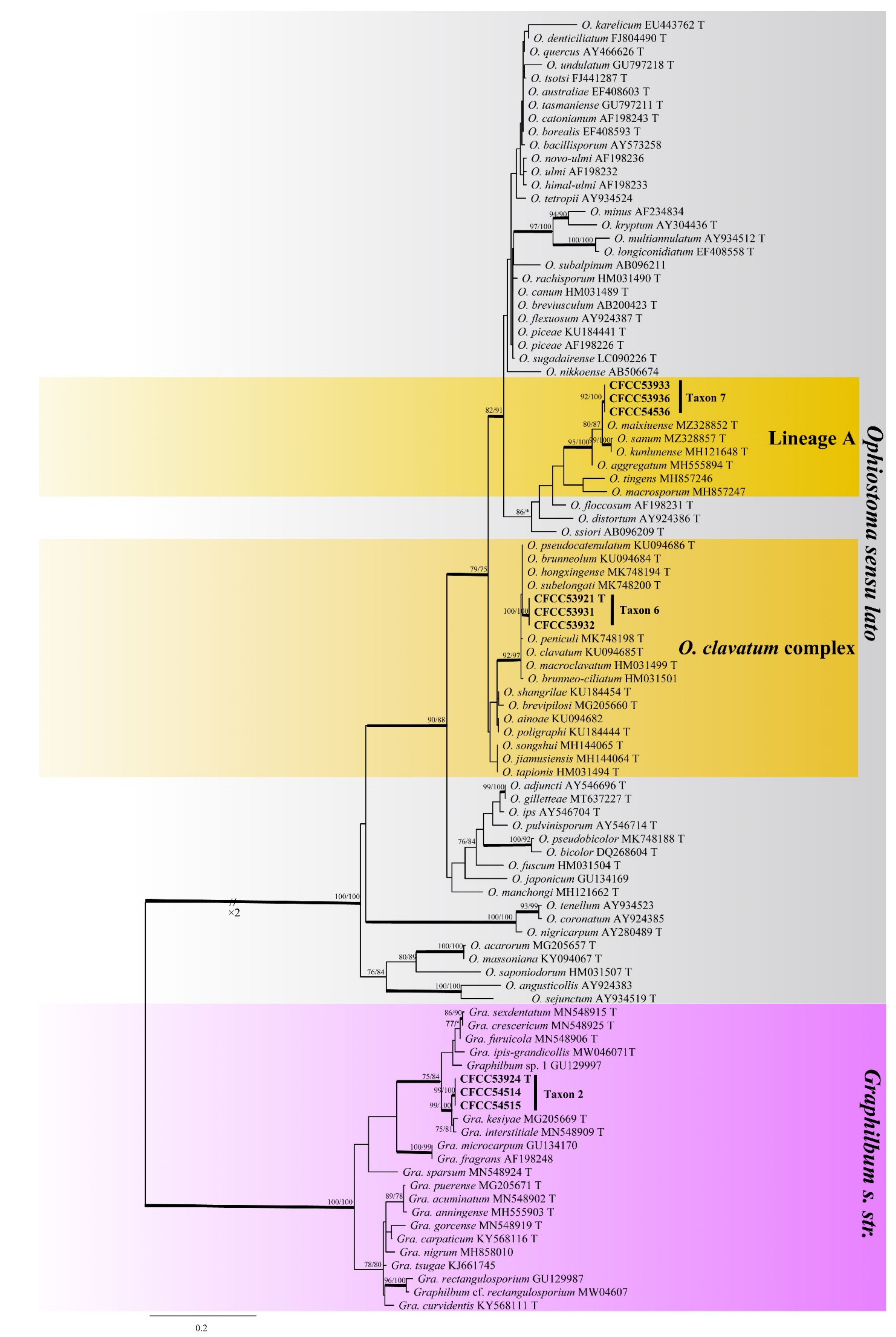
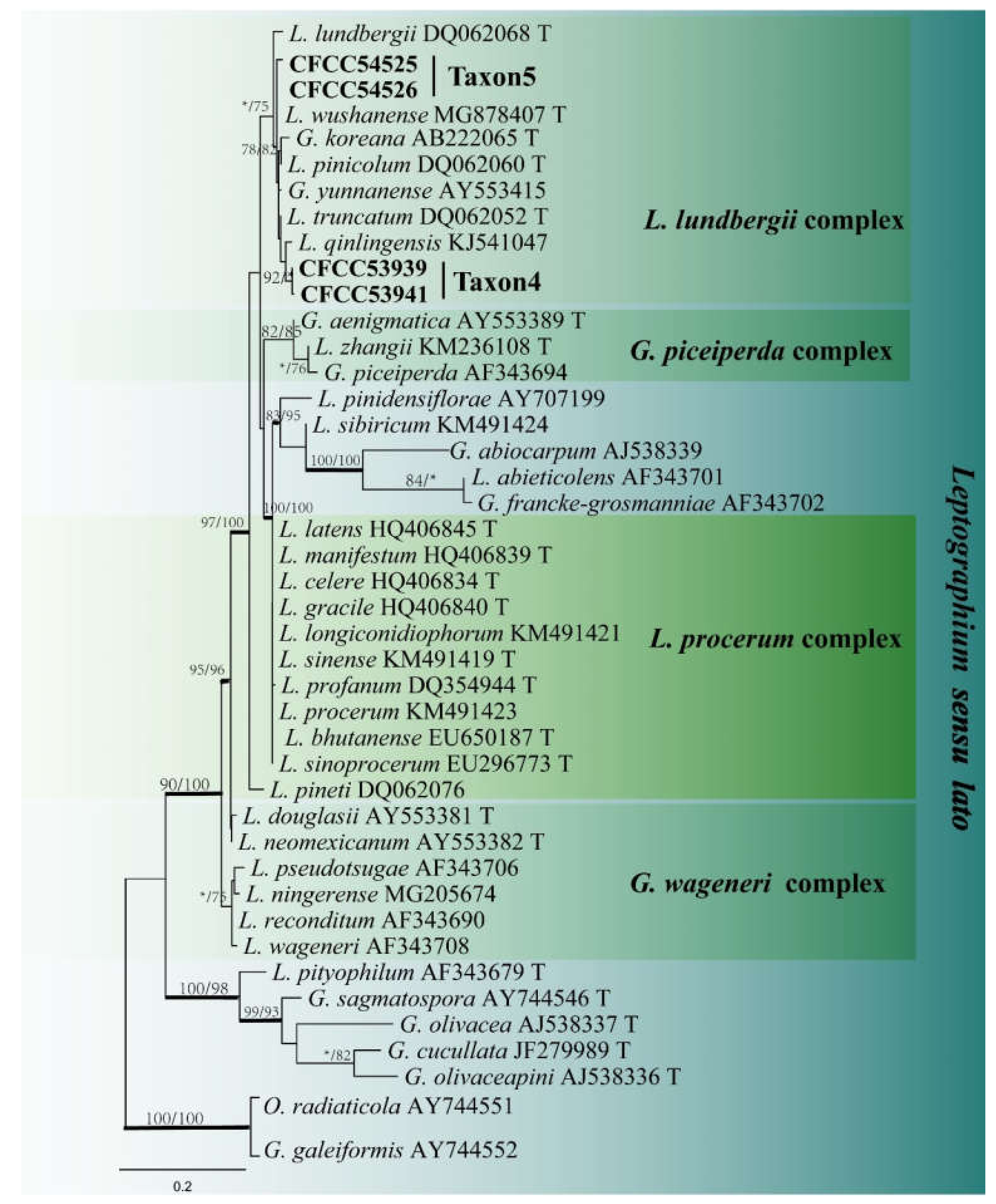
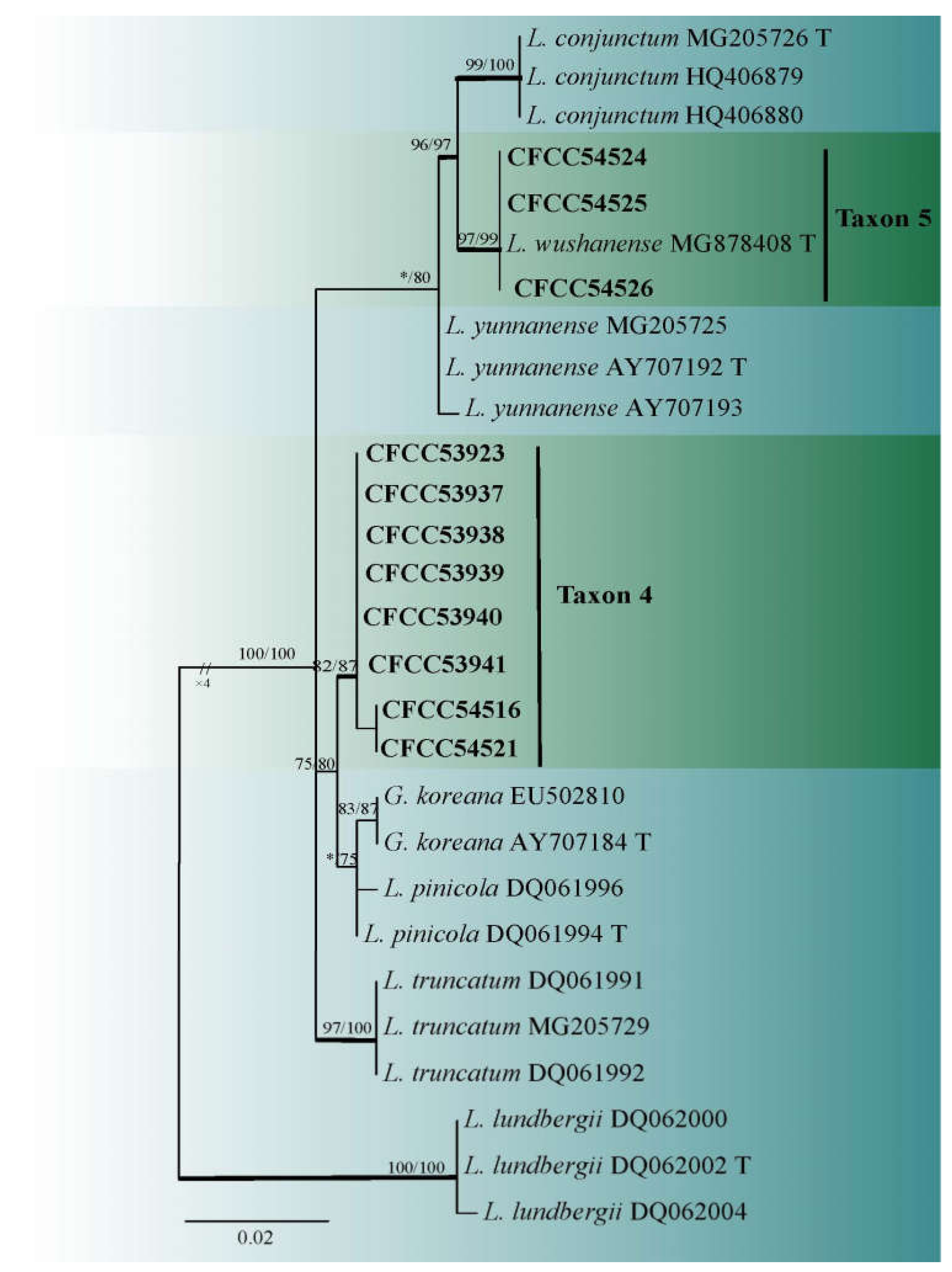
3.2. DNA Sequencing and Phylogenetic Analyses
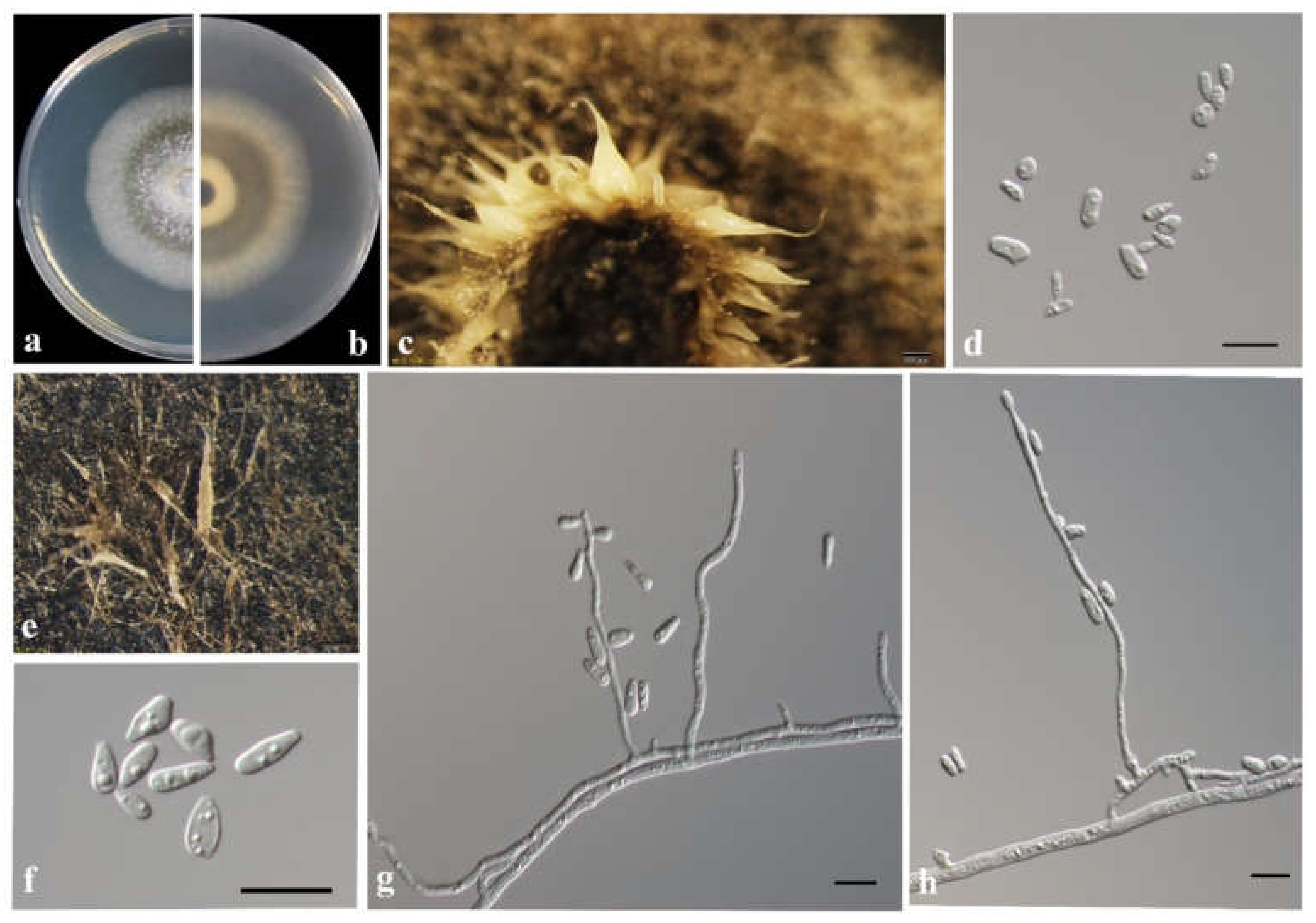
3.3. Morphology and Taxonomy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vega, F.E.; Blackwell, M. Insect-Fungal Associations: Ecology and Evolution; Oxford University Press: New York, NY, USA, 2005. [Google Scholar]
- Skelton, J.; Jusino, M.A.; Carlson, P.S.; Smith, K.; Banik, M.T.; Lindner, D.L.; Palmer, M.; Hulcr, J. Relationships among wood-boring beetles, fungi, and the decomposition of forest biomass. Mol. Ecol. 2019, 28, 4971–4986. [Google Scholar] [CrossRef] [PubMed]
- Biedermann, P.H.W.; Vega, F.E. Ecology and Evolution of Insect-Fungus Mutualisms. Annu. Rev. Entomol. 2020, 65, 431–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidberger, J. Naturgeschichte des Apfelborkenkäfers Apate dispar. Beiträge zur Obstbaumzucht und zur Naturgeschichte der den Obstbäumen Schädlichen Insekten 1836, 4, 213–230. [Google Scholar]
- Hartig, T. Ambrosia des Bostrichus dispar. Allg. Forst-Und Jagdztg. 1844, 13, 73. [Google Scholar]
- Kile, G.A. Plant diseases caused by species of Ceratocystis sensu stricto and Chalara. In Ceratocystis and Ophiostoma: Taxonomy, Ecology and Pathogenicity; Wingfield, M.J., Seifert, K.A., Webber, J., Eds.; APS Press: St. Paul, MN, USA, 1993; pp. 173–184. [Google Scholar]
- Kirisits, T. Fungal associates of European bark beetles with special emphasis on the ophiostomatoid fungi. In Bark and Wood Boring Insects in Living Trees in Europe, A Synthesis; Lieutier, F., Day, K.R., Battisti, A., Gregoire, J.C., Evans, H.F., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2004; pp. 181–235. [Google Scholar]
- Kirisits, T. Dutch Elm Disease and Other Ophiostoma Diseases; CAB International: Wallingford, UK, 2013; pp. 256–282. [Google Scholar]
- Lu, M.; Wingfield, M.J.; Gillette, N.; Sun, J.H. Do novel genotypes drive the success of an invasive bark beetle–fungus complex? Implications for potential reinvasion. Ecology 2011, 92, 2013–2019. [Google Scholar] [CrossRef] [Green Version]
- Bracewell, R.R.; Vanderpool, D.; Good, J.M.; Six, D. Cascading speciation among mutualists and antagonists in a tree-beetle-fungi interaction. Proc. R. Soc. B 2018, 285, 20180694. [Google Scholar] [CrossRef]
- Hulcr, J.; Stelinski, L.L. The Ambrosia Symbiosis: From Evolutionary Ecology to Practical Management. Annu. Rev. Entomol. 2017, 62, 285–303. [Google Scholar] [CrossRef] [Green Version]
- Biedermann, P.H.W.; Rohlfs, M. Evolutionary feedbacks between insect sociality and microbial management. Curr. Opin. Insect Sci. 2017, 22, 92–100. [Google Scholar] [CrossRef]
- Li, H.; Young, S.E.; Poulsen, M.; Currie, C.R. Symbiont-mediated digestion of plant biomass in fungus-farming insects. Annu. Rev. Entomol. 2020, 66, 297–316. [Google Scholar] [CrossRef]
- Li, X.J.; Gao, J.M.; Chen, H.; Zhang, A.L. Toxins from a symbiotic fungus, Leptographium qinlingensis associated with Dendroctonus armandi and their in vitro toxicities to Pinus armandii seedlings. Eur. J. Plant Pathol. 2012, 134, 239–247. [Google Scholar] [CrossRef]
- Cai, B.H. The distribution characteristics of bark beetles and stem-boring pests in China. Shaanxi For. Sci. Technol. 1980, 1, 1–3. [Google Scholar]
- Chen, H.; Tang, M.; Ye, H.M. Niche of bark beetles within Pinus armandii ecosystem in inner Qinling Mountains. Sci. Silvae Sin. 1999, 35, 40–44. [Google Scholar]
- Zhang, Z.Y.; Cha, Y.P.; Wang, S.M. Bionomics of Dendroctonus armandi in Shennongjia forestry district. For. Pest Dis. 2015, 34, 1–4. [Google Scholar]
- Tang, M.; Chen, H. Effect of symbiotic fungi of Dendroctonus armandi on host trees. Sci. Silvae Sin. 1999, 35, 63–66. [Google Scholar]
- Hong, C.; Zeng, B.; Zha, Y.; Hua, X.; Wang, S.; Chen, J. Study on Dendroctonus armandi Diffusion Regular Pattern in Shennongjia Forestry District. Hubei For. Sci. Technol. 2017, 4, 231. [Google Scholar]
- Guerrero, R.; Margulis, L.; Berlanga, M. Symbiogenesis: The holobiont as a unit of evolution. Int. Microbiol. 2013, 16, 133–143. [Google Scholar]
- Hulcr, J.; Barnes, I.; De Beer, Z.W.; Duong, T.A.; Gazis, R.; Johnson, A.J.; Jusino, M.A.; Kasson, M.T.; Li, Y.; Lynch, S.; et al. Bark beetle mycobiome: Collaboratively defined research priorities on a widespread insect-fungus symbiosis. Symbiosis 2020, 81, 101–113. [Google Scholar] [CrossRef]
- Xie, S.A.; Shu, L.J.; Axel, S.; Ding, Y.; Hou, Q.S.; Cai, M. Anatomical characteristics in xylem tissue of Pinus armandii infected by the bark beetle Dendroctonus armandi (Coleoptera: Scolytidae) and its associated blue-stain fungus Ceratocystis polonica. Acta Entomol. Sin. 2008, 51, 1327–1333. [Google Scholar]
- Skelton, J.; Jusino, M.A.; Li, Y.; Bateman, C.; Thai, P.H.; Wu, C.X.; Lindner, D.L.; Hulcr, J. Detecting symbioses in complex communities: The Fungal Symbionts of Bark and Ambrosia Beetles Within Asian Pines. Microb. Ecol. 2018, 76, 839–850. [Google Scholar] [CrossRef]
- Tang, M.; Chen, H.; Zhao, J.P.; Zhu, C.J. Leptographium qinlingensis sp. nov. associated with Dendroctonus armandi in Pinus armandii. J. Huazhong Central China Agric. Univ. 2004, 23, 5–6. [Google Scholar]
- Hu, X.; Li, M.; Chen, H. Community structure of gut fungi during different developmental stages of the Chinese white pine beetle (Dendroctonus armandi). Sci. Rep. 2015, 5, 8411. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, K.; Wingfield, M.J. Leptographium Species: Tree Pathogens, Insect Associates and Agents of Blue-Stain; American Phytopathological Society Press: St Paul, MN, USA, 2001; pp. 164–167. [Google Scholar]
- Marin, M.; Preisig, O.; Wingfield, B.D.; Kirisits, T.; Wingfield, M.J. Phenotypic and DNA sequence data comparisons reveal three discrete species in the Ceratocystis polonica species complex. Mycol. Res. 2005, 109, 1137–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marincowitz, S.; Duong, T.A.; Taerum, S.J.; de Beer, Z.W.; Wingfield, M.J. Fungal associates of an invasive pine-infesting bark beetle, Dendroctonus valens, including seven new Ophiostomatalean fungi. Pers.—Mol. Phylogeny Evol. Fungi 2020, 45, 177–195. [Google Scholar] [CrossRef] [PubMed]
- De Beer, Z.W.; Wingfield, M.J. Emerging lineages in the Ophiostomatales. In The Ophiostomatoid Fungi: Expanding Frontiers; Seifert, K.A., de Beer, Z.W., Wingfield, M.J., Eds.; CBS-KNAW Fungal Biodiversity Centre: Utrecht, The Netherlands, 2013; pp. 21–46. [Google Scholar]
- Seifert, K.A.; Webber, J.F.; Wingfield, M.J. Methods for Studying Species of Ophiostoma and Ceratocystis. In Ceratocystis and Ophiostoma: Taxonomy, Ecology and Pathogenicity; The American Phytopathological Society Press: St. Paul, MN, USA, 1993; pp. 255–259. [Google Scholar]
- Rayner, R.W. A Mycological Colour Chart; CMI: Egham, UK; British Mycological Society: London, UK, 1970. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Application; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [Green Version]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar]
- Jacobs, K.; Bergdahl, D.R.; Wingfield, M.J.; Halik, S.; Seifert, K.A.; Bright, D.E.; Wingfield, B.D. Leptographium wingfieldii introduced into North America and found associated with exotic Tomicus piniperda and native bark beetles. Mycol. Res. 2004, 108, 411–418. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony; Version 4; Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- De Beer, Z.W.; Duong, T.A.; Wingfield, M.J. The divorce of Sporothrix and Ophiostoma: Solution to a problematic relationship. Stud. Mycol. 2016, 83, 165–191. [Google Scholar] [CrossRef] [Green Version]
- De Beer, Z.W.; Seifert, K.A.; Wingfield, M.J. A nomenclator for ophiostomatoid genera and species in the Ophiostomatales and Microascales. In The Ophiostomatoid Fungi: Expanding Frontiers; Seifert, K.A., de Beer, Z.W., Wingfield, M.J., Eds.; CBS-KNAW Fungal Biodiversity Centre: Utrecht, The Netherlands, 2013; pp. 245–322. [Google Scholar]
- Pan, Y.; Lu, J.; Zhou, X.D.; Chen, P.; Zhang, H.; Ye, H. Leptographium wushanense sp. nov. associated with Tomicus armandii on Pinus armandii in Southwestern China. Mycoscience 2018, 61, 43–48. [Google Scholar] [CrossRef]
- Chang, R.L.; Duong, T.A.; Taerum, S.J.; Wingfield, M.J.; Zhou, X.D.; de Beer, Z.W. Ophiostomatoid fungi associated with conifer-infesting beetles and their phoretic mites in Yunnan, China. MycoKeys 2017, 115, 317–318. [Google Scholar] [CrossRef] [Green Version]
- Masuya, H.; Kim, J.J.; Wingfield, M.J.; Yamaoka, Y.; Kim, G.H. Discovery and description of a teleomorph for Leptographium koreanum. Mycotaxon 2005, 94, 159–173. [Google Scholar]
- Jacobs, K.; Solheim, H.; Wingfield, B.D.; Wingfield, M.J. Taxonomic re-evaluation of Leptographium lundbergii based on DNA sequence comparisons and morphology. Myco. Res. 2005, 109, 1149–1161. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.J.; Lim, Y.W.; Breuil, C.; Wingfield, M.; Zhou, X.D.; Kim, G.H. A new Leptographium species associated with Tomicus piniperda infesting pine logs in Korea. Mycol. Res. 2005, 109, 275–284. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.; Decock, C.; Zhang, X.Y.; Maraite, H. Ophiostomatoid fungi (Ascomycota) associated with Pinus tabuliformis infested by Dendroctonus valens (Coleoptera) in northern China and an assessment of their pathogenicity on mature trees. Antonie Van Leeuwenhoek 2009, 96, 275–293. [Google Scholar] [CrossRef]
- Linnakoski, R.; Jankowiak, R.; Villari, C.; Kirisits, T.; Wingfieldet, M.J. The Ophiostoma clavatum species complex: A newly defined group in the Ophiostomatales including three novel taxa. Antonie Van Leeuwenhoek 2016, 109, 987–1018. [Google Scholar] [CrossRef] [Green Version]
- Mathiesen, A. Einige neue Ophiostoma-arten in Schweden. Sven Bot. Tidskr 1951, 45, 203–232. [Google Scholar]
- Chen, H.; Tang, M.; Zhu, C.J.; Hu, J.J. The Enzymes in the Secretions of Dendroctonus armandi (Scolytidae) and Their Symbiotic Fungus of Leptographium qinlingensis. Sci. Silvae Sin. 2004, 40, 123–126. [Google Scholar]
- Pu, X.J.; Chen, H.; Wang, S.J. Influences of Leptographium qinglingensis on Metabolism of Pinus armandi. J. Northwest For. Univ. 2008, 23, 109–112. [Google Scholar]
- Pham, T.; Chen, H.; Yu, J.; Dai, L.; Zhang, R.; Vu, T.Q.T. The Differential effects of the blue-stain fungus Leptographium qinlingensis on monoterpenes and sesquiterpenes in the stem of Chinese white pine (Pinus armandii) saplings. Forests 2014, 5, 2730–2749. [Google Scholar] [CrossRef] [Green Version]
- Liou, J.Y.; Shih, J.Y.; Tzean, S.S. Esteya, a new nematophagous genus from Taiwan, attacking the pinewood nematode (Bursaphelenchus xylophilus). Mycol. Res. 1999, 103, 242–248. [Google Scholar] [CrossRef]
- Kubátová, A.; Novotný, D.; Prášil, K.; Mrácek, Z. The nematophagous hyphomycete Esteya vermicola found in the Czech Republic. Czech Mycol. 2000, 52, 227–235. [Google Scholar] [CrossRef]
- Wang, H.M.; Wang, Z.; Liu, F.; Wu, C.X.; Zhang, S.F.; Kong, X.B.; Decock, C.; Lu, Q.; Zhang, Z. Differential patterns of ophiostomatoid fungal communities associated with three sympatric Tomicus species infesting pines in southwestern China, with description of four new species. MycoKeys 2019, 50, 93–133. [Google Scholar]
- Wang, X.; Wang, T.; Wang, J.; Guan, T.; Li, H. Morphological, molecular and biological characterization of Esteya vermicola, a nematophagous fungus isolated from intercepted wood packing materials exported from Brazil. Mycoscience 2014, 55, 367–377. [Google Scholar] [CrossRef]
- Wang, C.Y.; Fang, Z.M.; Wang, Z.; Gu, J.; Chang, K.S. High infection activities of two Esteya vermicola isolates against pinewood nematode. Afr. J. Microbiol. Res. 2009, 3, 581–584. [Google Scholar]
- Li, Y.; Yu, H.; Araújo, J.P.M.; Zhang, X.; Hulcr, J. Esteya floridanum sp. nov.: An Ophiostomatalean Nematophagous Fungus and its Potential to Control the Pine Wood Nematode. Phytopathology 2020, 111, 304–311. [Google Scholar] [CrossRef]
- Mouton, M.; Wingfield, M.J.; Van Wyk, P.S.; Van Wyk, P.W.J. Graphium pseudormiticum sp. nov.: A new hyphomycete with unusual conidiogenesis. Mycol. Res. 1994, 98, 1272–1276. [Google Scholar] [CrossRef]
- Lackner, M.; de Hoog, G.S. Parascedosporium and its relatives: Phylogeny and ecological trends. IMA Fungus 2011, 2, 39–48. [Google Scholar] [CrossRef]
- Paciura, D.; Zhou, X.D.; De Beer, Z.W.; Jacobs, K.; Ye, H.; Wingfield, M.J. Characterisation of synnematous bark beetle-associated fungi from China, including Graphium carbonarium sp. nov. Fungal Divers. 2010, 40, 75–88. [Google Scholar] [CrossRef] [Green Version]
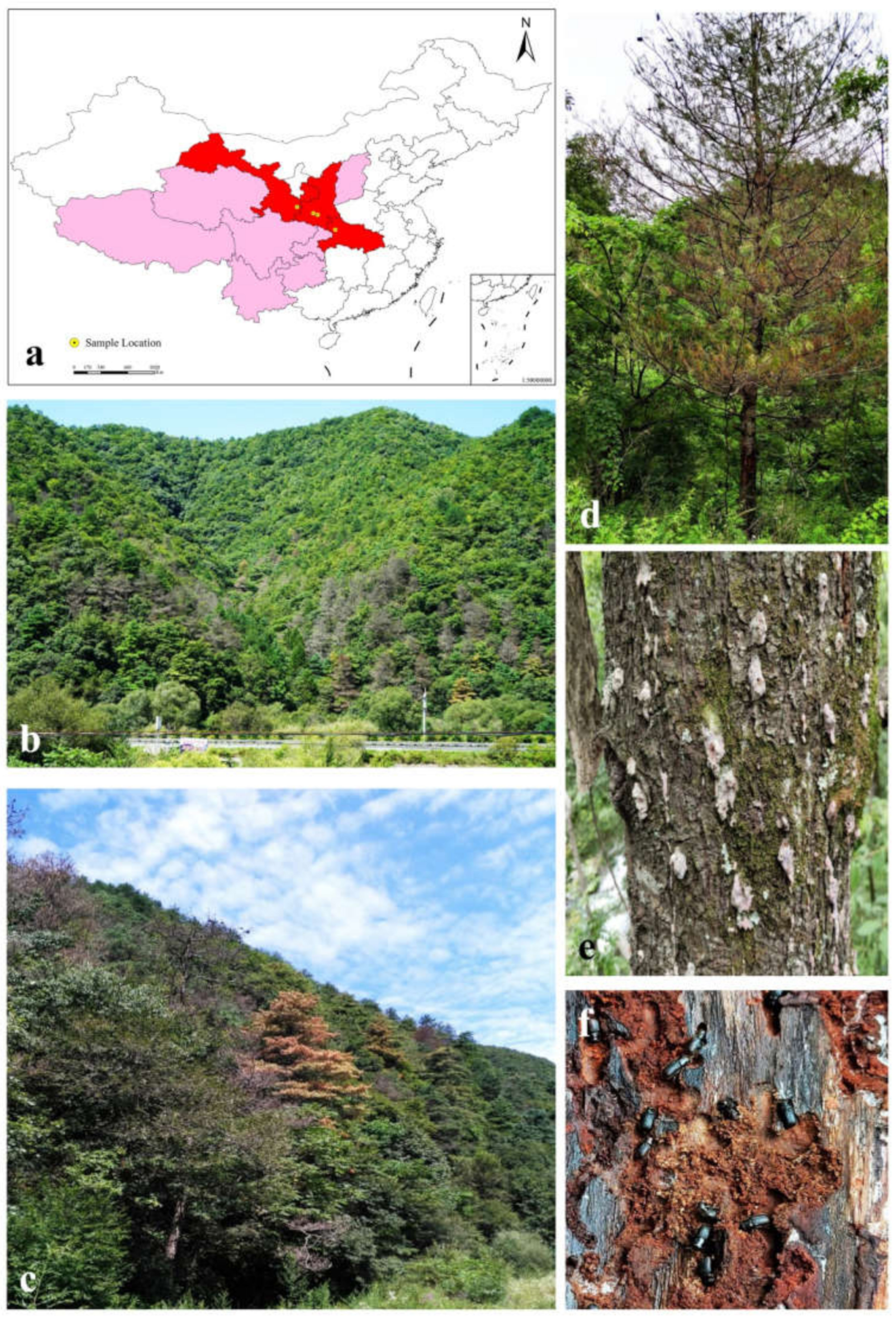




| Location | Longitude\Latitude | Altitude\m | No. of Hosts | No. of Tissue Pieces | No. of Adult Beetles/Galleries |
|---|---|---|---|---|---|
| Shennongjia Forest Area, Hubei Province | 31°45′9″ N, 110°28′34″ E | 1821 | 5 | 381 | 30 |
| Dangchuan Forest Farm, Gansu Province | 34°20′53″ N, 106°7′56″ E | 1482 | 2 | 158 | 10 |
| Foping county, Shaanxi Province | 33°38′22″ N, 107°58′26″ E | 1769 | 2 | 131 | 12 |
| Huoditang Forest Form, Shaanxi Province | 33°27′56″ N, 108°28′27″ E | 2356 | 4 | 370 | 37 |
| Group Taxon | Name | Strain No. | Location | GenBank No. | ||
|---|---|---|---|---|---|---|
| LSU/ITS/ITS2-LSU | TUB2 | TEF1-α | ||||
| Taxon 1 | Esteya vermicola | CFCC53942, CXY2518 | Shennongjia Forest Area, Hubei Province | MW465992 | MW690920 | - |
| Taxon 2 | Graphilbum parakesiyea sp. nov. | CFCC53924, CXY2516 T | Shennongjia Forest Area, Hubei Province | MW459985 | MW770444 | - |
| CFCC54514, CXY2539 | Foping County, Shaanxi Province | MW459986 | MW770445 | - | ||
| CFCC54515, CXY2540 | Foping County, Shaanxi Province | MW459987 | MW770446 | - | ||
| Taxon 3 | Graphium pseudormiticum | CFCC53943, CXY2519 | Shennongjia Forest Area, Hubei Province | MW459988 | - | MW690919 |
| Taxon 4 | Leptographium qinlingense | CFCC53937, CXY2510 | Dangchuan Forest Farm, Gansu Province | MW463377 | MW723023 | MW677124 |
| CFCC53938, CXY2511 | Dangchuan Forest Farm, Gansu Province | MW463378 | MW723024 | MW677125 | ||
| CFCC53923, CXY2512 | Dangchuan Forest Farm, Gansu Province | MW463379 | MW723025 | MW677126 | ||
| CFCC53939, CXY2513 | Dangchuan Forest Farm, Gansu Province | MW463380 | MW723026 | MW677128 | ||
| CFCC53940, CXY2514 | Dangchuan Forest Farm, Gansu Province | MW463381 | MW723027 | MW677129 | ||
| CFCC53941, CXY2515 | Foping County, Shaanxi Province | MW463382 | MW723028 | MW677130 | ||
| CFCC54521, CXY2541 | Foping County, Shaanxi Province | MW463383 | MW723029 | MW677131 | ||
| CFCC54516, CXY2542 | Foping County, Shaanxi Province | MW463384 | MW723030 | MW677127 | ||
| Taxon 5 | L. wushanense | CFCC54524, CXY2543 | Huoditang Forest Farm, Shaanxi Province | MW463385 | MW690921 | - |
| CFCC54525, CXY2544 | Huoditang Forest Farm, Shaanxi Province | MW463386 | MW690922 | - | ||
| CFCC54526, CXY2545 | Huoditang Forest Farm, Shaanxi Province | MW463387 | MW690923 | - | ||
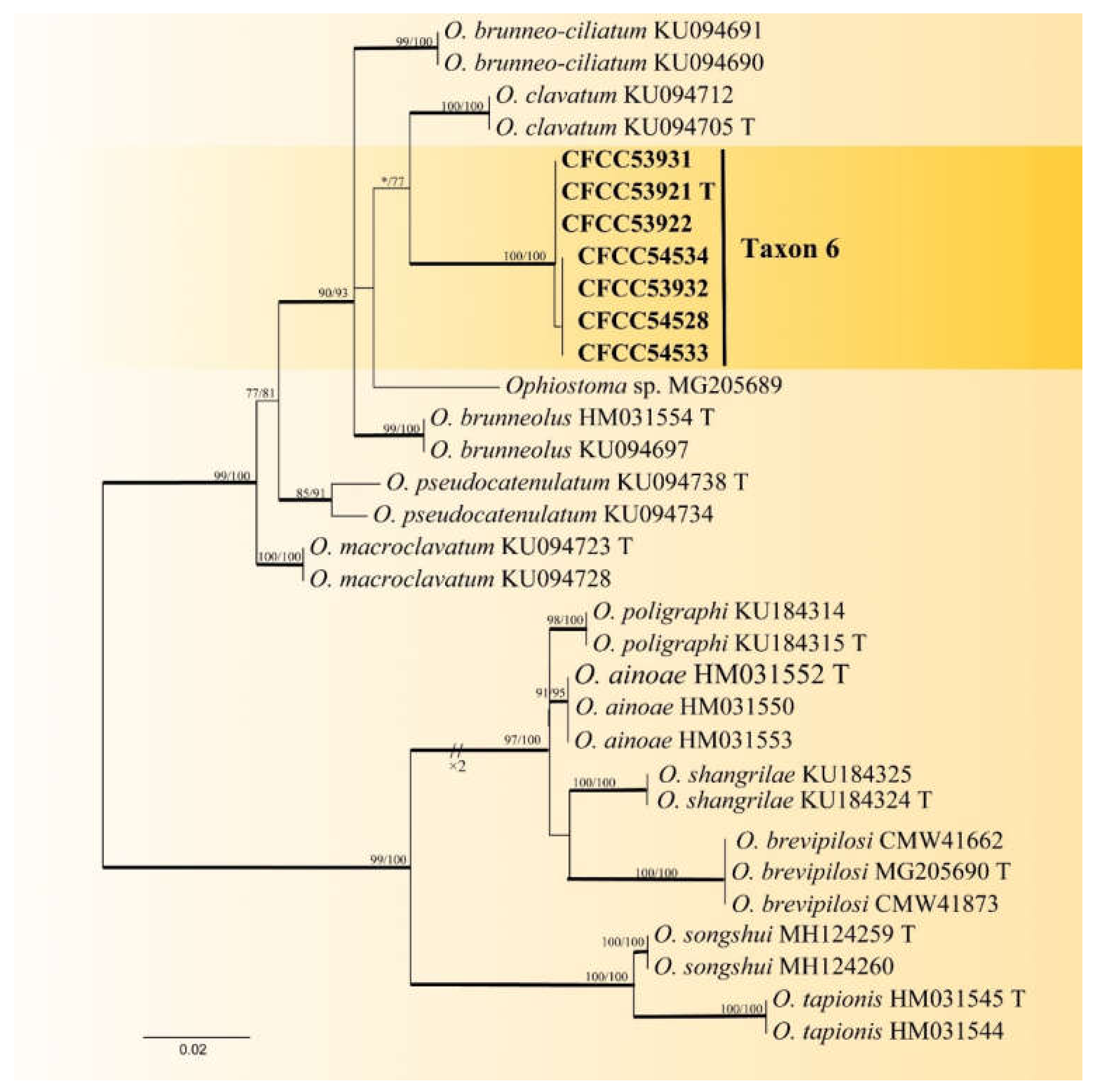
| Taxon 6 | Ophiostoma shennongense sp. nov. | CFCC53921, CXY2501 T | Shennongjia Forest Area, Hubei Province | MW459989 | MW741822 | - |
| CFCC53922, CXY2502 | Shennongjia Forest Area, Hubei Province | MW459990 | MW741823 | - | ||
| CFCC53931, CXY2503 | Dangchuan Forest Farm, Gansu Province | MW459991 | MW741824 | - | ||
| CFCC53932, CXY2505 | Foping County, Shaanxi Province | MW459992 | MW741825 | - | ||
| CFCC54528, CXY2534 | Dangchuan Forest Farm, Gansu Province | MW459993 | MW741826 | - | ||
| CFCC54534, CXY2535 | Huoditang Forest Farm, Shaanxi Province | MW459994 | MW741827 | - | ||
| CFCC54533, CXY2536 | Huoditang Forest Farm, Shaanxi Province | MW459995 | MW741828 | - | ||
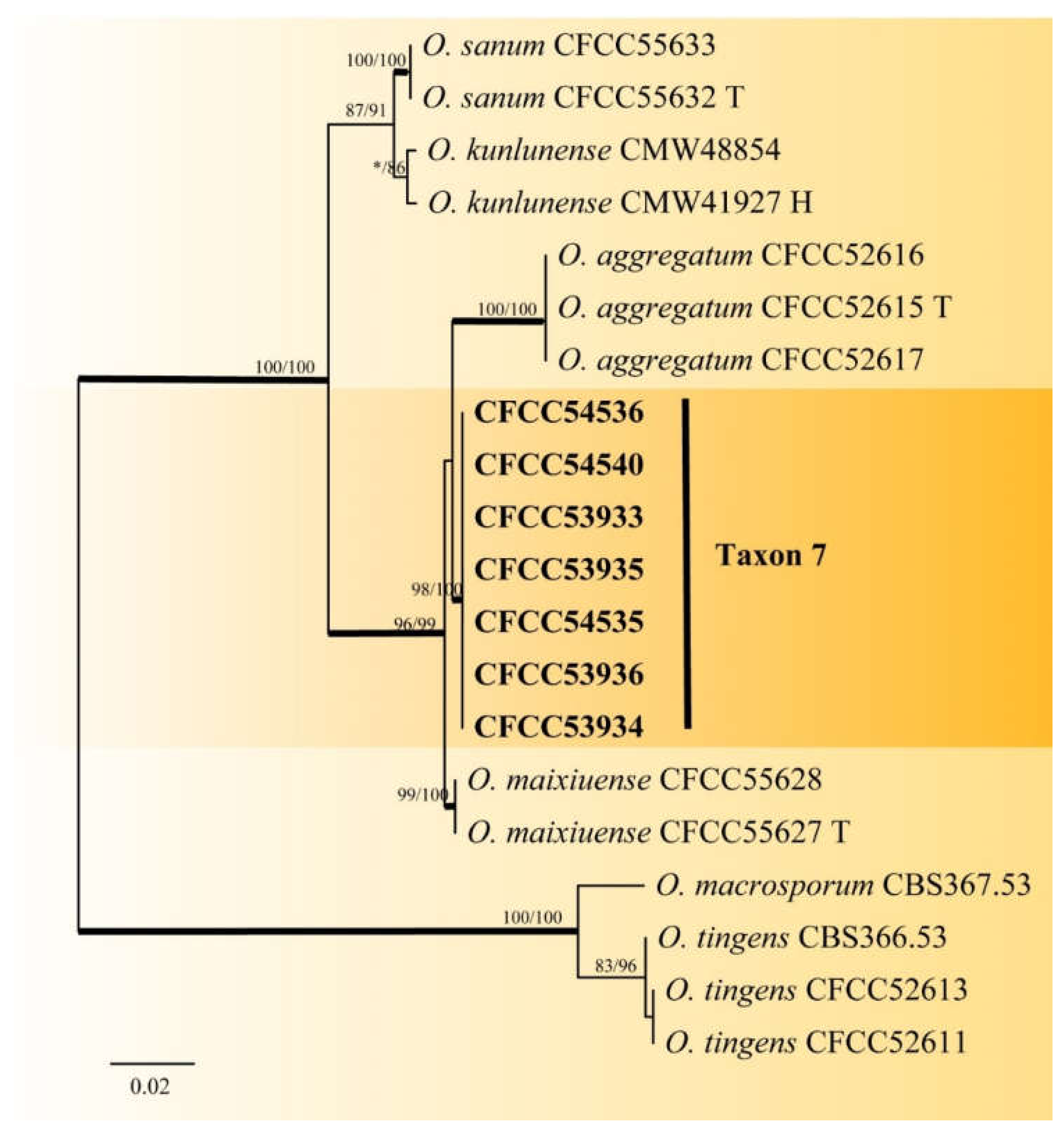
| Taxon 7 | Ophiostoma sp. 1 | CFCC53933, CXY2506 | Shennongjia Forest Area, Hubei Province | MW459996 | MW759042 | - |
| CFCC53934, CXY2507 | Shennongjia Forest Area, Hubei Province | MW459997 | MW759043 | - | ||
| CFCC53935, CXY2508 | Shennongjia Forest Area, Hubei Province | MW459998 | MW759044 | - | ||
| CFCC53936, CXY2509 | Shennongjia Forest Area, Hubei Province | MW459999 | MW759045 | - | ||
| CFCC54535, CXY2531 | Foping County, Shaanxi Province | MW460000 | MW759046 | - | ||
| CFCC54536, CXY2532 | Foping County, Shaanxi Province | MW460001 | MW759047 | - | ||
| CFCC54540, CXY2533 | Huoditang Forest Farm, Shaanxi Province | MW460002 | MW759048 | - | ||
| Taxon | Fungi Species | No. of Isolates 2018 | No. of Isolates 2019 | Total No. Strains | Percentage(%) | ||
|---|---|---|---|---|---|---|---|
| Shennongjia Forest Area | Dangchuan Forest Farm | Foping County | Huoditang Forest Farm | ||||
| Taxon 1 | Esteyea vermicola | 1 | 0 | 0 | 0 | 1 | 0.14 |
| Taxon 2 | Graphilbum parakesiyea | 2 | 0 | 15 | 8 | 25 | 3.60 |
| Taxon 3 | Graphium pseudormiticum | 1 | 0 | 0 | 0 | 1 | 0.14 |
| Taxon 4 | Leptographium qinlingensis | 2 | 24 | 31 | 5 | 62 | 8.92 |
| Taxon 5 | L. wushanense | 0 | 0 | 0 | 34 | 34 | 4.89 |
| Taxon 6 | Ophiostoma shennongensis | 266 | 42 | 27 | 215 | 549 | 78.99 |
| Taxon 7 | Ophiostoma sp. 1 | 6 | 3 | 4 | 9 | 22 | 3.17 |
| Total no. strains | 280 | 69 | 77 | 269 | 695 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Wang, T.; Liu, Y.; Zeng, F.; Zhang, H.; Decock, C.; Zhang, X.; Lu, Q. Diversity of Ophiostomatoid Fungi Associated with Dendroctonus armandi Infesting Pinus armandii in Western China. J. Fungi 2022, 8, 214. https://doi.org/10.3390/jof8030214
Wang H, Wang T, Liu Y, Zeng F, Zhang H, Decock C, Zhang X, Lu Q. Diversity of Ophiostomatoid Fungi Associated with Dendroctonus armandi Infesting Pinus armandii in Western China. Journal of Fungi. 2022; 8(3):214. https://doi.org/10.3390/jof8030214
Chicago/Turabian StyleWang, Huimin, Tiantian Wang, Ya Liu, Fanyong Zeng, Haifeng Zhang, Cony Decock, Xingyao Zhang, and Quan Lu. 2022. "Diversity of Ophiostomatoid Fungi Associated with Dendroctonus armandi Infesting Pinus armandii in Western China" Journal of Fungi 8, no. 3: 214. https://doi.org/10.3390/jof8030214
APA StyleWang, H., Wang, T., Liu, Y., Zeng, F., Zhang, H., Decock, C., Zhang, X., & Lu, Q. (2022). Diversity of Ophiostomatoid Fungi Associated with Dendroctonus armandi Infesting Pinus armandii in Western China. Journal of Fungi, 8(3), 214. https://doi.org/10.3390/jof8030214






