FFGA1 Protein Is Essential for Regulating Vegetative Growth, Cell Wall Integrity, and Protection against Stress in Flammunina filiformis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
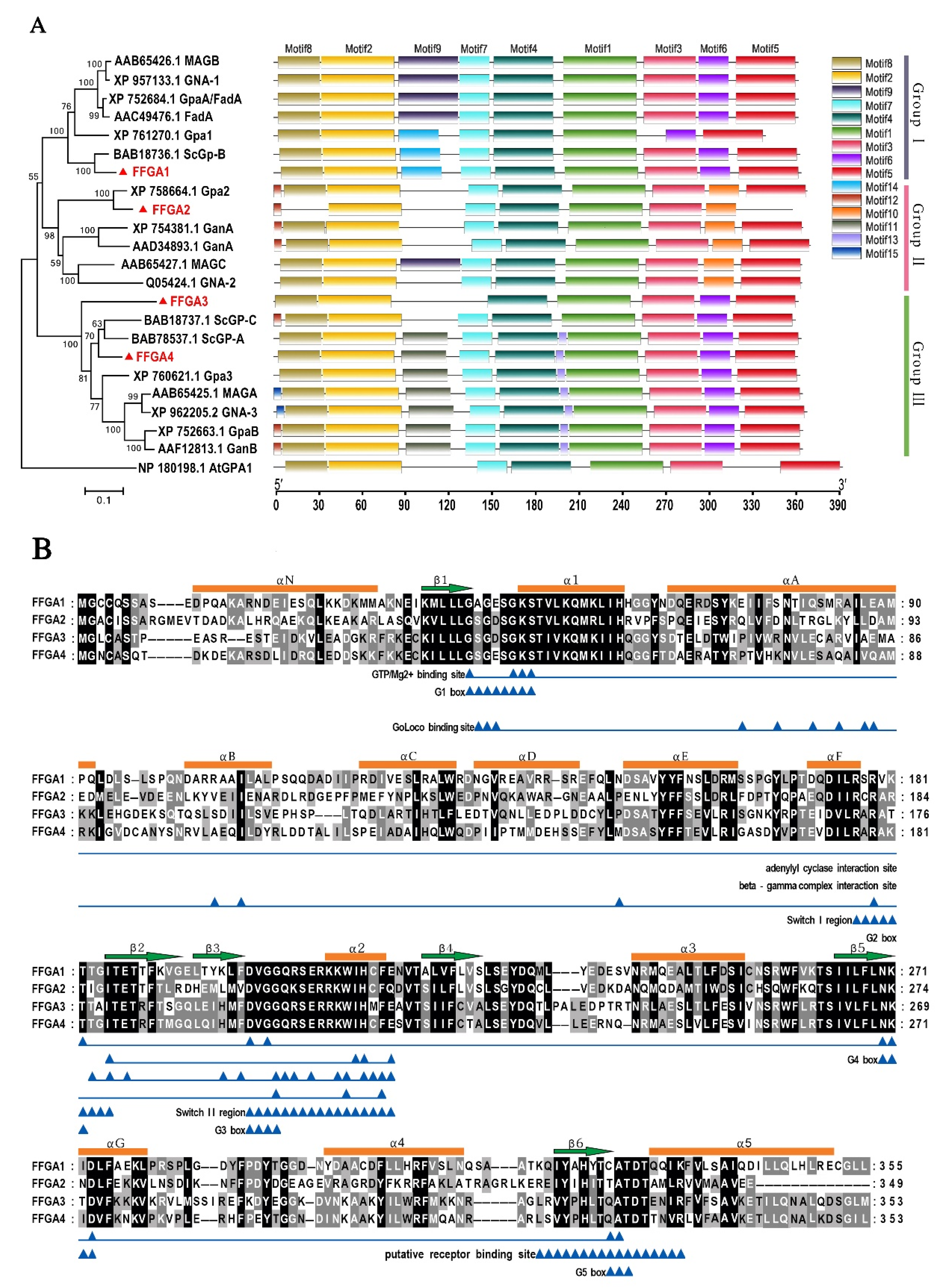
2.2. Phylogenetic Analysis of Gα Sequences
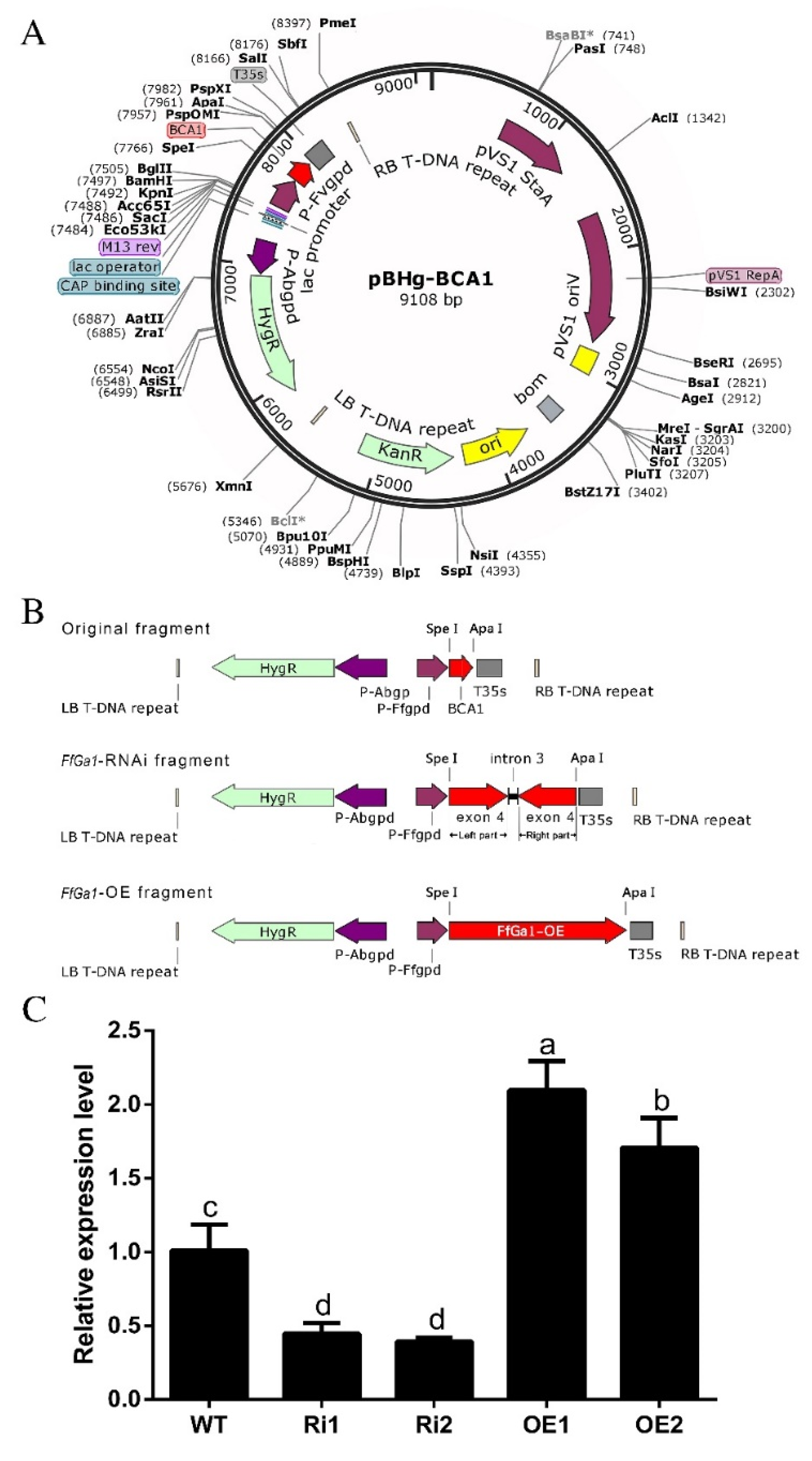
2.3. Generation of Overexpression and Knockdown Strains
2.4. Fungal Transformation and Screening of Positive Transgenic Strains
2.5. Phenotypic Characterization of OE and RNAi Transformants
2.6. Resistance of F. filiformis Transformants against Trichoderma
2.7. RNA Isolation, Complementary DNA (cDNA) Synthesis, and Expression Analysis of FFGA, Hydrophobin, and Chitin Synthetase
2.8. Statistical Analysis
3. Results
3.1. Identification and Characterization of Gα Subunit
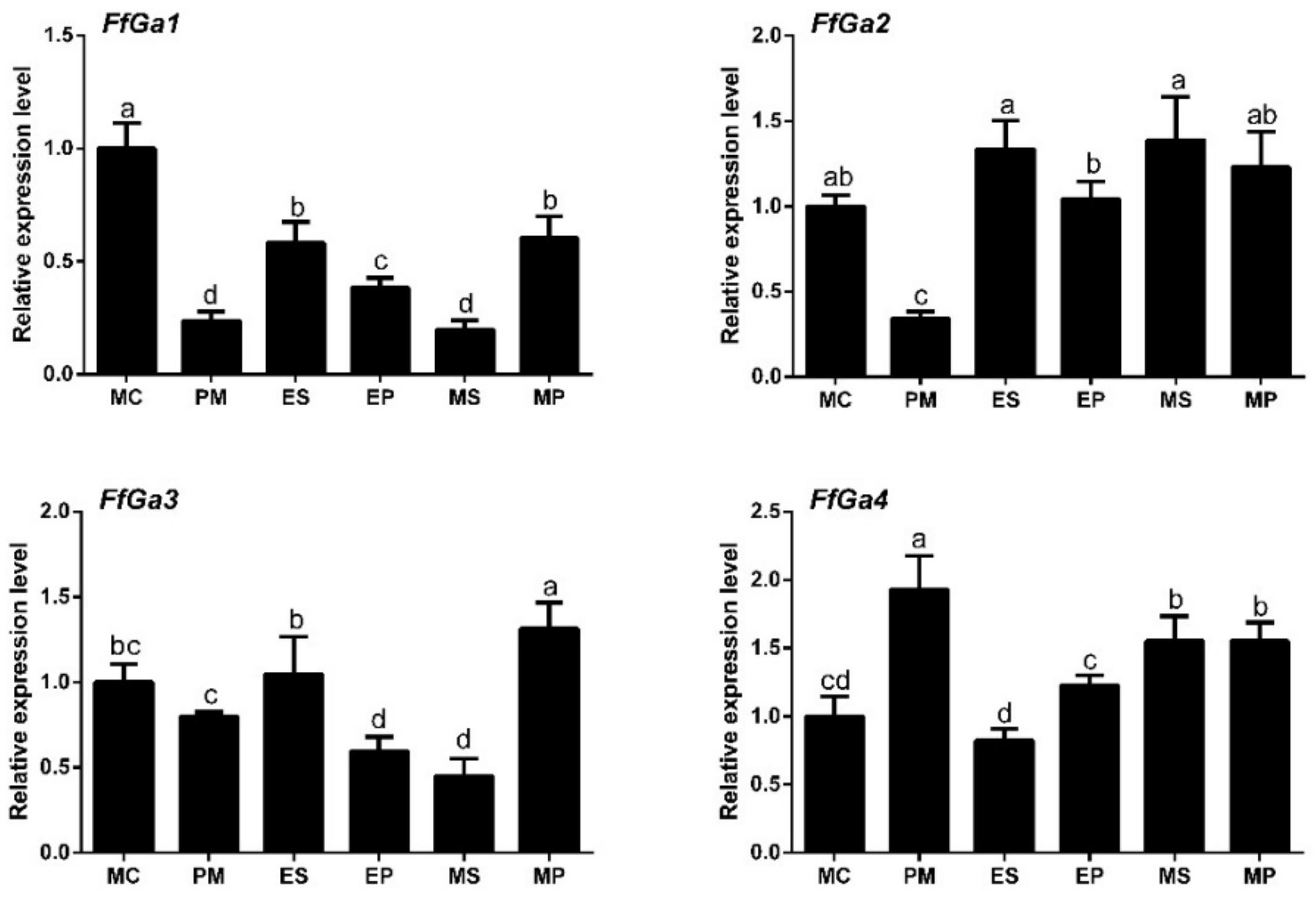
3.2. Expression of Gα Subunit at Different Stages of F. filiformis
3.3. Generation of Overexpression and Knockdown Transformants
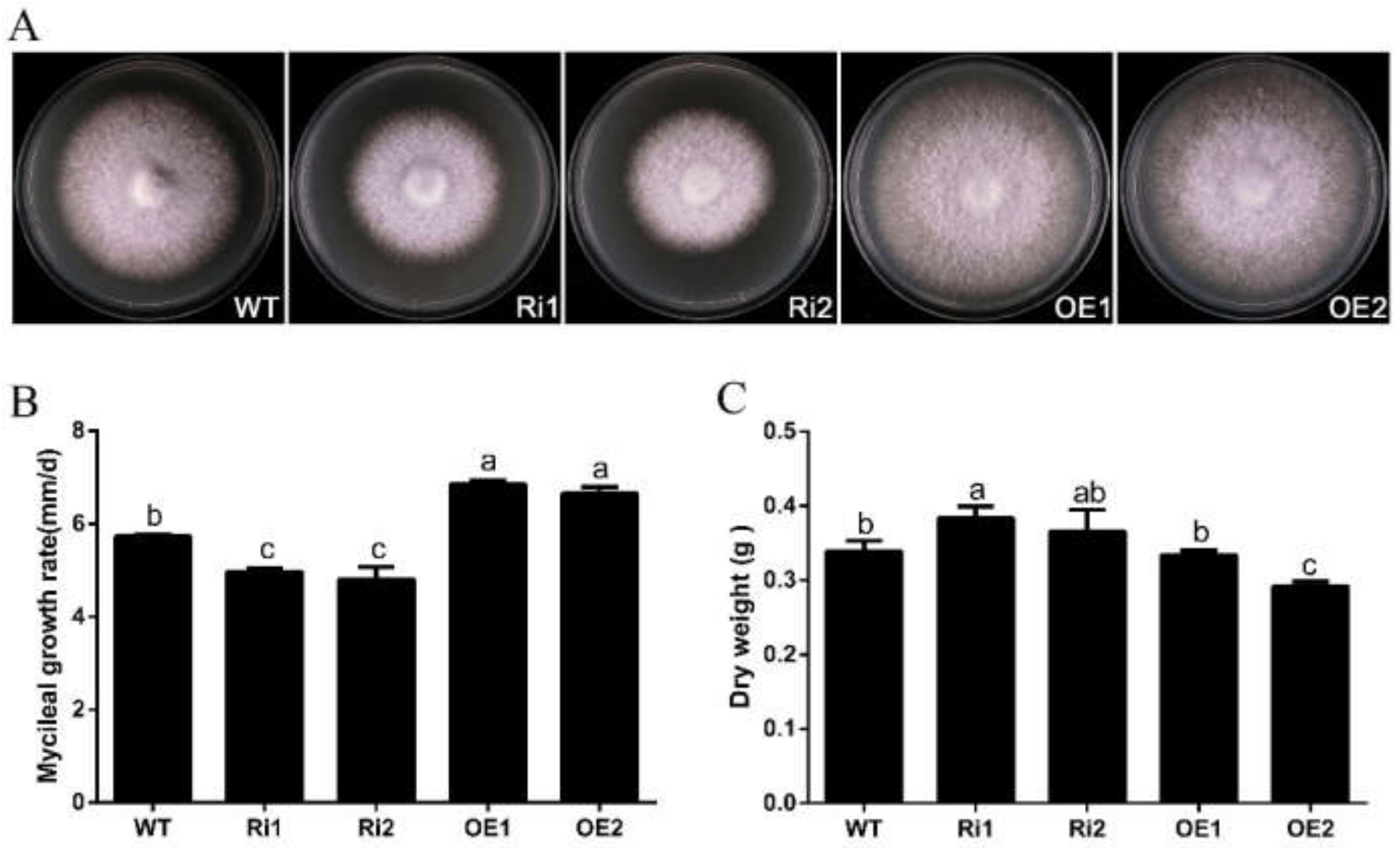
3.4. FfGa1 Positively Regulates Vegetative Growth in F. filiformis
3.5. FfGa1 Facilitates Thermoresistance
3.6. FfGa1 Is Required for Maintenance of Cell Wall Integrity and Hypertonic Stress Response in F. filiformis
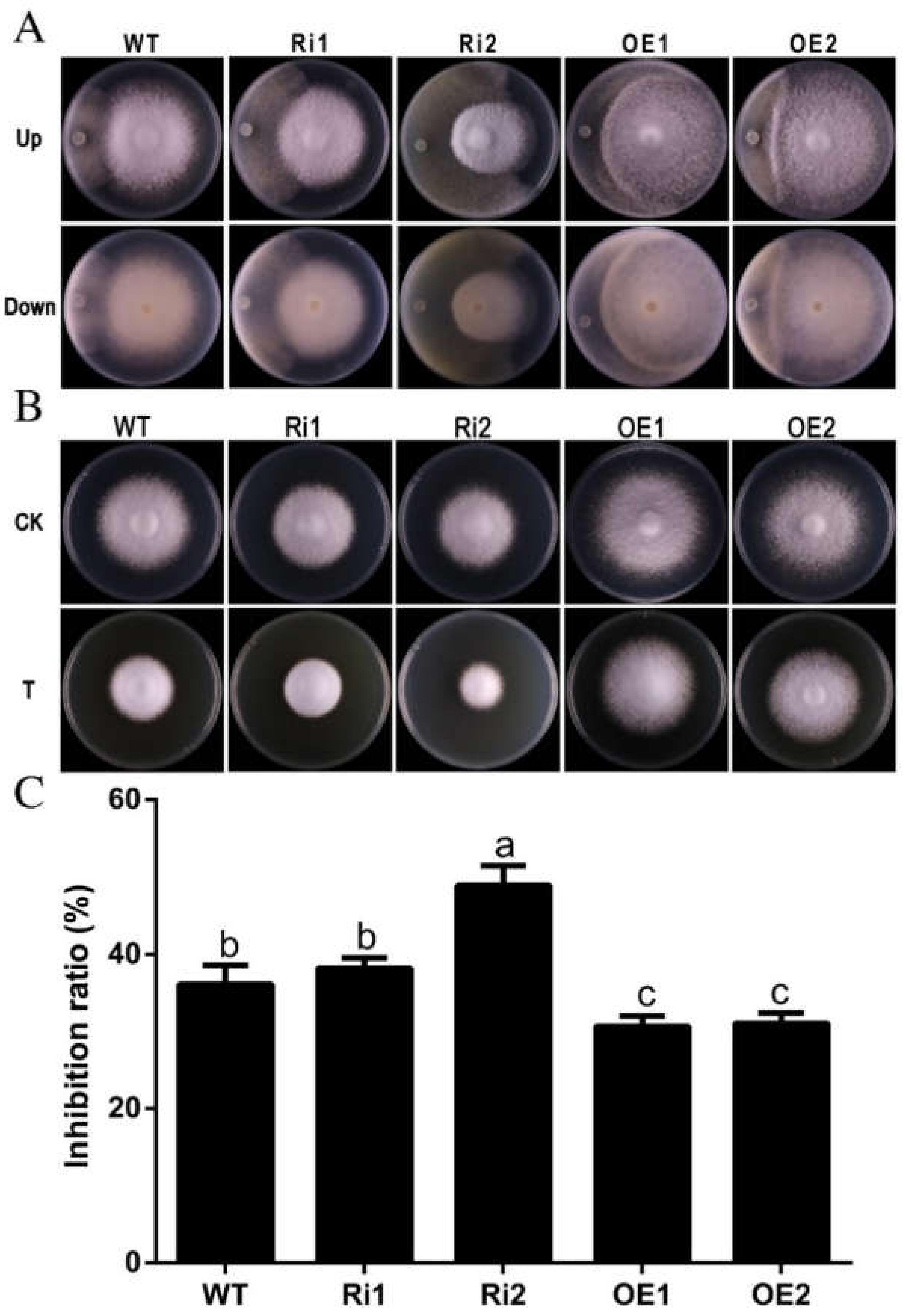
3.7. FfGa1 Involved in Resistance to Trichoderma sp.0018
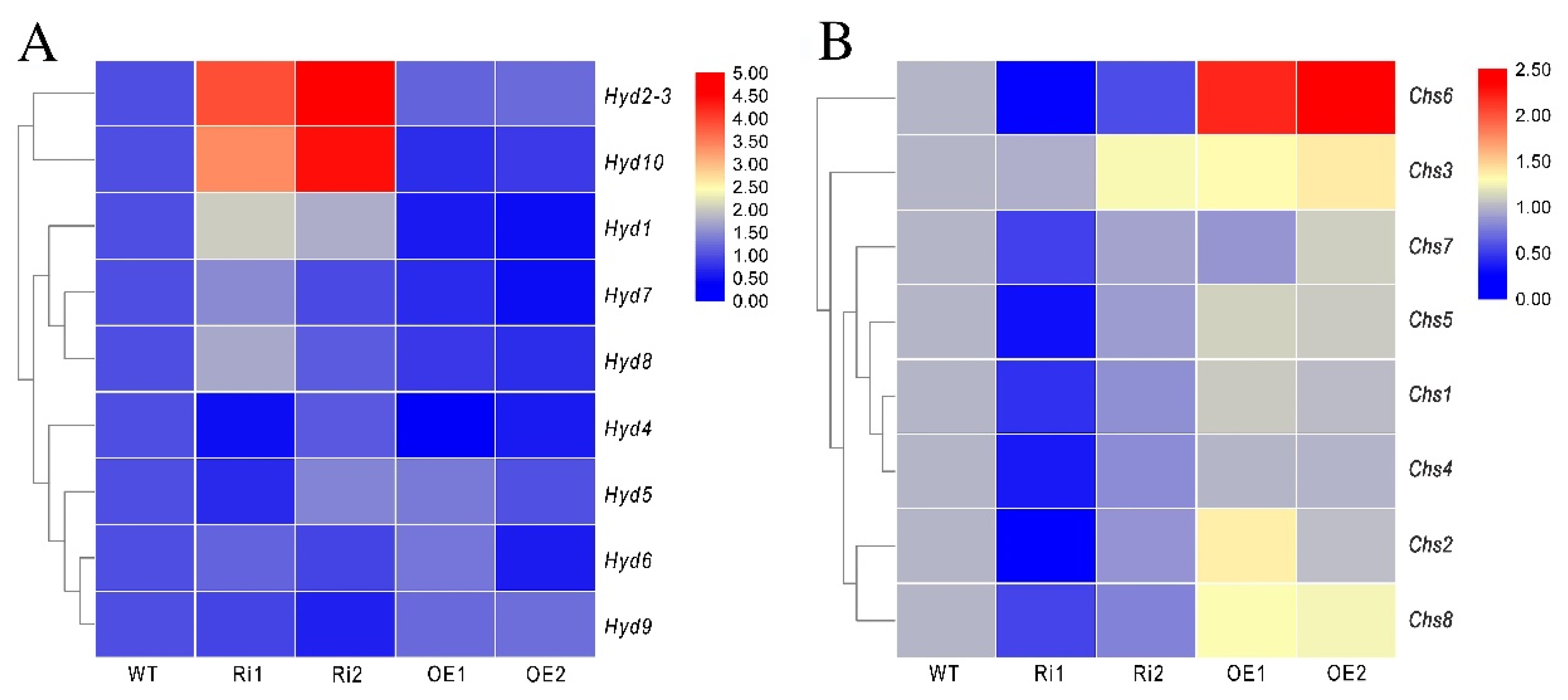
3.8. FfGa1 Regulates the Expression of the Hydrophobin and Chitin Synthase Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tao, Y.; Chen, R.; Yan, J.; Long, Y.; Tong, Z.; Song, H.; Xie, B. A hydrophobin gene, Hyd9, plays an important role in the formation of aerial hyphae and primordia in Flammulina filiformis. Gene 2019, 706, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-J.; Baek, J.H.; Lee, S.; Kim, C.; Rhee, H.; Kim, H.; Seo, J.-S.; Park, H.-R.; Yoon, D.-E.; Nam, J.-Y. Whole genome and global gene expression analyses of the model mushroom Flammulina velutipes reveal a high capacity for lignocellulose degradation. PLoS ONE 2014, 9, e93560. [Google Scholar] [CrossRef] [PubMed]
- Dowom, S.A.; Rezaeian, S.; Pourianfar, H.R. Agronomic and environmental factors affecting cultivation of the winter mushroom or Enokitake: Achievements and prospects. Appl. Microbiol. Biotechnol. 2019, 103, 2469–2481. [Google Scholar] [CrossRef] [PubMed]
- Kertesz, M.A.; Thai, M. Compost bacteria and fungi that influence growth and development of Agaricus bisporus and other commercial mushrooms. Appl. Microbiol. Biotechnol. 2018, 102, 1639–1650. [Google Scholar] [CrossRef]
- Liu, X.-B.; Xia, E.-H.; Li, M.; Cui, Y.-Y.; Wang, P.-M.; Zhang, J.-X.; Xie, B.-G.; Xu, J.-P.; Yan, J.-J.; Li, J. Transcriptome data reveal conserved patterns of fruiting body development and response to heat stress in the mushroom-forming fungus Flammulina filiformis. PLoS ONE 2020, 15, e0239890. [Google Scholar] [CrossRef]
- Liu, J.-Y.; Chang, M.-C.; Meng, J.-L.; Feng, C.-P.; Wang, Y. A comparative proteome approach reveals metabolic changes associated with Flammulina velutipes mycelia in response to cold and light stress. J. Agric. Food Chem. 2018, 66, 3716–3725. [Google Scholar] [CrossRef]
- Yamada, M.; Kurano, M.; Inatomi, S.; Taguchi, G.; Okazaki, M.; Shimosaka, M. Isolation and characterization of a gene coding for chitin deacetylase specifically expressed during fruiting body development in the basidiomycete Flammulina velutipes and its expression in the yeast Pichia pastoris. FEMS Microbiol. Lett. 2008, 289, 130–137. [Google Scholar] [CrossRef]
- Urano, D.; Jones, A.M. Heterotrimeric G protein–coupled signaling in plants. Annu. Rev. Plant Biol. 2014, 65, 365–384. [Google Scholar] [CrossRef]
- Pandey, S.; Vijayakumar, A. Emerging themes in heterotrimeric G-protein signaling in plants. Plant Sci. 2018, 270, 292–300. [Google Scholar] [CrossRef]
- Oldham, W.M.; Hamm, H.E. Structural basis of function in heterotrimeric G proteins. Q. Rev. Biophys. 2006, 39, 117–166. [Google Scholar] [CrossRef]
- Li, L.; Wright, S.J.; Krystofova, S.; Park, G.; Borkovich, K.A. Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 2007, 61, 423–452. [Google Scholar] [CrossRef]
- Garcia-Rico, R.O.; Gil-Duran, C.; Rojas-Aedo, J.F.; Vaca, I.; Figueroa, L.; Levican, G.; Chavez, R. Heterotrimeric G protein alpha subunit controls growth, stress response, extracellular protease activity, and cyclopiazonic acid production in Penicillium camemberti. Fungal Biol. 2017, 121, 754–762. [Google Scholar] [CrossRef]
- Yamagishi, K.; Kimura, T.; Suzuki, M.; Shinmoto, H. Suppression of fruit-body formation by constitutively active G-protein α-subunits ScGP-A and ScGP-C in the homobasidiomycete Schizophyllum commune. Microbiology 2002, 148, 2797–2809. [Google Scholar] [CrossRef]
- Baasiri, R.A.; Lu, X.; Rowley, P.S.; Turner, G.E.; Borkovich, K.A. Overlapping functions for two G protein α subunits in Neurospora crassa. Genetics 1997, 147, 137–145. [Google Scholar] [CrossRef]
- Kays, A.M.; Rowley, P.S.; Baasiri, R.A.; Borkovich, K.A. Regulation of conidiation and adenylyl cyclase levels by the Gα protein GNA-3 in Neurospora crassa. Mol. Cell. Biol. 2000, 20, 7693–7705. [Google Scholar] [CrossRef]
- Kays, A.M.; Borkovich, K.A. Severe impairment of growth and differentiation in a Neurospora crassa mutant lacking all heterotrimeric Gα proteins. Genetics 2004, 166, 1229–1240. [Google Scholar] [CrossRef]
- Park, H.S.; Kim, M.J.; Yu, J.H.; Shin, K.S. Heterotrimeric G-protein signalers and RGSs in Aspergillus fumigatus. Pathogens 2020, 9, 902. [Google Scholar] [CrossRef]
- Tong, Y.; Wu, H.; Liu, Z.; Wang, Z.; Huang, B. G-Protein Subunit Galphai in Mitochondria, MrGPA1, Affects Conidiation, Stress Resistance, and Virulence of Entomopathogenic Fungus Metarhizium robertsii. Front. Microbiol. 2020, 11, 1251. [Google Scholar] [CrossRef]
- Yang, Q.; Borkovich, K.A. Mutational activation of a Gαi causes uncontrolled proliferation of aerial hyphae and increased sensitivity to heat and oxidative stress in Neurospora crassa. Genetics 1999, 151, 107–117. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Latha, J.; Hadar, R.; Horwitz, B.A. Role of two G-protein alpha subunits, TgaA and TgaB, in the antagonism of plant pathogens by Trichoderma virens. Appl. Environ. Microbiol. 2004, 70, 542–549. [Google Scholar] [CrossRef]
- Nielsen, P.; Sørensen, J. Multi-target and medium-independent fungal antagonism by hydrolytic enzymes in Paenibacillus polymyxa and Bacillus pumilus strains from barley rhizosphere. FEMS Microbiol. Ecol. 1997, 22, 183–192. [Google Scholar] [CrossRef]
- Shi, L.; Fang, X.; Li, M.; Mu, D.; Ren, A.; Tan, Q.; Zhao, M. Development of a simple and efficient transformation system for the basidiomycetous medicinal fungus Ganoderma lucidum. World J. Microbiol. Biotechnol. 2012, 28, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, F.; Jiang, Y.; Wu, G.; Guo, L.; Chen, R.; Chen, B.; Lu, Y.; Dai, Y.; Xie, B. The multigene family of fungal laccases and their expression in the white rot basidiomycete Flammulina velutipes. Gene 2015, 563, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Chen, X.; Stone, M.; Schlagnhaufer, C.; Romaine, C.P. A fruiting body tissue method for efficient Agrobacterium-mediated transformation of Agaricus bisporus. Appl. Environ. Microbiol. 2000, 66, 4510–4513. [Google Scholar] [CrossRef]
- Hou, L.; Wang, L.; Wu, X.; Gao, W.; Zhang, J.; Huang, C. Expression patterns of two pal genes of Pleurotus ostreatus across developmental stages and under heat stress. BMC Microbiol. 2019, 19, 231. [Google Scholar] [CrossRef]
- Schouten, H.J.; vande Geest, H.; Papadimitriou, S.; Bemer, M.; Schaart, J.G.; Smulders, M.J.; Perez, G.S.; Schijlen, E. Re-sequencing transgenic plants revealed rearrangements at T-DNA inserts, and integration of a short T-DNA fragment, but no increase of small mutations elsewhere. Plant Cell Rep. 2017, 36, 493–504. [Google Scholar] [CrossRef]
- Thomy, J.; Sanchez, F.; Gut, M.; Cruz, F.; Alioto, T.; Piganeau, G.; Grimsley, N.; Yau, S. Combining Nanopore and Illumina sequencing permits detailed analysis of insertion mutations and structural variations produced by PEG-mediated transformation in Ostreococcus tauri. Cells 2020, 10, 664. [Google Scholar] [CrossRef]
- Wang, L.X.; Ren, L.L.; Liu, X.B.; Shi, J.; Wang, J.Z.; Luo, Y.Q. Effects of endophytic fungi in Mongolian pine on the selection behavior of woodwasp (Sirex noctilio) and the growth of its fungal symbiont. Pest Manag. Sci. 2019, 75, 492–505. [Google Scholar] [CrossRef]
- Tao, Y.; van Peer, A.F.; Huang, Q.; Shao, Y.; Zhang, L.; Xie, B.; Jiang, Y.; Zhu, J.; Xie, B. Identification of novel and robust internal control genes from Volvariella volvacea that are suitable for RT-qPCR in filamentous fungi. Sci. Rep. 2016, 6, 29236. [Google Scholar] [CrossRef]
- Taylor, S.; Wakem, M.; Dijkman, G.; Alsarraj, M.; Nguyen, M. A practical approach to RT-qPCR—Publishing data that conform to the MIQE guidelines. Methods 2010, 50, S1–S5. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hu, X.; Li, Y.; Li, C.; Fu, Y.; Cai, F.; Chen, Q.; Li, D. Combination of fucoxanthin and conjugated linoleic acid attenuates body weight gain and improves lipid metabolism in high-fat diet-induced obese rats. Arch. Biochem. Biophys. 2012, 519, 59–65. [Google Scholar] [CrossRef]
- Bölker, M. Sex and crime: Heterotrimeric G proteins in fungal mating and pathogenesis. Fungal Genet. Biol. 1998, 25, 143–156. [Google Scholar] [CrossRef]
- Yao, X.-Q.; Malik, R.U.; Griggs, N.W.; Skjærven, L.; Traynor, J.R.; Sivaramakrishnan, S.; Grant, B.J. Dynamic coupling and allosteric networks in the α subunit of heterotrimeric G proteins. J. Biol. Chem. 2016, 291, 4742–4753. [Google Scholar] [CrossRef]
- Garcia-Rico, R.O.; Martin, J.F.; Fierro, F. Heterotrimeric Ga protein Pga1 from Penicillium chrysogenum triggers germination in response to carbon sources and affects negatively resistance to different stress conditions. Fungal Genet. Biol. 2011, 48, 641–649. [Google Scholar] [CrossRef]
- Hong, Z.; Mann, P.; Brown, N.H.; Tran, L.E.; Shaw, K.J.; Hare, R.S.; DiDomenico, B. Cloning and characterization of KNR4, a yeast gene involved in (1, 3)-beta-glucan synthesis. Mol. Cell. Biol. 1994, 14, 1017–1025. [Google Scholar]
- Williams, J.; Clarkson, J.M.; Mills, P.R.; Cooper, R.M. Saprotrophic and mycoparasitic components of aggressiveness of Trichoderma harzianum groups toward the commercial mushroom Agaricus bisporus. Appl. Environ. Microbiol. 2003, 69, 4192–4199. [Google Scholar] [CrossRef]
- Segers, G.C.; Nuss, D.L. Constitutively activated Gα negatively regulates virulence, reproduction and hydrophobin gene expression in the chestnut blight fungus Cryphonectria parasitica. Fungal Genet. Biol. 2003, 38, 198–208. [Google Scholar] [CrossRef]
- Seibel, C.; Gremel, G.; do Nascimento Silva, R.; Schuster, A.; Kubicek, C.P.; Schmoll, M. Light-dependent roles of the G-protein α subunit GNA1 of Hypocrea jecorina (anamorph Trichoderma reesei). BMC Biol. 2009, 7, 58. [Google Scholar] [CrossRef]
- Gandía, M.; Harries, E.; Marcos, J.F. The myosin motor domain-containing chitin synthase PdChsVII is required for development, cell wall integrity and virulence in the citrus postharvest pathogen Penicillium digitatum. Fungal Genet. Biol. 2014, 67, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Ivey, F.; Hodge, P.; Turner, G.; Borkovich, K. The G alpha i homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol. Biol. Cell 1996, 7, 1283–1297. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, K.; Qin, Q.; Lin, G.; Hu, T.; Xu, Z.; Wang, S. G protein α subunit GpaB is required for asexual development, aflatoxin biosynthesis and pathogenicity by regulating cAMP signaling in Aspergillus flavus. Toxins 2018, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Harashima, T.; Heitman, J. The Gα protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gβ subunits. Mol. Cell 2002, 10, 163–173. [Google Scholar] [CrossRef]
- Guo, M.; Aston, C.; Burchett, S.A.; Dyke, C.; Fields, S.; Rajarao, S.J.R.; Uetz, P.; Wang, Y.; Young, K.; Dohlman, H.G. The yeast G protein α subunit Gpa1 transmits a signal through an RNA binding effector protein Scp160. Mol. Cell 2003, 12, 517–524. [Google Scholar] [CrossRef]
- Jain, S.; Akiyama, K.; Takata, R.; Ohguchi, T. Signaling via the G protein α subunit FGA2 is necessary for pathogenesis in Fusarium oxysporum. FEMS Microbiol. Lett. 2005, 243, 165–172. [Google Scholar] [CrossRef]
- Mah, J.-H.; Yu, J.-H. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot. Cell 2006, 5, 1585–1595. [Google Scholar] [CrossRef]
- García-Rico, R.O.; Martín, J.F.; Fierro, F. The pga1 gene of Penicillium chrysogenum NRRL 1951 encodes a heterotrimeric G protein alpha subunit that controls growth and development. Res. Microbiol. 2007, 158, 437–446. [Google Scholar] [CrossRef]
- Zhu, H.; Liu, D.; Zheng, L.; Chen, L.; Ma, A. Characterization of a G protein α subunit encoded gene from the dimorphic fungus-Tremella fuciformis. Antonie van Leeuwenhoek 2021, 114, 1949–1960. [Google Scholar] [CrossRef]
- Ivey, F.D.; Kays, A.M.; Borkovich, K.A. Shared and independent roles for a Gαi protein and adenylyl cyclase in regulating development and stress responses in Neurospora crassa. Eukaryot. Cell 2002, 1, 634–642. [Google Scholar] [CrossRef]
- Horwitz, B.A.; Sharon, A.; Lu, S.-W.; Ritter, V.; Sandrock, T.M.; Yoder, O.; Turgeon, B.G. A G Protein Alpha Subunit from Cochliobolus heterostrophus Involved in Mating and Appressorium Formation. Fungal Genet. Biol. 1999, 26, 19–32. [Google Scholar] [CrossRef]
- Zin, N.A.; Badaluddin, N.A. Biological functions of Trichoderma sp. for agriculture applications. Ann. Agric. Sci. 2020, 65, 168–178. [Google Scholar] [CrossRef]
- Wang, G.; Cao, X.; Ma, X.; Guo, M.; Liu, C.; Yan, L.; Bian, Y. Diversity and effect of Trichoderma sp. associated with green mold disease on Lentinula edodes in China. MicrobiologyOpen 2016, 5, 709–718. [Google Scholar] [CrossRef]
- Kosanovic, D.; Grogan, H.; Kavanagh, K. Exposure of Agaricus bisporus to Trichoderma aggressivum f. europaeum leads to growth inhibition and induction of an oxidative stress response. Fungal Biol. 2020, 124, 814–820. [Google Scholar] [CrossRef]
- Savoie, J.M.; Mata, G. The antagonistic action of Trichoderma sp. hyphae to Lentinula edodes hyphae changes lignocellulotytic activities during cultivation in wheat straw. World J. Microbiol. Biotechnol. 1999, 15, 369–373. [Google Scholar] [CrossRef]
- Savoie, J.M.; Mata, G. Trichoderma harzianum metabolites pre-adapt mushrooms to Trichoderma aggressivum antagonism. Mycologia 2003, 95, 191–199. [Google Scholar] [CrossRef]
- Ujor, V.C.; Monti, M.; Peiris, D.G.; Clements, M.O.; Hedger, J.N. The mycelial response of the white-rot fungus, Schizophyllum commune to the biocontrol agent, Trichoderma viride. Fungal Biol. 2012, 116, 332–341. [Google Scholar] [CrossRef]
- Lee, S.; Rojas, C.M.; Ishiga, Y.; Pandey, S.; Mysore, K.S. Arabidopsis heterotrimeric G-proteins play a critical role in host and nonhost resistance against Pseudomonas syringae pathogens. PLoS ONE 2013, 8, e82445. [Google Scholar] [CrossRef]
- Fuchs, B.B.; Mylonakis, E. Our paths might cross: The role of the fungal cell wall integrity pathway in stress response and cross talk with other stress response pathways. Eukaryot. Cell 2009, 8, 1616–1625. [Google Scholar] [CrossRef]
- Coca, M.A.; Damsz, B.; Yun, D.J.; Hasegawa, P.M.; Bressan, R.A.; Narasimhan, M.L. Heterotrimeric G-proteins of a filamentous fungus regulate cell wall composition and susceptibility to a plant PR-5 protein. Plant. J. 2000, 22, 61–69. [Google Scholar] [CrossRef]
- Zhang, J.; Jiang, H.; Du, Y.; Keyhani, N.O.; Xia, Y.; Jin, K. Members of chitin synthase family in Metarhizium acridum differentially affect fungal growth, stress tolerances, cell wall integrity and virulence. PLoS Pathog. 2019, 15, e1007964. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xia, Y.X.; Kim, B.; Keyhani, N.O. Two hydrophobins are involved in fungal spore coat rodlet layer assembly and each play distinct roles in surface interactions, development and pathogenesis in the entomopathogenic fungus, Beauveria bassiana. Mol. Microbiol. 2011, 80, 811–826. [Google Scholar] [CrossRef] [PubMed]
- Valsecchi, I.; Dupres, V.; Stephen-Victor, E.; Guijarro, J.I.; Gibbons, J.; Beau, R.; Bayry, J.; Coppee, J.-Y.; Lafont, F.; Latgé, J.-P. Role of hydrophobins in Aspergillus fumigatus. J. Fungi 2018, 4, 2. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, M.; Xie, Y.; Wang, M.; Yang, H.; Hu, B.; Mukhtar, I.; Liu, Y.; Tao, Y.; Liu, F.; Xie, B. FFGA1 Protein Is Essential for Regulating Vegetative Growth, Cell Wall Integrity, and Protection against Stress in Flammunina filiformis. J. Fungi 2022, 8, 401. https://doi.org/10.3390/jof8040401
Du M, Xie Y, Wang M, Yang H, Hu B, Mukhtar I, Liu Y, Tao Y, Liu F, Xie B. FFGA1 Protein Is Essential for Regulating Vegetative Growth, Cell Wall Integrity, and Protection against Stress in Flammunina filiformis. Journal of Fungi. 2022; 8(4):401. https://doi.org/10.3390/jof8040401
Chicago/Turabian StyleDu, Muyun, Yongbo Xie, Meng Wang, Huan Yang, Banghui Hu, Irum Mukhtar, Yuanyuan Liu, Yongxin Tao, Fang Liu, and Baogui Xie. 2022. "FFGA1 Protein Is Essential for Regulating Vegetative Growth, Cell Wall Integrity, and Protection against Stress in Flammunina filiformis" Journal of Fungi 8, no. 4: 401. https://doi.org/10.3390/jof8040401
APA StyleDu, M., Xie, Y., Wang, M., Yang, H., Hu, B., Mukhtar, I., Liu, Y., Tao, Y., Liu, F., & Xie, B. (2022). FFGA1 Protein Is Essential for Regulating Vegetative Growth, Cell Wall Integrity, and Protection against Stress in Flammunina filiformis. Journal of Fungi, 8(4), 401. https://doi.org/10.3390/jof8040401





