Hydrogenosome, Pairing Anaerobic Fungi and H2-Utilizing Microorganisms Based on Metabolic Ties to Facilitate Biomass Utilization
Abstract
:1. Introduction
2. An Overview of Anaerobic Fungi
2.1. Classification of Anaerobic Fungi
2.2. Digestion of Plant Fiber by Anaerobic Fungi
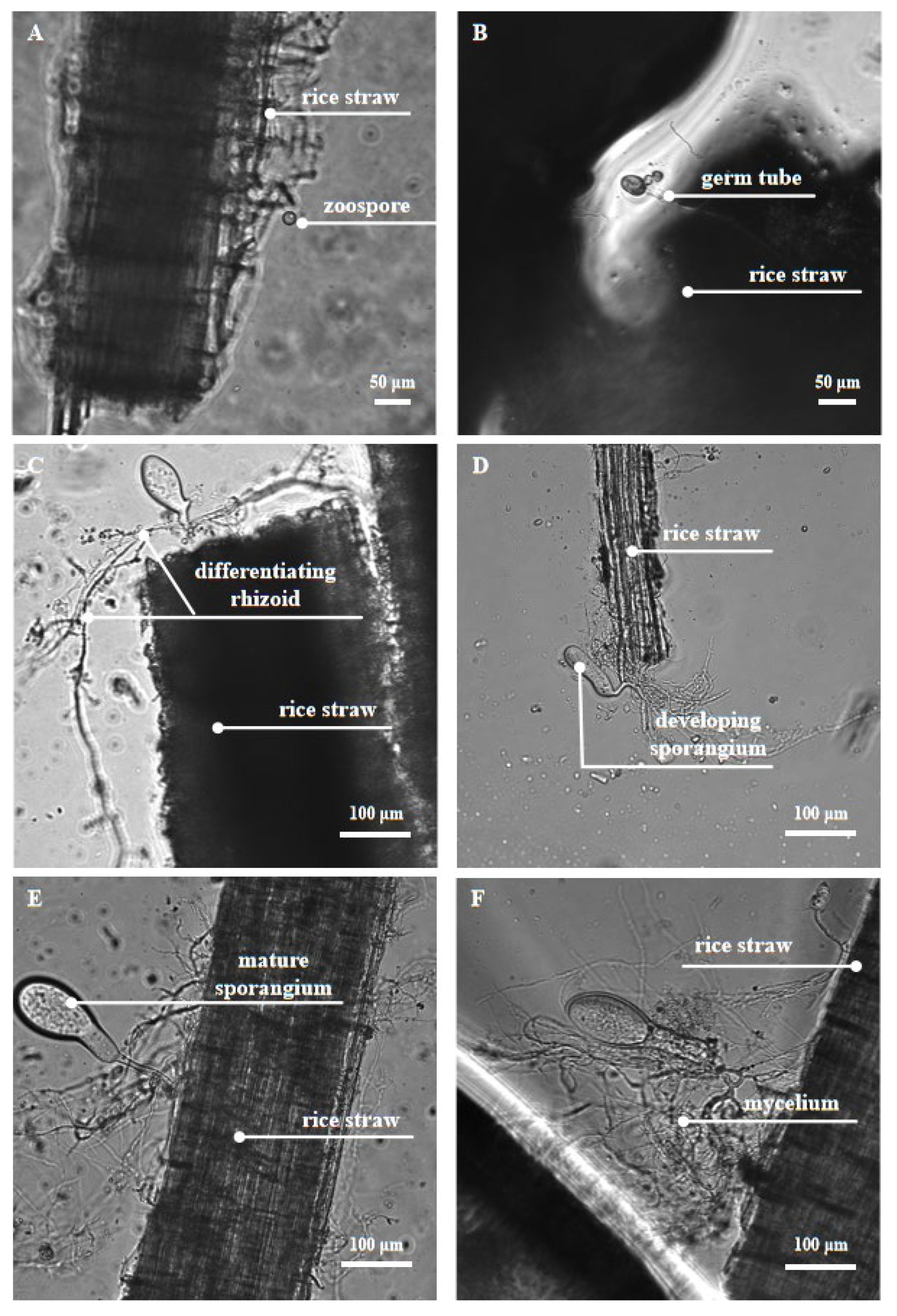
2.2.1. Physical Degradation with Fungal Rhizoids
2.2.2. Digestion by Diverse Plant Fiber-Degrading Enzymes
3. An Overview of the Hydrogenosome
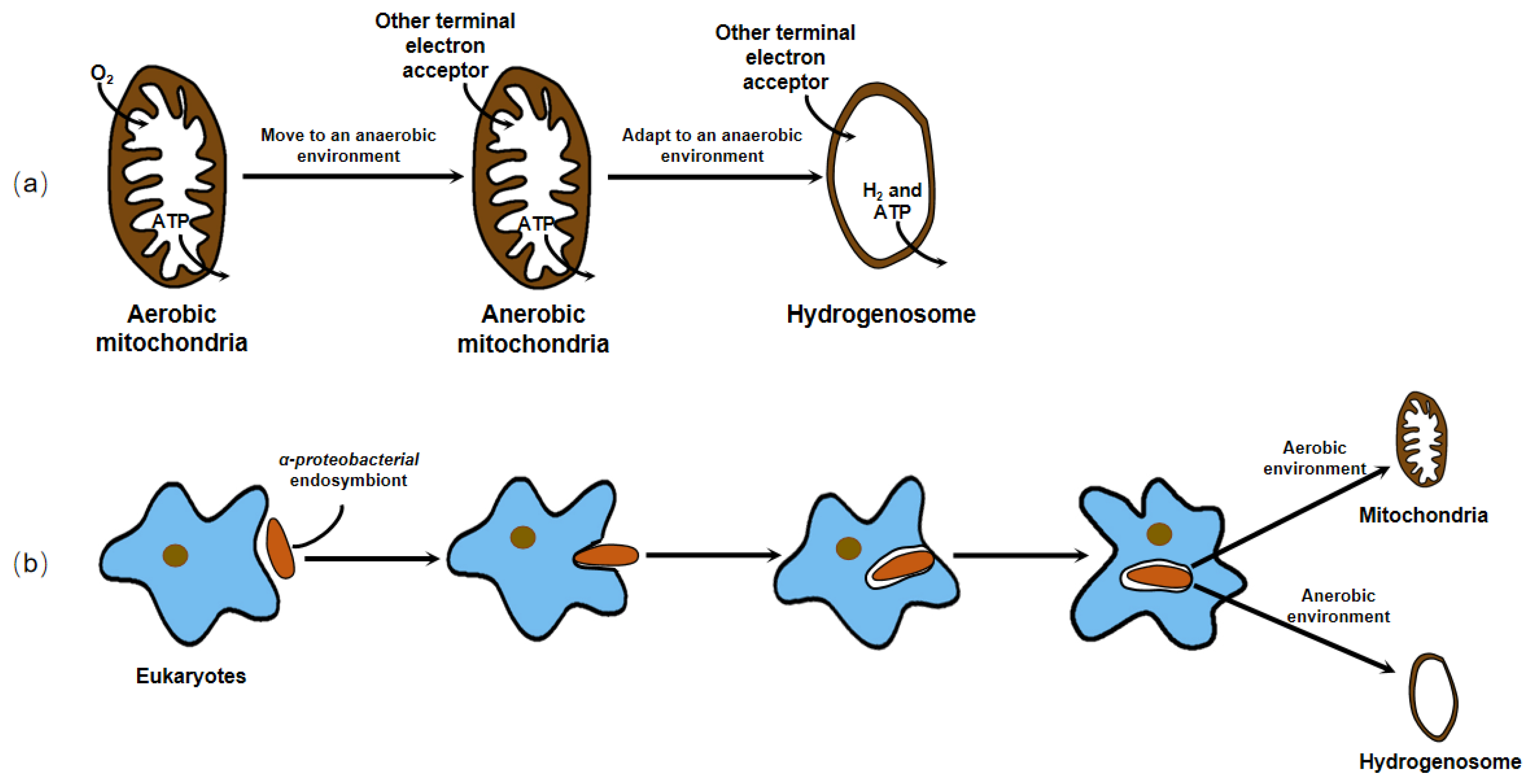
3.1. The Origin of the Hydrogenosome
3.2. Structure and Function of the Hydrogenosome
3.2.1. Structure of the Hydrogenosome
3.2.2. Function of the Hydrogenosome
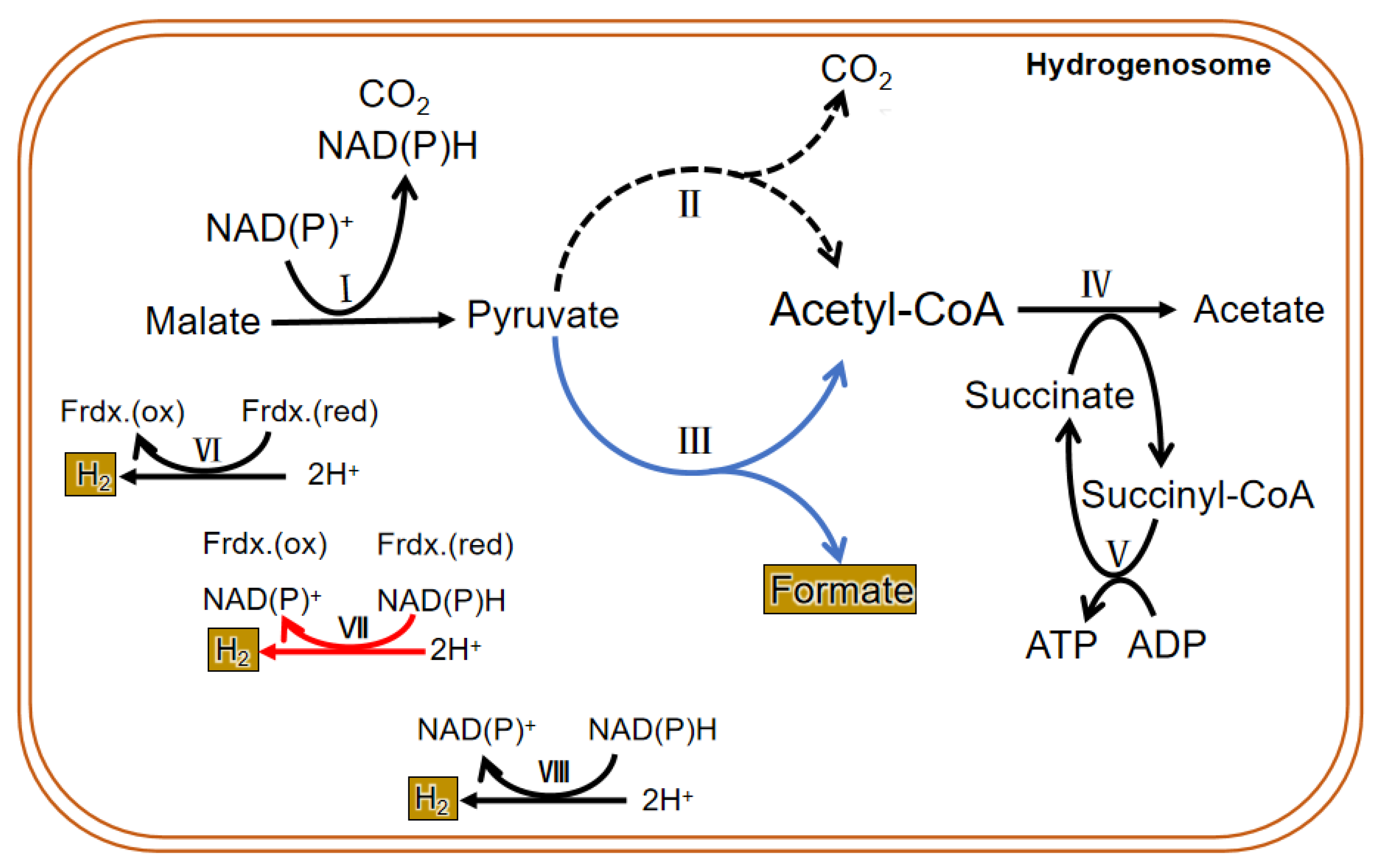
3.3. Metabolism in the Hydrogenosome
3.3.1. Carbohydrate Metabolism
3.3.2. Amino Acid Metabolism
3.3.3. The Unique Metabolism of the Hydrogenosome in Anaerobic Fungi
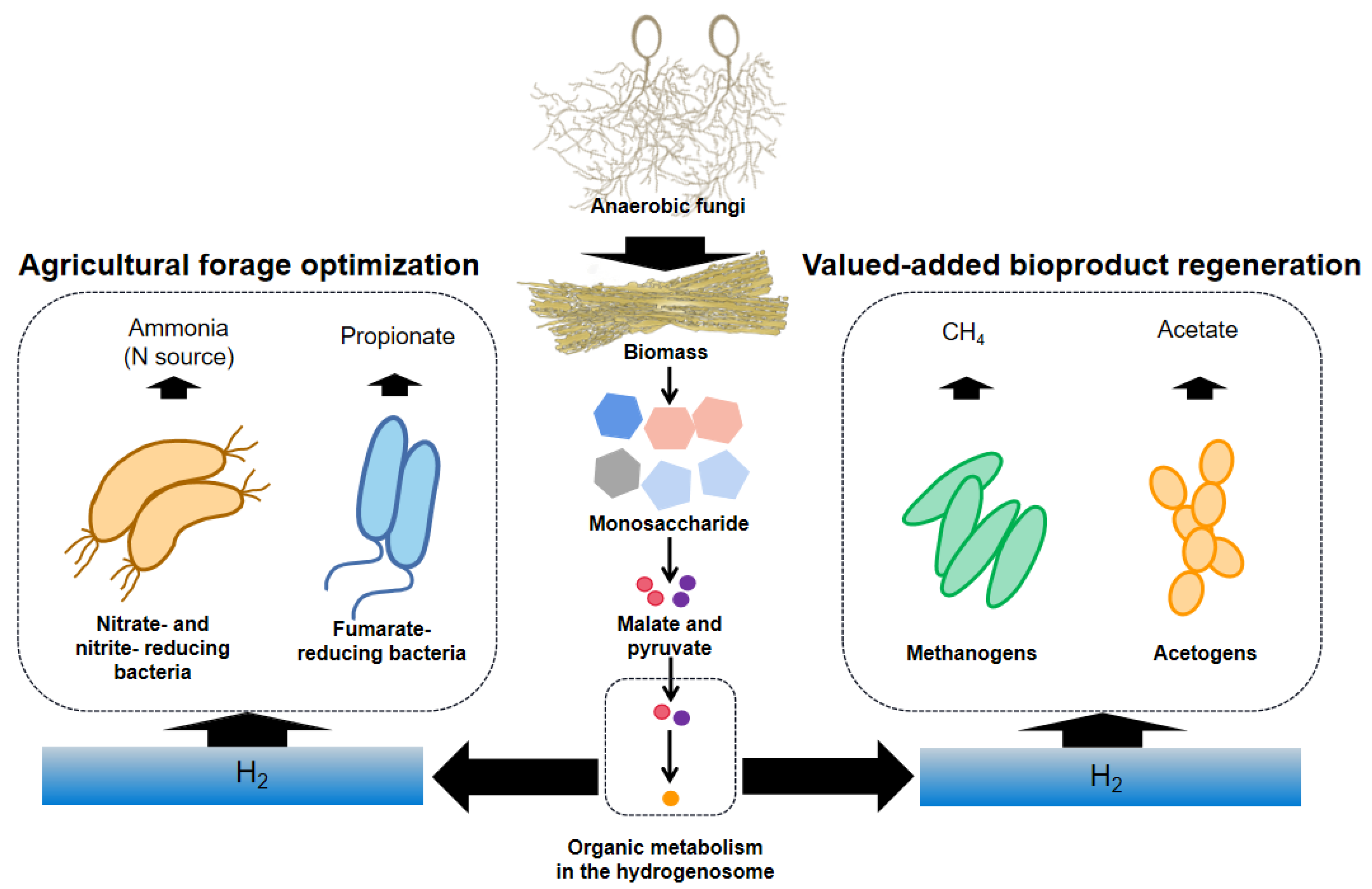
4. The Action of the Hydrogenosome Involved in Promoting the Utilization of Biomass
4.1. The Role of the Hydrogenosome towards CH4 Generation
4.2. The Role of the Hydrogenosome towards Acetate Generation
4.3. The Potential Role of the Hydrogenosome in Improving the Nutritional Value of Feed
4.4. Expansibility and Challenges
5. Conclusions and Expectation
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Creevey, C.J.; Kelly, W.J.; Henderson, G.; Leahy, S.C. Determining the culturability of the rumen bacterial microbiome. Microb. Biotechnol. 2014, 7, 467–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooker, C.A.; Lee, K.Z.; Solomon, K.V. Leveraging anaerobic fungi for biotechnology. Curr. Opin. Biotechnol. 2019, 59, 103–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akin, D.E.; Borneman, W.S.; Lyon, C.E. Degradation of leaf blades and stems by monocentric and polycentric isolates of ruminal fungi. Anim. Feed Sci. Technol. 1990, 31, 205–221. [Google Scholar] [CrossRef]
- Bauchop, T. Rumen anaerobic fungi of cattle and sheep. Appl. Environ. Microbiol. 1979, 38, 148–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kessler, D.; Leibrecht, I.; Knappe, J. Pyruvate-formate-lyase-deactivase and acetyl-coA reductase activities of Escherichia colireside on a polymeric protein particle encoded byadhe. FEBS Lett. 1991, 281, 59–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, M.; Mentel, M.; van Hellemond, J.J.; Henze, K.; Woehle, C.; Gould, S.B.; Yu, R.Y.; van der Giezen, M.; Tielens, A.G.; Martin, W.F. Biochemistry and evolution of anaerobic energy metabolism in mukaryotes. Microbiol. Mol. Biol. Rev. 2012, 76, 444–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McAllister, T.A.; Newbold, C.J. Redirecting rumen fermentation to reduce methanogenesis. Aust. J. Exp. Agric. 2008, 48, 7–13. [Google Scholar] [CrossRef]
- Bauchop, T.; Mountfort, D.O. Cellulose fermentation by a rumen anaerobic fungus in both the absence and the presence of rumen methanogens. Appl. Environ. Microbiol. 1981, 42, 1103–1110. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Jin, W.; Mu, C.; Cheng, Y.; Zhu, W. Indigenously associated methanogens intensified the metabolism in hydrogenosomes of anaerobic fungi with xylose as substrate. J. Basic Microbiol. 2017, 57, 933–940. [Google Scholar] [CrossRef]
- Li, Y.; Jin, W.; Cheng, Y.; Zhu, W. Effect of the associated methanogen Methanobrevibacter thaueri on the dynamic profile of end and intermediate metabolites of anaerobic fungus Piromyces sp. F1. Curr. Microbiol. 2017, 57, 933–940. [Google Scholar] [CrossRef]
- Munn, E.A.; Orpin, C.G. The fine structure of Eadie’s ovals isolated from sheep rumen. J. Gen. Microbiol. 1975, 90, 41–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruninger, R.J.; Puniya, A.K.; Callaghan, T.M.; Edwards, J.E.; Youssef, N.; Dagar, S.S.; Fliegerova, K.; Griffith, G.W.; Forster, R.; Tsang, A.; et al. Anaerobic fungi (phylum Neocallimastigomycota): Advances in understanding their taxonomy, life cycle, ecology, role and biotechnological potential. FEMS Microbiol. Ecol. 2014, 90, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Orpin, C.G. Studies on the rumen flagellate Neocallimastix frontalis. J. Gen. Microbiol. 1975, 91, 249–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orpin, C.G. The occurrence of chitin in the cell walls of the rumen organisms Neocallimastix frontalis, Piromonas communis and Sphaeromonas communis. J. Gen. Microbiol. 1977, 99, 215–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heath, I.B.; Bauchop, T.; Skipp, R.A. Assignment of the rumen anaerobe Neocallimastix frontalis to the Spizellomycetales (Chytridiomycetes) on the basis of its polyflagellate zoospore ultrastructure. Can. J. Bot. 1983, 61, 295–307. [Google Scholar] [CrossRef]
- Gold, J.J.; Heath, I.B.; Bauchop, T. Ultrastructural description of a new Chytrid genus of Caecum anaerobe, Caecomyces equi gen. nov., sp. nov., assigned to the Neocallimasticaceae. Biosystems 1988, 21, 403–415. [Google Scholar] [CrossRef]
- Barr, D.J.S.; Kudo, H.; Jakober, K.D.; Cheng, K.J. Morphology and development of rumen fungi: Neocallimastix sp., Piromyces communis, and Orpinomyces bovis gen. nov., sp. nov. Can. J. Bot. 1989, 67, 2815–2824. [Google Scholar] [CrossRef]
- Breton, A.; Bernalier, A.; Dusser, M.; Fonty, G.; Gaillard-Martinie, B.; Guillot, J. Anaeromyces mucronatus nov. gen., nov. sp. a new strictly anaerobic rumen fungus with polycentric thallus. FEMS Microbiol. Lett. 1990, 70, 177–182. [Google Scholar] [CrossRef]
- Ozkose, E.; Thomas, B.J.; Davies, D.R.; Griffith, G.W.; Theodorou, M.K. Cyllamyces aberensis gen. nov. sp. nov., a new anaerobic gut fungus with branched sporangiophores isolated from cattle. Can. J. Bot. 2001, 79, 666–673. [Google Scholar] [CrossRef]
- Callaghan, T.M.; Podmirseg, S.M.; Hohlweck, D.; Edwards, J.E.; Puniya, A.K.; Dagar, S.S.; Griffith, G.W. Buwchfawromyces eastonii gen. nov., sp. nov.: A new anaerobic fungus (Neocallimastigomycota) isolated from buffalo faeces. MycoKeys 2015, 9, 11–28. [Google Scholar] [CrossRef]
- Dagar, S.S.; Kumar, S.; Griffith, G.W.; Edwards, J.E.; Callaghan, T.M.; Singh, R.; Nagpal, A.K.; Puniya, A.K. A new anaerobic fungus (Oontomyces anksri gen. nov., sp. nov.) from the digestive tract of the indian camel (Camelus dromedarius). Fungal Biol. 2015, 119, 731–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanafy, R.A.; Elshahed, M.S.; Liggenstoffer, A.S.; Griffith, G.W.; Youssef, N.H. Pecoramyces ruminantium, gen. nov., sp. nov., an anaerobic gut fungus from the feces of cattle and sheep. Mycologia 2017, 109, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Hanafy, R.A.; Elshahed, M.S.; Youssef, N.H. Feramyces austinii, gen. nov., sp. nov., an anaerobic gut fungus from rumen and fecal samples of wild barbary sheep and fallow deer. Mycologia 2018, 110, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.; Lanjekar, V.B.; Dhakephalkar, P.K.; Callaghan, T.M.; Griffith, G.W.; Dagar, S.S. Liebetanzomyces polymorphus gen. et sp. nov., a new anaerobic fungus (Neocallimastigomycota) isolated from the rumen of a goat. MycoKeys 2018, 40, 89–110. [Google Scholar] [CrossRef]
- Hanafy, R.A.; Lanjekar, V.B.; Dhakephalkar, P.K.; Callaghan, T.M.; Dagar, S.S.; Griffith, G.W.; Elshahed, M.S.; Youssef, N.H. Seven new Neocallimastigomycota genera from wild, zoo-housed, and domesticated herbivores greatly expand the taxonomic diversity of the phylum. Mycologia 2020, 112, 1212–1239. [Google Scholar] [CrossRef]
- Stabel, M.; Hanafy, R.A.; Schweitzer, T.; Greif, M.; Aliyu, H.; Flad, V.; Young, D.; Lebuhn, M.; Elshahed, M.S.; Ochsenreither, K.; et al. Aestipascuomyces dupliciliberans gen. nov., sp. nov., the first cultured representative of the uncultured sk4 clade from aoudad sheep and alpaca. Microorganisms 2020, 8, 1734. [Google Scholar] [CrossRef]
- Hanafy, R.A.; Elshahed, M.S.; Youssef, N.H. Paucimyces polynucleatus gen. nov., sp. nov., a novel polycentric genus of anaerobic gut fungi from the faeces of a wild blackbuck antelope. Int. J. Syst. Evol. Microbiol. 2021, 71, 004832. [Google Scholar] [CrossRef]
- Mura, E.; Edwards, J.; Kittelmann, S.; Kaerger, K.; Voigt, K.; Mrázek, J.; Moniello, G.; Fliegerova, K. Anaerobic fungal communities differ along the horse digestive tract. Fungal Biol. 2019, 123, 240–246. [Google Scholar] [CrossRef] [Green Version]
- Bowman, B.H.; Taylor, J.W.; White, T.J. Molecular evolution of the fungi: Human pathogens. Mol. Biol. Evol. 1992, 9, 893–904. [Google Scholar]
- Li, J.L.; Heath, I.B. Chytridiomycetous gut fungi, oft overlooked contributors to herbivore digestion. Can. J. Microbiol. 1993, 39, 1003–1013. [Google Scholar] [CrossRef]
- Dore, J.; Stahl, D.A. Phylogeny of anaerobic rumen Chytridiomycetes inferred from small subunit ribosomal-RNA sequence comparisons. Can. J. Bot. 1991, 69, 1964–1971. [Google Scholar] [CrossRef]
- Li, J.; Heath, I.B.; Packer, L. The Phylogenetic relationships of the anaerobic Chytridiomycetous gut fungi (Neocallimasticaceae) and the Chytridiomycota. II. Cladistic analysis of structural data and description of Neocallimasticales ord.nov. Can. J. Bot. 1993, 71, 393–407. [Google Scholar] [CrossRef]
- Brookman, J.L. Identification and Characterization of Anaerobic Gut Fungi Using Molecular Methodologies Based on Ribosomal Its1 and 18s rRNA. Microbiology 2000, 146, 393–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirohi, S.K.; Choudhury, P.K.; Puniya, A.K. Ribosomal ITS1 sequence-based diversity analysis of anaerobic rumen fungi in cattle fed on high fiber diet. Ann. Microbiol. 2013, 63, 1571–1577. [Google Scholar] [CrossRef]
- Nicholson, M.J.; McSweeney, C.S.; Mackie, R.I.; Brookman, J.L.; Theodorou, M.K. Diversity of anaerobic gut fungal populations analysed using ribosomal ITS1 sequences in faeces of wild and domesticated herbivores. Anaerobe 2010, 16, 66–73. [Google Scholar] [CrossRef]
- Edwards, J.E.; Hermes, G.D.A.; Kittelmann, S.; Nijsse, B.; Smidt, H. Assessment of the accuracy of high-throughput sequencing of the ITS1 region of Neocallimastigomycota for community composition analysis. Front. Microbiol. 2019, 10, 2370. [Google Scholar] [CrossRef]
- Vinzelj, J.; Joshi, A.; Insam, H.; Podmirseg, S.M. Employing anaerobic fungi in biogas production: Challenges & opportunities. Bioresour. Technol. 2020, 300, 122687. [Google Scholar] [CrossRef]
- Dollhofer, V.; Callaghan, T.M.; Dorn-In, S.; Bauer, J.; Lebuhn, M. Development of three specific PCR-based tools to determine quantity, cellulolytic transcriptional activity and phylogeny of anaerobic fungi. J. Microbiol. Methods 2016, 127, 28–40. [Google Scholar] [CrossRef]
- Haitjema, C.H.; Gilmore, S.P.; Henske, J.K.; Solomon, K.V.; de Groot, R.; Kuo, A.; Mondo, S.J.; Salamov, A.A.; LaButti, K.; Zhao, Z.; et al. A parts list for fungal cellulosomes revealed by comparative genomics. Nat. Microbiol. 2017, 2, 655–681. [Google Scholar] [CrossRef] [Green Version]
- Henske, J.K.; Gilmore, S.P.; Knop, D.; Cunningham, F.J.; Sexton, J.A.; Smallwood, C.R.; Shutthanandan, V.; Evans, J.E.; Theodorou, M.K.; O’Malley, M.A. Transcriptomic characterization of Caecomyces churrovis: A novel, non-rhizoid-forming lignocellulolytic anaerobic fungus. Biotechnol. Biofuels 2017, 10, 305. [Google Scholar] [CrossRef] [Green Version]
- Wilken, S.E.; Monk, J.M.; Leggieri, P.A.; Lawson, C.E.; Lankiewicz, T.S.; Seppälä, S.; Daum, C.G.; Jenkins, J.; Lipzen, A.M.; Mondo, S.J.; et al. Experimentally validated reconstruction and analysis of a genome-scale metabolic model of an anaerobic Neocallimastigomycota fungus. mSystems 2021, 6, e00002–e00021. [Google Scholar] [CrossRef] [PubMed]
- Youssef, N.H.; Couger, M.B.; Struchtemeyer, C.G.; Liggenstoffer, A.S.; Prade, R.A.; Najar, F.Z.; Atiyeh, H.K.; Wilkins, M.R.; Elshahed, M.S. The genome of the anaerobic fungus Orpinomyces sp. strain C1A reveals the unique evolutionary history of a remarkable plant biomass degrader. Appl. Environ. Microbiol. 2013, 79, 4620–4634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Li, Y.; Jin, W.; Sharpton, T.J.; Mackie, R.I.; Cann, I.; Cheng, Y.; Zhu, W. Combined genomic, transcriptomic, proteomic, and physiological characterization of the growth of Pecoramyces sp. F1 in monoculture and co-culture with a syntrophic methanogen. Front. Microbiol. 2019, 10, 435. [Google Scholar] [CrossRef] [Green Version]
- Gruninger, R.J.; Nguyen, T.T.M.; Reid, I.D.; Yanke, J.L.; Wang, P.; Abbott, D.W.; Tsang, A.; McAllister, T. Application of transcriptomics to compare the carbohydrate active enzymes that are expressed by diverse genera of anaerobic fungi to degrade plant cell wall carbohydrates. Front. Microbiol. 2018, 9, 1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orpin, C.G.; Bountiff, L. Zoospore chemotaxis in the rumen phycomycete Neocallimastix frontalis. J. Gen. Microbiol. 1978, 104, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.E.; Kingston-Smith, A.H.; Jimenez, H.R.; Huws, S.A.; Skøt, K.P.; Griffith, G.W.; McEwan, N.R.; Theodorou, M.K. Dynamics of Initial Colonization of Nonconserved Perennial Ryegrass by Anaerobic Fungi in the Bovine Rumen. FEMS Microbiol. Ecol. 2008, 66, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Haitjema, C.H.; Solomon, K.V.; Henske, J.K.; Theodorou, M.K.; O’Malley, M.A. Anaerobic gut Fungi: Advances in isolation, culture, and cellulolytic enzyme discovery for biofuel production. Biotechnol. Bioeng. 2014, 111, 1471–1482. [Google Scholar] [CrossRef]
- Cheng, Y.; Shi, Q.; Sun, R.; Liang, D.; Li, Y. The biotechnological potential of anaerobic fungi on fiber degradation and methane production. World J. Microbiol. Biotechnol. 2018, 34, 155. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Gilmore, S.P.; Henske, J.K.; O’Malley, M.A. Driving biomass breakdown through engineered cellulosomes. Bioengineered 2015, 6, 204–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mountfort, D.O.; Asher, R.A. Production and regulation of cellulase by two strains of the rumen anaerobic fungus Neocallimastix frontalis. Appl. Environ. Microbiol. 1985, 49, 1314–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, T.M.; Wilson, C.A.; McCrae, S.I.; Joblin, K.N. A highly active extracellular cellulase from the anaerobic rumen fungus Neocallimastix frontalis. FEMS Microbiol. Lett. 1986, 34, 37–40. [Google Scholar] [CrossRef]
- Williams, A.G.; Orpin, C.G. Polysaccharide-degrading enzymes formed by three species of anaerobic rumen fungi grown on a range of carbohydrate substrates. Can. J. Microbiol. 1987, 33, 418–426. [Google Scholar] [CrossRef]
- Borneman, W.S.; Akin, D.E.; Ljungdahl, L.G. Fermentation products and plant cell wall-degrading enzymes produced by monocentric and polycentric anaerobic ruminal fungi. Appl. Environ. Microbiol. 1989, 55, 1066–1073. [Google Scholar] [CrossRef] [Green Version]
- Ljungdahl, L.G. The cellulase/hemicellulase system of the anaerobic fungus Orpinomyce sp C-2 and aspects of its applied use. Ann. N. Y. Acad. Sci. 2008, 1125, 308–321. [Google Scholar] [CrossRef]
- Kameshwar, A.K.S.; Qin, W. Genome wide analysis reveals the extrinsic cellulolytic and biohydrogen generating abilities of Neocallimastigomycota fungi. J. Genom. 2018, 6, 74–87. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Meng, Z.; Xu, Y.; Shi, Q.; Ma, Y.; Min, A.; Cheng, Y.; Zhu, W. Interactions between anaerobic fungi and methanogens in the rumen and their biotechnological potential in biogas production from lignocellulosic materials. Microorganisms 2021, 9, 190. [Google Scholar] [CrossRef]
- Couger, M.B.; Youssef, N.H.; Struchtemeyer, C.G.; Liggenstoffer, A.S.; Elshahed, M.S. Transcriptomic analysis of lignocellulosic biomass degradation by the anaerobic fungal isolate Orpinomyces sp. strain C1A. Biotechnol. Biofuels. 2015, 8, 208. [Google Scholar] [CrossRef] [Green Version]
- Pérez, J.; Muñoz-Dorado, J.; de la Rubia, T.; Martínez, J. Biodegradation and biological treatments of cellulose, hemicellulose and lignin: An overview. Int. Microbiol. 2002, 5, 53–63. [Google Scholar]
- Purwantini, E.; Torto-Alalibo, T.; Lomax, J.; Setubal, J.C.; Tyler, B.M.; Mukhopadhyay, B. Genetic resources for methane production from biomass described with the gene ontology. Front. Microbiol. 2014, 5, 634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, S.S. Fermentative characteristics and fibrolytic activities of anaerobic gut fungi isolated from wild and domestic ruminants. Arch. Anim. Nutr. 2010, 64, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Zkse, E. Effects of essential oils supplementation on survival rate and lignocellulolytic enzyme activities of rumen fungi isolated from cattle. Agric. Nat. Resour. 2017, 20, 235–241. [Google Scholar] [CrossRef] [Green Version]
- Rabee, A.E.; Forster, R.J.; Elekwachi, C.O.; Kewan, K.Z.; Sabra, E.A.; Shawket, S.M.; Mahrous, H.A.; Khamiss, O.A. Community structure and fibrolytic activities of anaerobic rumen fungi in dromedary camels. J. Basic Microbiol. 2019, 59, 101–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, G.P.; Denman, S.E.; Glassop, D.; Johnson, J.S.; Dierens, L.M.; Gobius, K.S.; Aylward, J.H. Modification of a xylanase c-DNA isolated from an anaerobic fungus Neocallimastix patriciarum for high-level expression in Escherichia coli. J. Biotechnol. 1995, 38, 269–277. [Google Scholar] [CrossRef]
- Akyol, I.; Comlekcioglu, U.; Kar, B.; Ekinci, M.S.; Ozkose, E. Cloning of a xylanase gene xyn2a from rumen fungus Neocallimastix sp Gmlf2 in Escherichia coli and its partial characterization. Biologia 2009, 64, 664–670. [Google Scholar] [CrossRef] [Green Version]
- Juturu, V.; Wu, J.C. Microbial xylanases: Engineering, production and industrial applications. Biotechnol. Adv. 2012, 30, 1219–1227. [Google Scholar] [CrossRef]
- Li, X.L.; Skory, C.D.; Ximenes, E.A.; Jordan, D.B.; Dien, B.S.; Hughes, S.R.; Cotta, M.A. Expression of an AT-rich xylanase gene from the anaerobic fungus Orpinomyces sp. strain PC-2 in and secretion of the heterologous enzyme by Hypocrea jecorina. Appl. Microbiol. Biotechnol. 2007, 74, 1264–1275. [Google Scholar] [CrossRef]
- Hou, P.; Li, Y.; Wu, B.; Yan, Z.; Yan, B.; Gao, P. Cellulolytic complex exists in cellulolytic myxobacterium Sorangium. Enzyme Microb. Technol. 2004, 38, 273–278. [Google Scholar]
- Eibinger, M.; Ganner, T.; Plank, H.; Nidetzky, B. A biological nanomachine at work: Watching the cellulosome degrade crystalline cellulose. ACS Cent. Sci. 2020, 6, 739–746. [Google Scholar] [CrossRef]
- Steenbakkers, P.J.; Harhangi, H.R.; Bosscher, M.W.; van der Hooft, M.M.; Keltjens, J.T.; van der Drift, C.; Vogels, G.D.; op den Camp, H.J. beta-Glucosidase in cellulosome of the anaerobic fungus Piromyces sp. strain E2 is a family 3 glycoside hydrolase. Biochem. J. 2003, 370, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Dyall, S.D.; Johnson, P.J. Origins of hydrogenosomes and mitochondria: Evolution and organelle biogenesis. Curr. Opin. Microbiol. 2000, 3, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Stairs, C.W.; Leger, M.M.; Roger, A.J. Diversity and origins of anaerobic metabolism in mitochondria and related organelles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, M.W. The pre-endosymbiont hypothesis: A new perspective on the origin and evolution of mitochondria. Cold Spring Harb. Perspect. Biol. 2014, 6, a016097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boxma, B.; de Graaf, R.M.; van der Staay, G.W.; van Alen, T.A.; Ricard, G.; Gabaldón, T.; van Hoek, A.H.; Moon-van der Staay, S.Y.; Koopman, W.J.; van Hellemond, J.J.; et al. An anaerobic mitochondrion that produces hydrogen. Nature 2005, 434, 74–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hackstein, J.H.P.; de Graaf, R.M.; van Hellemond, J.J.; Tielens, A.G.M. Hydrogenosomes of anaerobic ciliates. In Hydrogenosomes and Mitosomes: Mitochondria of Anaerobic Eukaryotes; Tachezy, J., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 97–112. [Google Scholar]
- Van der Giezen, M.; Kiel, J.A.; Sjollema, K.A.; Prins, R.A. The hydrogenosomal malic enzyme from the anaerobic fungus Neocallimastix frontalis is targeted to mitochondria of the methylotrophic yeast hansenula polymorpha. Curr. Genet. 1998, 33, 131–135. [Google Scholar] [CrossRef]
- Dacks, J.B.; Dyal, P.L.; Embley, T.M.; van der Giezen, M. Hydrogenosomal succinyl-CoA synthetase from the rumen-dwelling fungus Neocallimastix patriciarum; An energy-producing enzyme of mitochondrial origin. Gene 2006, 373, 75–82. [Google Scholar] [CrossRef]
- van Grinsven, K.W.A.; Rosnowsky, S.; van Weelden, S.W.H.; Pütz, S.; van der Giezen, M.; Martin, W.; van Hellemond, J.J.; Tielens, A.G.M.; Henze, K. Acetate:succinate CoA-transferase in the hydrogenosomes of Trichomonas vaginalis: Identification and characterization. J. Biol. Chem. 2008, 283, 1411–1418. [Google Scholar] [CrossRef] [Green Version]
- Lewis, W.H.; Lind, A.E.; Sendra, K.M.; Onsbring, H.; Williams, T.A.; Esteban, G.F.; Hirt, R.P.; Ettema, T.J.G.; Embley, T.M. Convergent evolution of hydrogenosomes from mitochondria by gene transfer and loss. Mol. Biol. Evol. 2020, 37, 524–539. [Google Scholar] [CrossRef]
- Diniz, J.A.P.; Benchimol, M. Monocercomonas sp.: Cytochemistry and fine structure of freeze-fractured membranes. J. Eukaryot. Microbiol. 1998, 45, 314–322. [Google Scholar]
- Benchimol, M.; Durand, R.; Almeida, J.C. A double membrane surrounds the hydrogenosomes of the anaerobic fungus Neocallimastix frontalis. FEMS Microbiol. Lett. 1997, 154, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Marvin-Sikkema, F.D.; Lahpor, G.A.; Kraak, M.N.; Gottschal, J.C.; Prins, R.A. Characterization of an anaerobic fungus from llama faeces. J. Gen. Microbiol. 1992, 138, 2235–2241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, A.; Hann, A.C.; Linstead, D.; Lloyd, D. Energy-dispersive X-ray microanalysis of membrane-associated inclusions in hydrogenosomes isolated from Trichomonas vaginalis. J. Gen. Microbiol. 1985, 131, 2933–2939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindmark, D.G.; Müller, M. Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism. J. Biol. Chem. 1973, 248, 7724–7728. [Google Scholar]
- Bingham, A.S.; Smith, P.R.; Swartz, J.R. Evolution of an [Fefe] hydrogenase with decreased oxygen sensitivity. Int. J. Hydrogrn Energy 2011, 37, 2965–2976. [Google Scholar]
- Lindmark, D.G.; Müller, M. Superoxide dismutase in the anaerobic flagellates, Tritrichomonas foetus and Monocercomonas sp. J. Biol. Chem. 1974, 249, 4634–4637. [Google Scholar]
- Coombs, G.H.; Westrop, G.D.; Suchan, P.; Puzova, G.; Hirt, R.P.; Embley, T.M.; Mottram, J.C.; Müller, S. The amitochondriate eukaryote Trichomonas vaginalis contains a divergent thioredoxin-linked peroxiredoxin antioxidant system. J. Biol. Chem. 2004, 279, 5249–5256. [Google Scholar] [CrossRef] [Green Version]
- Biagini, G.A.; Finlay, B.J.; Lloyd, D. Evolution of the hydrogenosome. FEMS Microbiol. Lett. 1997, 155, 133–140. [Google Scholar] [CrossRef] [Green Version]
- Do, P.M.; Angerhofer, A.; Hrdy, I.; Bardonova, L.; Ingram, L.O.; Shanmugam, K.T. Engineering Escherichia coli for fermentative dihydrogen production: Potential role of NADH-ferredoxin oxidoreductase from the hydrogenosome of anaerobic protozoa. Appl. Biochem. Biotechnol. 2009, 153, 21–33. [Google Scholar] [CrossRef]
- Gawryluk, R.M.R.; Kamikawa, R.; Stairs, C.W.; Silberman, J.D.; Brown, M.W.; Roger, A.J. The Earliest Stages of Mitochondrial Adaptation to Low Oxygen Revealed in a Novel Rhizarian. Curr. Biol. 2016, 26, 2729–2738. [Google Scholar]
- Petrin, D.; Delgaty, K.; Bhatt, R.; Garber, G. Clinical and microbiological aspects of Trichomonas vaginalis. Clin. Microbiol. Rev. 1998, 11, 300–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, M.; Brown, M.T.; McArthur, A.G.; Johnson, P.J. Proteins of the glycine decarboxylase complex in the hydrogenosome of Trichomonas vaginalis. Eukaryot. Cell. 2006, 5, 2062–2071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, R.E.; Brown, M.T.; Shiflett, A.M.; Dyall, S.D.; Hayes, R.D.; Xie, Y.; Loo, J.A.; Johnson, P.J. The Trichomonas vaginalis hydrogenosome proteome is highly reduced relative to mitochondria, yet complex compared with mitosomes. Int. J. Parasitol. 2011, 41, 1421–1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, K.Y.; Ong, S.C.; Wu, C.C.; Hsu, C.W.; Lin, H.C.; Fang, Y.K.; Cheng, W.H.; Huang, P.J.; Chiu, C.H.; Tang, P. Metabolic reprogramming of hydrogenosomal amino acids in Trichomonas vaginalis under glucose restriction. J. Microbiol. Immunol. Infect. 2019, 52, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Boxma, B.; Voncken, F.; Jannink, S.; van Alen, T.; Akhmanova, A.; van Weelden, S.W.; van Hellemond, J.J.; Ricard, G.; Huynen, M.; Tielens, A.G.; et al. The anaerobic chytridiomycete fungus Piromyces sp. E2 produces ethanol via pyruvate:formate lyase and an alcohol dehydrogenase E. Mol. Microbiol. 2004, 51, 1389–1399. [Google Scholar] [CrossRef]
- Ivarsson, M.; Schnürer, A.; Bengtson, S.; Neubeck, A. Anaerobic fungi: A potential source of biological H2 in the oce anic crust. Front. Microbiol. 2016, 7, 674. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.; Khan, M.D.; Sabir, S.; Nizami, A.S.; Anwer, A.H.; Rehan, M.; ZainKhan, M. Deciphering the effects of temperature on bio-methane generation through anaerobic digestion. Environ. Sci. Pollut. Res. Int. 2020, 27, 29766–29777. [Google Scholar] [CrossRef]
- Nitsos, C.; Matsakas, L.; Triantafyllidis, K.; Rova, U.; Christakopoulos, P. Evaluation of mediterranean agricultural residues as a potential feedstock for the production of biogas via anaerobic fermentation. Biomed. Res. Int. 2015, 2015, 171635. [Google Scholar] [CrossRef] [Green Version]
- Mortreuil, P.; Baggio, S.; Lagnet, C.; Schraauwers, B.; Monlau, F. Fast prediction of organic wastes methane potential by near infrared reflectance spectroscopy: A successful tool for farm-scale biogas plant monitoring. Waste Manag. Res. 2018, 36, 800–809. [Google Scholar] [CrossRef]
- Shekar, N.R.C.; Ramaraju, H.K. Biomethanisation of municipal solid waste by anaerobic fungi co-culture with methanogens: An approach to energy recovery. J. Environ. Sci. Eng. 2010, 4, 60–66. [Google Scholar]
- Ferraro, A.; Dottorini, G.; Massini, G.; Mazzurco Miritana, V.; Signorini, A.; Lembo, G.; Fabbricino, M. Combined bioaugmentation with anaerobic ruminal fungi and fermentative bacteria to enhance biogas production from wheat straw and mushroom spent straw. Bioresour. Technol. 2018, 260, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Shetty, D.; Joshi, A.; Dagar, S.S.; Kshirsagar, P.; Dhakephalkar, P.K. Bioaugmentation of anaerobic fungus Orpinomyces joyonii boosts sustainable biomethanation of rice straw without pretreatment. Biomass Bioenergy 2020, 138, 105546. [Google Scholar] [CrossRef]
- Di Marco, A.A.; Bobik, T.A.; Wolfe, R.S. Unusual coenzymes of methanogenesis. Annu. Rev. Biochem. 1990, 59, 355–394. [Google Scholar] [CrossRef]
- Cheng, Y.F.; Edwards, J.E.; Allison, G.G.; Zhu, W.Y.; Theodorou, M.K. Diversity and activity of enriched ruminal cultures of anaerobic fungi and methanogens grown together on lignocellulose in consecutive batch culture. Bioresour. Technol. 2009, 100, 4821–4828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales-Vera, R.; Crawford, J.; Dou, C.; Bura, R.; Gustafson, R. Techno-economic analysis of producing glacial acetic acid from poplar biomass via bioconversion. Molecules 2020, 25, 4328. [Google Scholar] [CrossRef]
- Wang, J.; Lin, M.; Xu, M.; Yang, S.T. Anaerobic fermentation for production of carboxylic acids as bulk chemicals from renewable biomass. Adv. Biochem. Eng. Biotechnol. 2016, 156, 323–361. [Google Scholar] [CrossRef]
- Amidon, T.E.; Liu, S. Water-based woody biorefinery. Biotechnol. Adv. 2009, 27, 542–550. [Google Scholar] [CrossRef]
- Budsberg, E.; Morales-Vera, R.; Crawford, J.T.; Bura, R.; Gustafson, R. Production routes to bio-acetic acid: Life cycle assessment. Biotechnol. Biofuels 2020, 13, 154. [Google Scholar] [CrossRef]
- Ni, B.J.; Liu, H.; Nie, Y.Q.; Zeng, R.J.; Du, G.C.; Chen, J.; Yu, H.Q. Coupling glucose fermentation and homoacetogenesis for elevated acetate production: Experimental and mathematical approaches. Biotechnol. Bioeng. 2011, 108, 345–353. [Google Scholar] [CrossRef]
- Zhou, F.; Wang, C.; Wei, J. Simultaneous acetic acid separation and monosaccharide concentration by reverse osmosis. Bioresour. Technol. 2013, 131, 349–356. [Google Scholar] [CrossRef]
- GarcíA, N.; Caballero, J.A. Economic and Environmental Assessment of Alternatives to the Extraction of Acetic Acid from Water. Ind. Eng. Chem. Res. 2011, 50, 10717–10729. [Google Scholar] [CrossRef]
- Wang, P.; Tan, Z. Ammonia assimilation in rumen bacteria: A review. Anim. Biotechnol. 2013, 24, 107–128. [Google Scholar] [CrossRef]
- Lopes, A.S.M.; de Oliveira, J.S.; Santos, E.M.; Medeiros, A.N.; Givisiez, P.E.N.; Lemos, M.L.P.; Santos, F.N.S.; Silva, N.M.V.; Azevedo, P.S.; Sousa, L.S.; et al. Goats fed with non-protein nitrogen: Ruminal bacterial community and ruminal fermentation, intake, digestibility and nitrogen balance. J. Agric. Sci. 2020, 158, 781–790. [Google Scholar] [CrossRef]
- Iwamoto, M.; Asanuma, N.; Hino, T. Ability of Selenomonas ruminantium, Veillonella parvula, and Wolinella succinogenes to reduce nitrate and nitrite with special reference to the suppression of ruminal methanogenesis. Anaerobe 2002, 8, 209–215. [Google Scholar]
- Yang, C.; Rooke, J.A.; Cabeza, I.; Wallace, R.J. Nitrate and inhibition of ruminal methanogenesis: Microbial ecology, obstacles, and opportunities for lowering methane emissions from ruminant livestock. Front. Microbiol. 2016, 7, 132. [Google Scholar] [CrossRef] [PubMed]
- van Zijderveld, S.M.; Gerrits, W.J.; Apajalahti, J.A.; Newbold, J.R.; Dijkstra, J.; Leng, R.A.; Perdok, H.B. Nitrate and sulfate: Effective alternative hydrogen sinks for mitigation of ruminal methane production in sheep. J. Dairy Sci. 2010, 93, 5856–5866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nolan, J.V.; Hegarty, R.S.; Hegarty, J.; Godwin, I.R.; Woodgate, R. Effects of dietary nitrate on fermentation, methane production and digesta kinetics in sheep. Anim. Prod. Sci. 2010, 50, 801–806. [Google Scholar] [CrossRef]
- Matthews, C.; Crispie, F.; Lewis, E.; Reid, M.; O’Toole, P.W.; Cotter, P.D. The rumen microbiome: A crucial consideration when optimising milk and meat production and nitrogen utilisation efficiency. Gut Microbes 2019, 10, 115–132. [Google Scholar] [CrossRef]
- Newbold, C.J.; Ramos-Morales, E. Review: Ruminal microbiome and microbial metabolome: Effects of diet and ruminant host. Animal 2020, 14, s78–s86. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, J.; Ikeda, M.; Matsuoka, S.; Fujita, H. Prophylactic effect of L-cysteine to acute and subclinical nitrate toxicity in sheep. Anim. Feed Sci. Technol. 1998, 74, 273–280. [Google Scholar] [CrossRef]
- Lundberg, J.O.; Weitzberg, E.; Gladwin, M.T. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 2008, 7, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Sar, C.; Mwenya, B.; Pen, B.; Takaura, K.; Morikawa, R.; Tsujimoto, A.; Kuwaki, K.; Isogai, N.; Shinzato, I.; Asakura, Y.; et al. Effect of ruminal administration of Escherichia coli wild type or a genetically modified strain with enhanced high nitrite reductase activity on methane emission and nitrate toxicity in nitrate-infused sheep. Br. J. Nutr. 2005, 94, 691–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, Q.; Shi, C.; Hou, X.; Ren, L.; Zhou, Z. Response of Ruminal Fermentation, Methane Production and Dry Matter Digestibility to Microbial Source and Nitrate Addition Level in an in Vitro Incubation with Rumen Microbes Obtained from Wethers. J. Anim. Vet. Adv. 2012, 11, 3334–3341. [Google Scholar]
- Mamvura, C.I.; Cho, S.; Mbiriri, D.T.; Lee, H.G.; Choi, N.J. Effect of encapsulating nitrate in sesame gum on in vitro rumen fermentation parameters. Asian-Australas. J. Anim. Sci. 2014, 27, 1577–1583. [Google Scholar] [CrossRef] [Green Version]
- El-Zaiata, H.M.; Araujob, R.C.; Louvandinic, H.; Patiñoe, H.O.; Abdalla, A.L. Effects of dietary replacement of urea with encapsulated nitrate and cashew nut shell liquid on nutrient digestibility, nitrogen balance, and carcass characteristics in growing lambs. Anim. Feed Sci. Technol. 2020, 266, 114515. [Google Scholar] [CrossRef]
- Ungerfeld, E.M. Shifts in metabolic hydrogen sinks in the methanogenesis-inhibited ruminal fermentation: A meta-analysis. Front. Microbiol. 2015, 6, 37. [Google Scholar] [CrossRef]
- Ungerfeld, E.M. A theoretical comparison between two ruminal electron sinks. Front. Microbiol. 2013, 4, 319. [Google Scholar] [CrossRef] [Green Version]
- Mamuad, L.; Kim, S.H.; Jeong, C.D.; Choi, Y.J.; Jeon, C.O.; Lee, S.S. Effect of fumarate reducing bacteria on in vitro rumen fermentation, methane mitigation and microbial diversity. J. Microbiol. 2014, 52, 120–128. [Google Scholar] [CrossRef]
- Jin, W.; Xue, C.; Liu, J.; Yin, Y.; Zhu, W.; Mao, S. Effects of disodium fumarate on in vitro rumen fermentation, the production of lipopolysaccharide and biogenic amines, and the rumen bacterial community. Curr. Microbiol. 2017, 74, 1337–1342. [Google Scholar] [CrossRef]
- Pal, K.; Patra, A.K.; Sahoo, A.; Soren, N.M. Effects of nitrate and fumarate in tree leaves-based diets on nutrient utilization, rumen fermentation, microbial protein supply and blood profiles in sheep. Livest. Sci. 2015, 172, 5–15. [Google Scholar] [CrossRef]
- Eruden, B.; Syuhei, K.; Toshihiko, K.; Hisao, I.; Sada, A.; Takehiro, N.; Motohiko, I.; Toshio, I.; Kunihiko, N.; Yoshio, I. Effect of fumaric acid on methane production, rumen fermentation and digestibility of cattle fed roughage alone. Anim. Sci. J. 2001, 72, 139–146. [Google Scholar]
- Abrar, A.; Kondo, M.; Kitamura, T.; Ban-Tokuda, T.; Matsui, H. Effect of supplementation of rice bran and fumarate alone or in combination on in vitro rumen fermentation, methanogenesis and methanogens. Anim. Sci. J. 2016, 87, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, N.; Cao, Y.; Jin, C.; Li, F.; Cai, C.; Yao, J. Effects of fumaric acid supplementation on methane production and rumen fermentation in goats fed diets varying in forage and concentrate particle size. J. Anim. Sci. Biotechnol. 2018, 9, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hattori, K.; Matsui, H. Diversity of fumarate reducing bacteria in the bovine rumen revealed by culture dependent and independent approaches. Anaerobe 2008, 14, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Mamuad, L.L.; Kim, S.H.; Lee, S.S.; Cho, K.K.; Jeon, C.O.; Lee, S.S. Characterization, metabolites and gas formation of fumarate reducing bacteria isolated from Korean native goat (Capra hircus coreanae). J. Microbiol. 2012, 50, 925–931. [Google Scholar] [CrossRef]
- Kim, S.H.; Mamuad, L.L.; Kim, D.W.; Kim, S.K.; Lee, S.S. Fumarate reductase-producing Enterococci reduce methane production in rumen fermentation in vitro. J. Microbiol. Biotechnol. 2016, 26, 558–566. [Google Scholar] [CrossRef]
- Podolsky, I.A.; Seppala, S.; Lankiewicz, T.S.; Brown, J.L.; Swift, C.L.; O’Malley, M.A. Harnessing Nature’s Anaerobes for Biotechnology and Bioprocessing. Annu. Rev. Chem. Biomol. Eng. 2019, 10, 105–128. [Google Scholar] [CrossRef]
- Shi, Q.; Li, Y.; Li, Y.; Cheng, Y.; Zhu, W. Effects of steam explosion on lignocellulosic degradation of, and methane production from, corn stover by a co-cultured anaerobic fungus and methanogen. Bioresour. Technol. 2019, 290, 121796. [Google Scholar] [CrossRef]
- Ma, Y.; Li, Y.; Li, Y.; Cheng, Y.; Zhu, W. The enrichment of anaerobic fungi and methanogens showed higher lignocellulose degrading and methane producing ability than that of bacteria and methanogens. World J. Microbiol. Biotechnol. 2020, 36, 125. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, W.; Meng, Q. Fermentation of plant cell walls by ruminal bacteria, protozoa and fungi and their interaction with fibre particle size. Arch. Anim. Nutr. 2007, 61, 114–125. [Google Scholar] [CrossRef]
- Lee, S.S.; Ha, J.K.; Cheng, K. Relative contributions of bacteria, protozoa, and fungi to in vitro degradation of orchard grass cell walls and their interactions. Appl. Environ. Microbiol. 2000, 66, 3807–3813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Li, D.; Wang, Q.; Wu, S. Improved hydrogen production and biomass through the co-cultivation of Chlamydomonas reinhardtii and Bradyrhizobium japonicum. Int. J. Hydrogen Energy 2016, 41, 9276–9283. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Xi, L.; Sun, X.; Ge, B.; Liu, D.; Han, Z.; Pu, X.; Huang, F. Enhanced hydrogen production through co-cultivation of Chlamydomonas reinhardtii CC-503 and a facultative autotrophic sulfide-oxidizing bacterium under sulfurated conditions. Int. J. Hydrog. Energy 2018, 43, 15005–15013. [Google Scholar] [CrossRef]
- Li, X.; Huang, S.; Yu, J.; Wang, Q.; Wu, S. Improvement of hydrogen production of Chlamydomonas reinhardtii by co-cultivation with isolated bacteria. Int. J. Hydrog. Energy 2013, 38, 10779–10787. [Google Scholar] [CrossRef]
- Lee, S.M.; Guan, L.L.; Eun, J.S.; Kim, C.H.; Lee, S.J.; Kim, E.T.; Lee, S.S. The effect of anaerobic fungal inoculation on the fermentation characteristics of rice straw silages. J. Appl. Microbiol. 2015, 118, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhao, C.; Liu, S.; Zhang, T.; Yao, J.; Cao, Y. Effects of Piromyces sp. CN6 CGMCC 14449 on fermentation quality, nutrient composition and the in vitro degradation rate of whole crop maize silage. AMB Express 2019, 9, 121. [Google Scholar] [CrossRef] [Green Version]
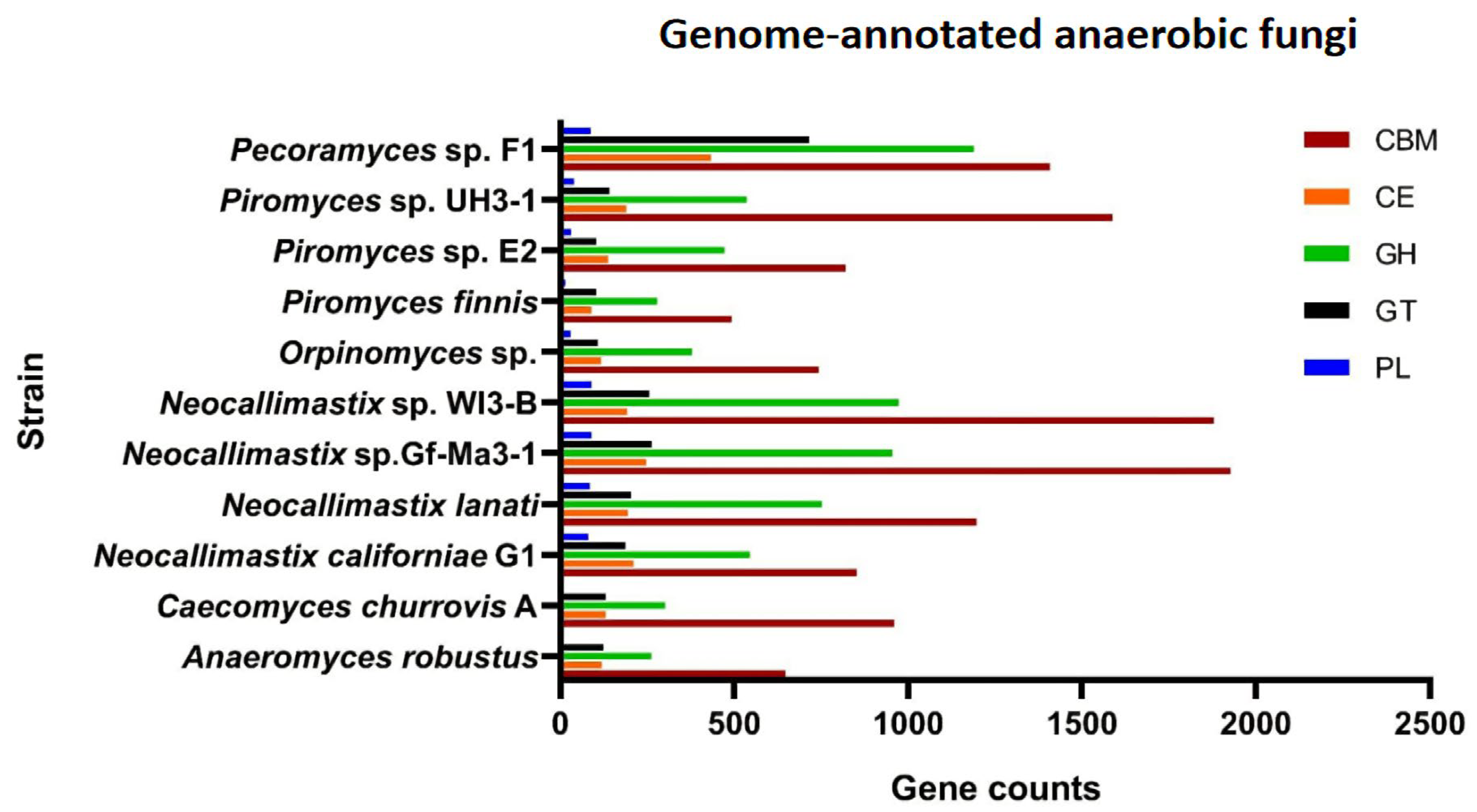

| Organism | Assembly Length | Genes Count | Isolation Source | Sample | Published |
|---|---|---|---|---|---|
| Piromyces sp. UH3-1 | 84,096,456 | 16,867 | Donkey | Feces | - |
| Piromyces sp. E2 | 71,019,055 | 14,648 | Elephant | Feces | [39] |
| Piromyces finnis | 56,455,805 | 10,992 | Horse | Feces | [39] |
| Caecomyces churrovis A | 165,495,782 | 15,009 | Sheep | Feces | [40] |
| Anaeromyces robustus | 71,685,009 | 12,832 | Sheep | Feces | [39] |
| Neocallimastix sp. Gf-Ma3-1 | 209,503,801 | 28,646 | Giraffe | Feces | - |
| Neocallimastix sp. WI3-B | 206,810,295 | 28,960 | Wildebeest | Feces | - |
| Neocallimastix lanati | 200,974,851 | 27,677 | Sheep | Feces | [41] |
| Neocallimastix californiae G1 | 193,032,486 | 20,219 | Goat | Feces | [39] |
| Pecoramyces ruminantium C1A | 100,954,185 | 18,936 | Angus steer | Feces | [42] |
| Pecoramyces sp. F1 | 106,834,627 | 17,740 | Goat | Rumen sample | [43] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Zhong, P.; Li, Y.; Sun, Z.; Sun, X.; Aung, M.; Hao, L.; Cheng, Y.; Zhu, W. Hydrogenosome, Pairing Anaerobic Fungi and H2-Utilizing Microorganisms Based on Metabolic Ties to Facilitate Biomass Utilization. J. Fungi 2022, 8, 338. https://doi.org/10.3390/jof8040338
Ma J, Zhong P, Li Y, Sun Z, Sun X, Aung M, Hao L, Cheng Y, Zhu W. Hydrogenosome, Pairing Anaerobic Fungi and H2-Utilizing Microorganisms Based on Metabolic Ties to Facilitate Biomass Utilization. Journal of Fungi. 2022; 8(4):338. https://doi.org/10.3390/jof8040338
Chicago/Turabian StyleMa, Jing, Pei Zhong, Yuqi Li, Zhanying Sun, Xiaoni Sun, Min Aung, Lizhuang Hao, Yanfen Cheng, and Weiyun Zhu. 2022. "Hydrogenosome, Pairing Anaerobic Fungi and H2-Utilizing Microorganisms Based on Metabolic Ties to Facilitate Biomass Utilization" Journal of Fungi 8, no. 4: 338. https://doi.org/10.3390/jof8040338
APA StyleMa, J., Zhong, P., Li, Y., Sun, Z., Sun, X., Aung, M., Hao, L., Cheng, Y., & Zhu, W. (2022). Hydrogenosome, Pairing Anaerobic Fungi and H2-Utilizing Microorganisms Based on Metabolic Ties to Facilitate Biomass Utilization. Journal of Fungi, 8(4), 338. https://doi.org/10.3390/jof8040338









